Structure Prediction and Analysis of Hepatitis E Virus Non-Structural Proteins from the Replication and Transcription Machinery by AlphaFold2
Abstract
:1. Introduction
2. Materials and Methods
3. Results
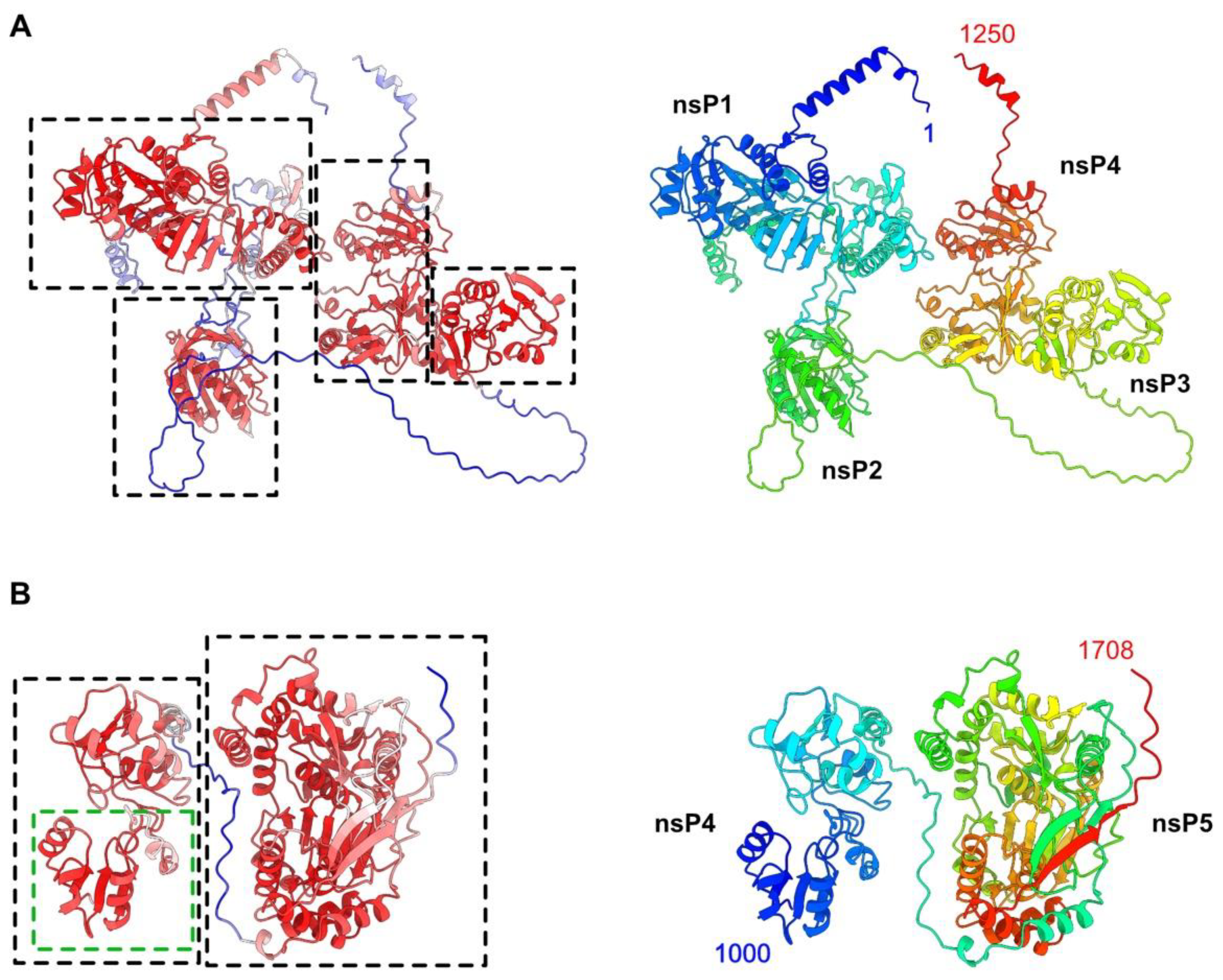
3.1. AlphaFold2 Structure Prediction of the HEV-3 Polyprotein pORF1
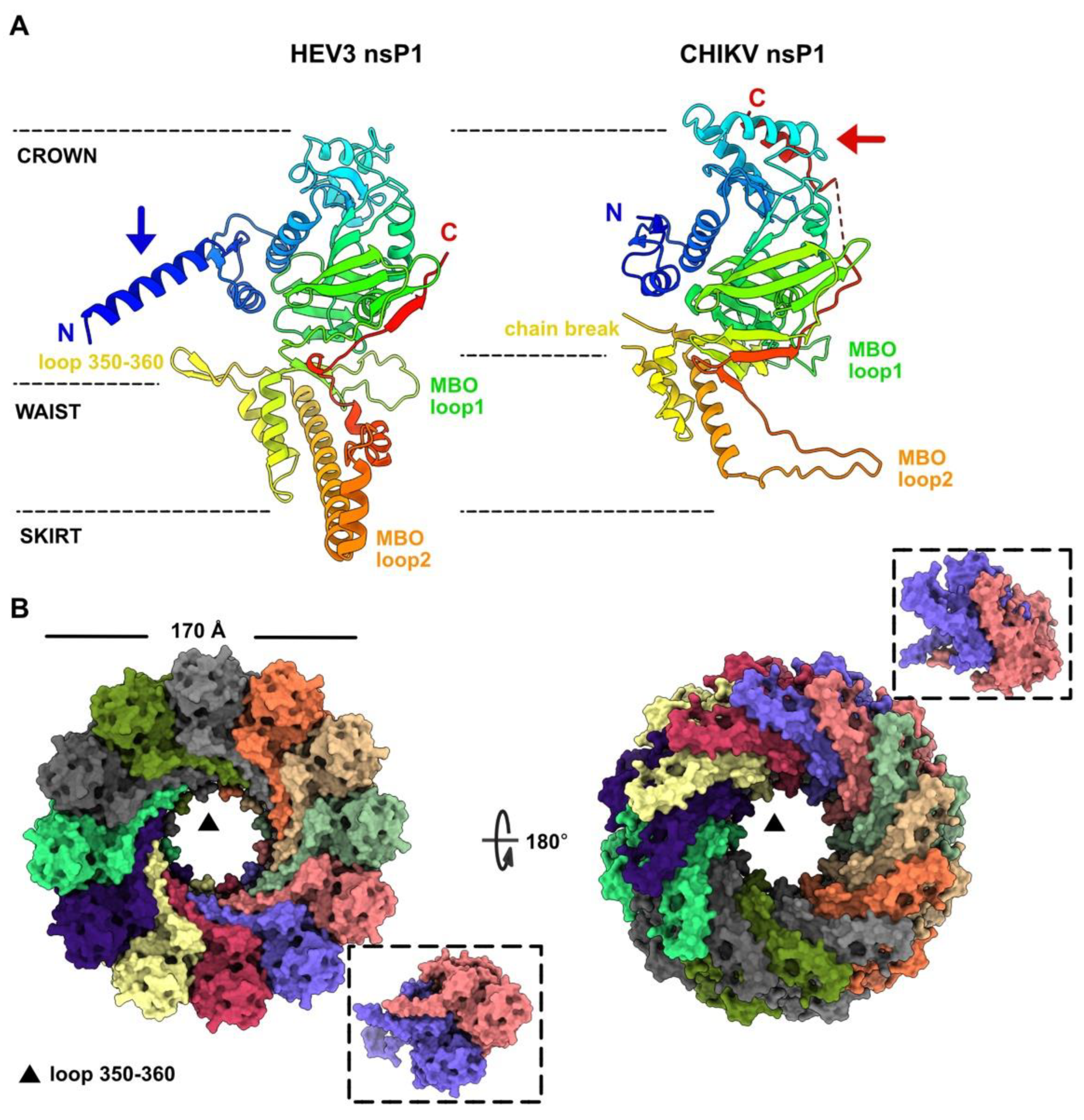
3.2. HEV-3 nsP1: A Putative Capping Pore of the Viral Replication Factory
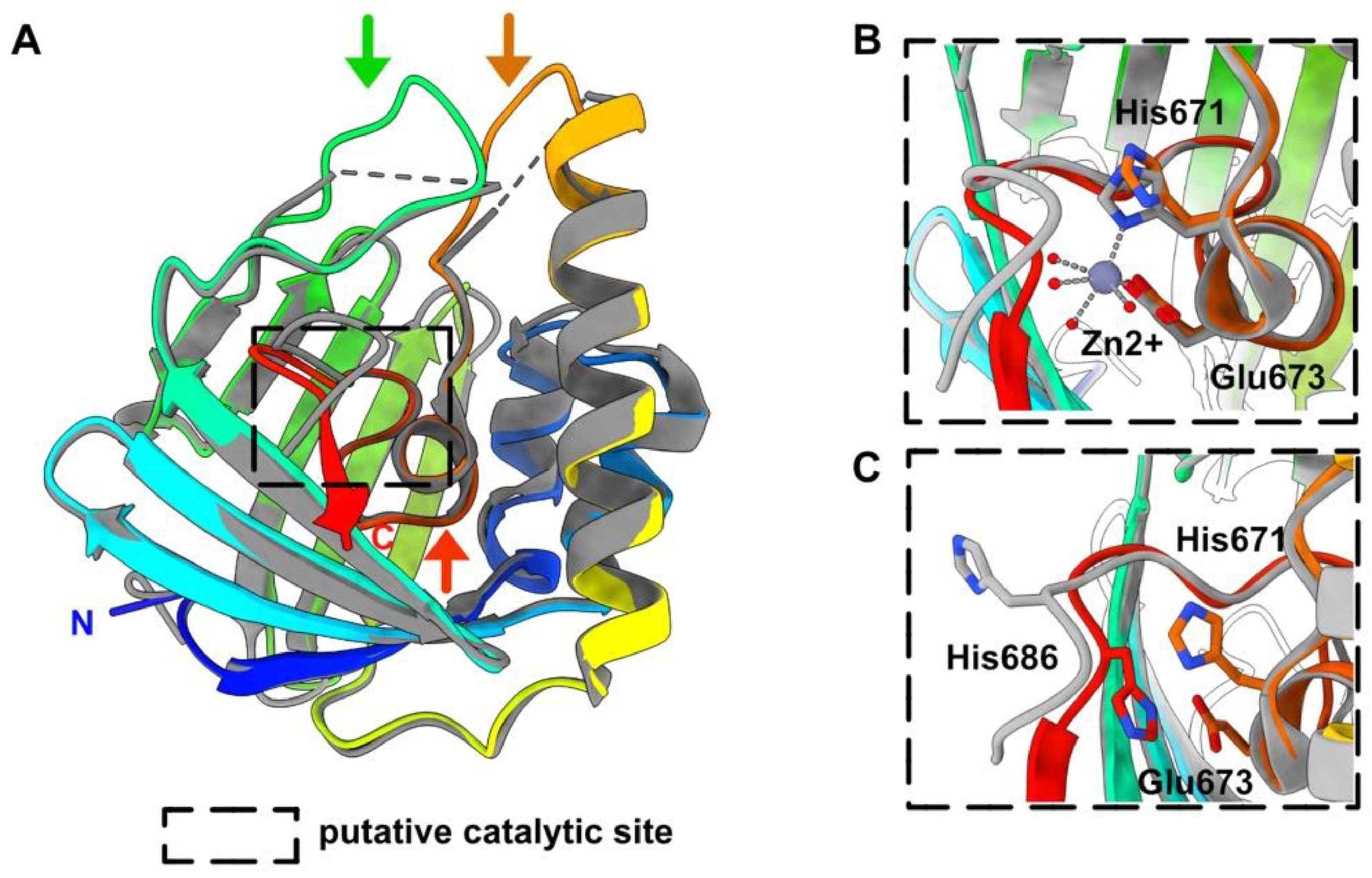
3.3. HEV-3 nsP2: A Metal Binding Protein
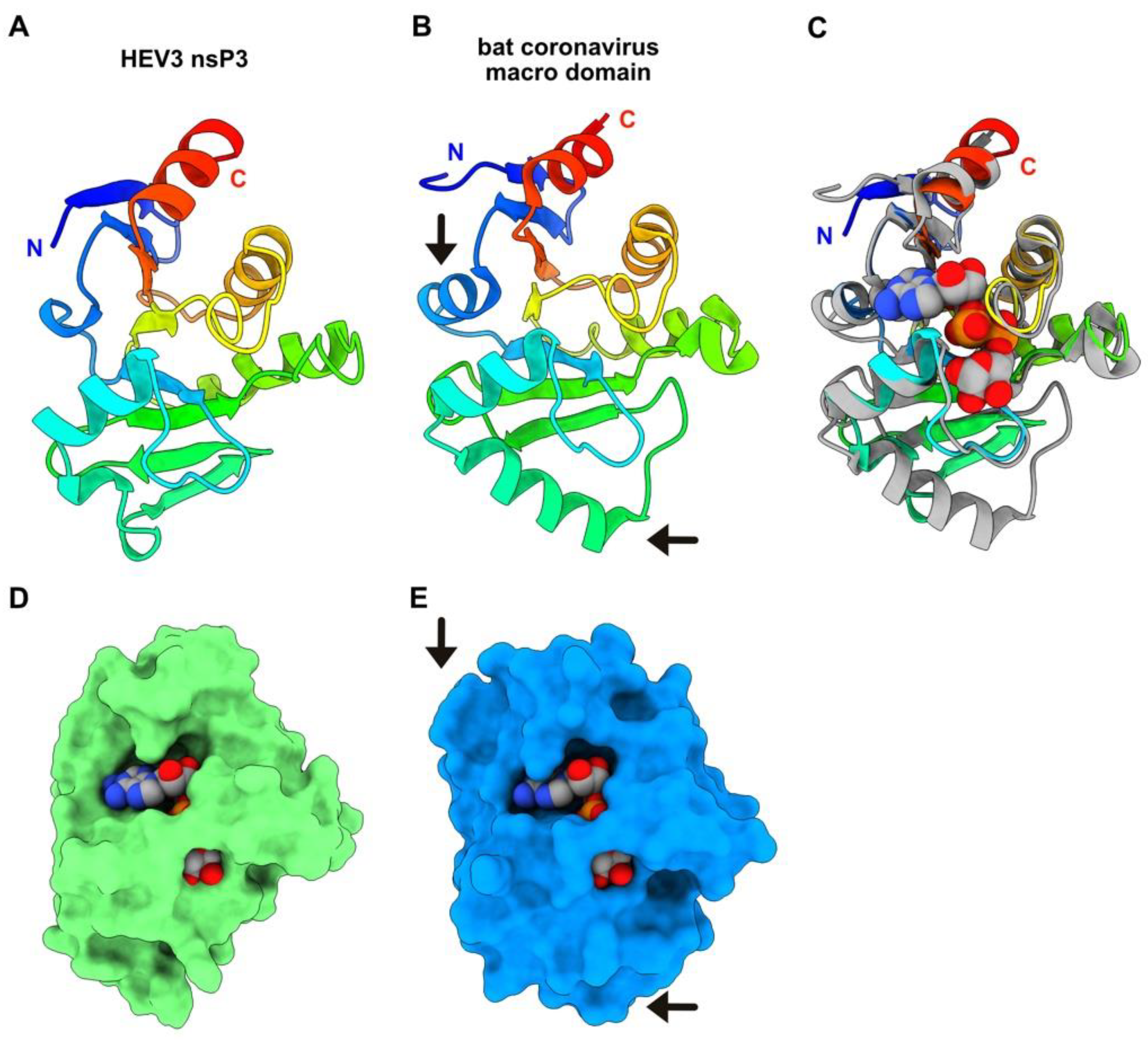
3.4. HEV-3 nsP3 Is a Macro Domain
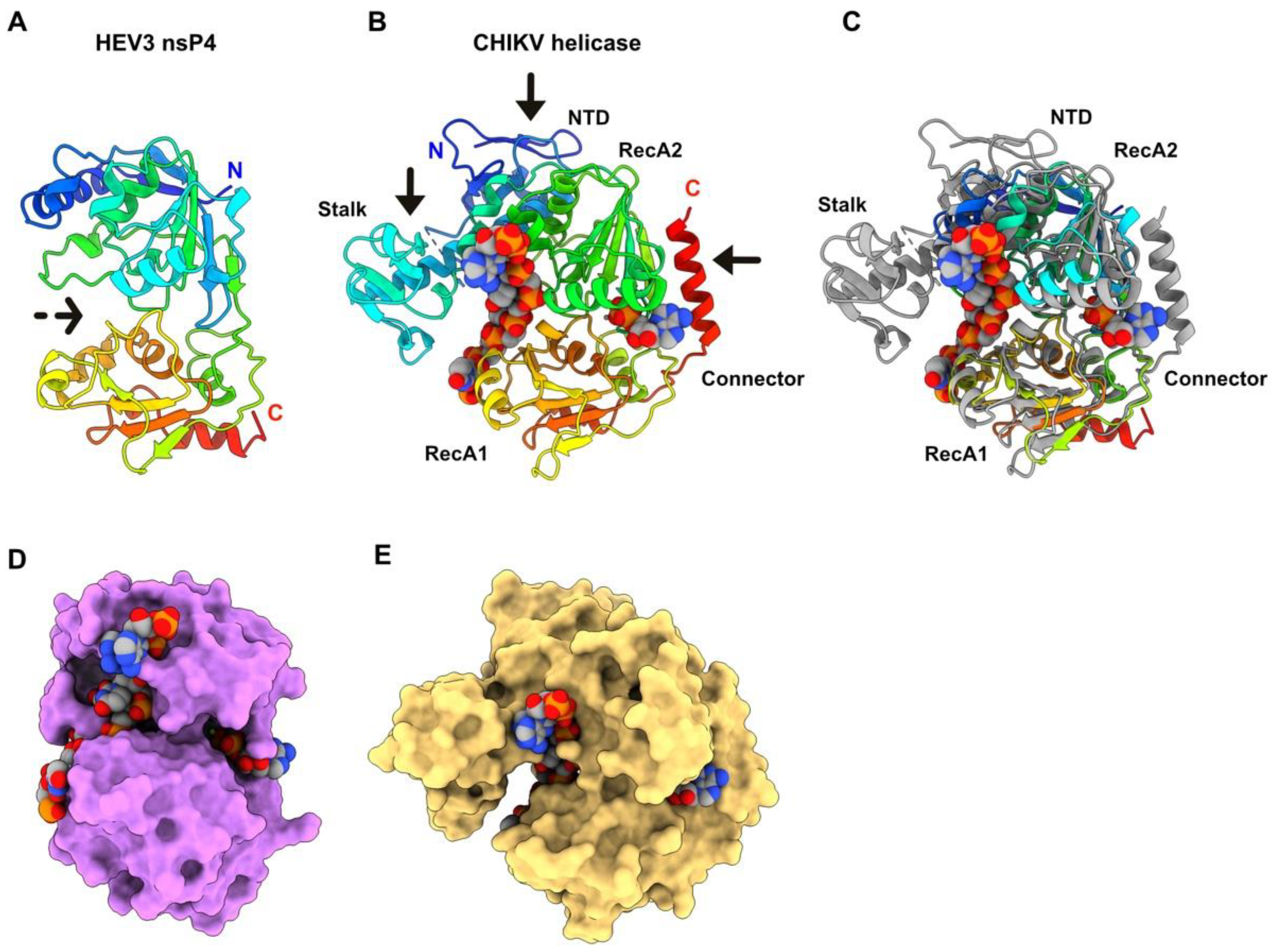
3.5. HEV-3 nsP4: A Helicase
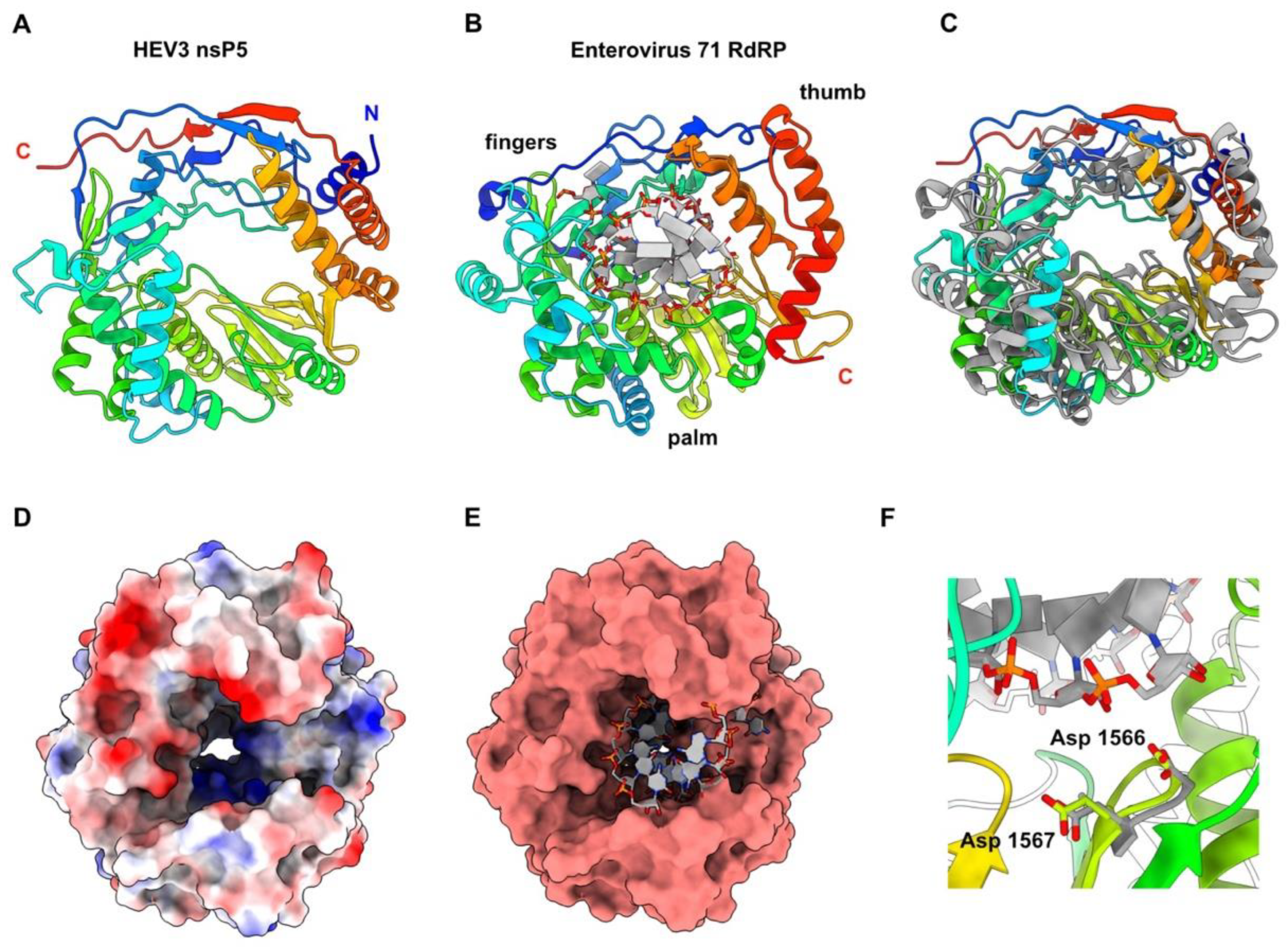
3.6. HEV-3 nsP5: The RNA-Dependent RNA Polymerase
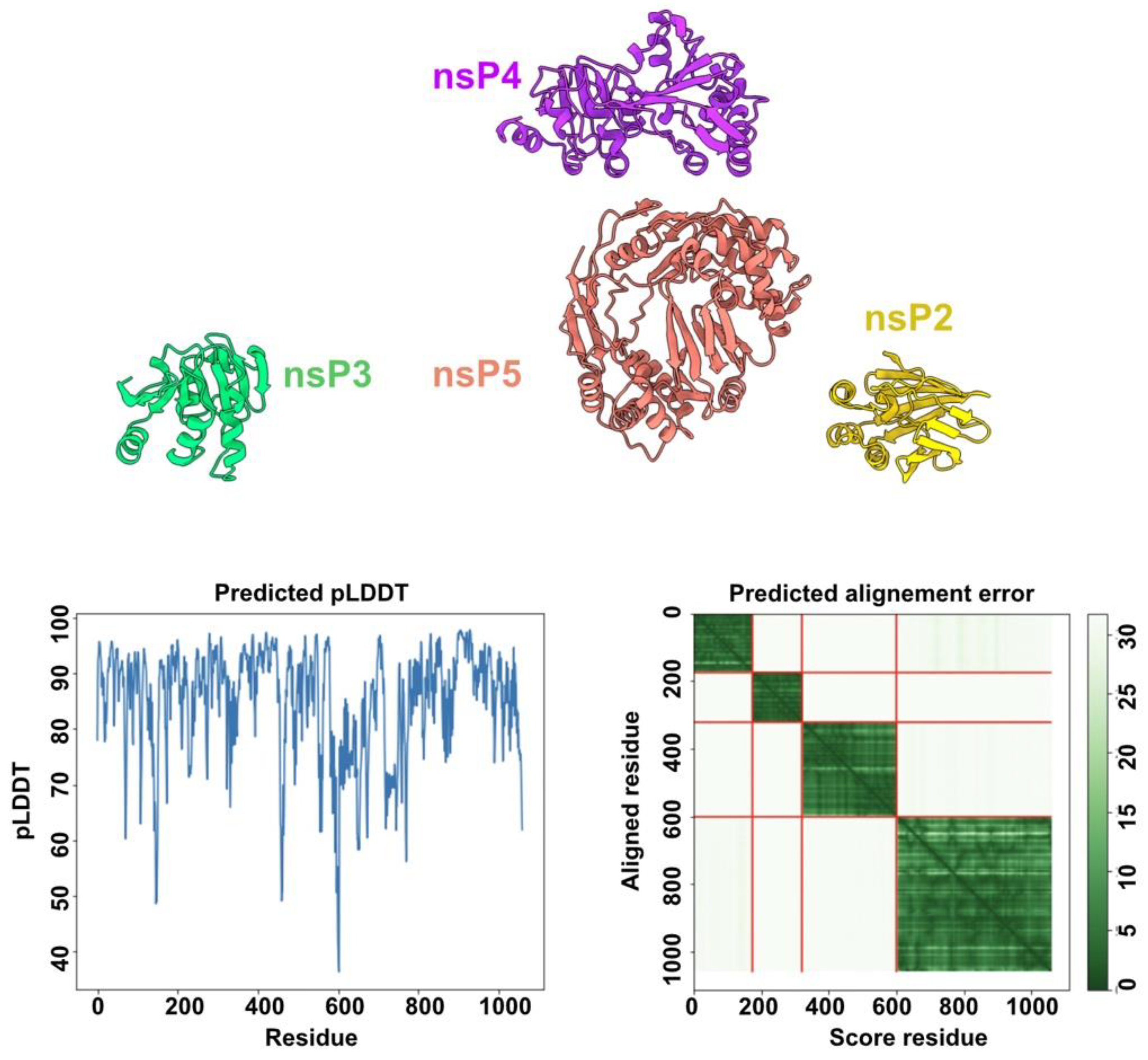
3.7. HEV-3 nsPs May Not Assemble Pre-Formed Replicative Complexes in the Absence of RNA Substrates
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hoofnagle, J.H.; Nelson, K.E.; Purcell, R.H. Hepatitis E. N. Engl. J. Med. 2012, 367, 1237–1244. [Google Scholar] [CrossRef] [PubMed]
- Khuroo, M.S.; Teli, M.R.; Skidmore, S.; Sofi, M.A.; Khuroo, M.I. Incidence and Severity of Viral Hepatitis in Pregnancy. Am. J. Med. 1981, 70, 252–255. [Google Scholar] [CrossRef]
- Dalton, H.R.; Saunders, M.; Woolson, K.L. Hepatitis E Virus in Developed Countries: One of the Most Successful Zoonotic Viral Diseases in Human History? J. Virus Erad. 2015, 1, 23–29. [Google Scholar] [CrossRef]
- Purdy, M.A.; Harrison, T.J.; Jameel, S.; Meng, X.-J.; Okamoto, H.; Van der Poel, W.H.M.; Smith, D.B. Ictv Report Consortium, null ICTV Virus Taxonomy Profile: Hepeviridae. J. Gen. Virol. 2017, 98, 2645–2646. [Google Scholar] [CrossRef]
- Khuroo, M.S.; Khuroo, M.S.; Khuroo, N.S. Hepatitis E: Discovery, Global Impact, Control and Cure. World J. Gastroenterol. 2016, 22, 7030–7045. [Google Scholar] [CrossRef]
- Reyes, G.R.; Purdy, M.A.; Kim, J.P.; Luk, K.C.; Young, L.M.; Fry, K.E.; Bradley, D.W. Isolation of a CDNA from the Virus Responsible for Enterically Transmitted Non-A, Non-B Hepatitis. Science 1990, 247, 1335–1339. [Google Scholar] [CrossRef]
- Yin, X.; Ambardekar, C.; Lu, Y.; Feng, Z. Distinct Entry Mechanisms for Nonenveloped and Quasi-Enveloped Hepatitis E Viruses. J. Virol. 2016, 90, 4232–4242. [Google Scholar] [CrossRef] [Green Version]
- Koonin, E.V.; Gorbalenya, A.E.; Purdy, M.A.; Rozanov, M.N.; Reyes, G.R.; Bradley, D.W. Computer-Assisted Assignment of Functional Domains in the Nonstructural Polyprotein of Hepatitis E Virus: Delineation of an Additional Group of Positive-Strand RNA Plant and Animal Viruses. Proc. Natl. Acad. Sci. USA 1992, 89, 8259–8263. [Google Scholar] [CrossRef] [Green Version]
- Nimgaonkar, I.; Ding, Q.; Schwartz, R.E.; Ploss, A. Hepatitis E Virus: Advances and Challenges. Nat. Rev. Gastroenterol. Hepatol. 2017, 15, 96–110. [Google Scholar] [CrossRef]
- Ansari, I.H.; Nanda, S.K.; Durgapal, H.; Agrawal, S.; Mohanty, S.K.; Gupta, D.; Jameel, S.; Panda, S.K. Cloning, Sequencing, and Expression of the Hepatitis E Virus (HEV) Nonstructural Open Reading Frame 1 (ORF1). J. Med. Virol. 2000, 60, 275–283. [Google Scholar] [CrossRef]
- Ju, X.; Xiang, G.; Gong, M.; Yang, R.; Qin, J.; Li, Y.; Nan, Y.; Yang, Y.; Zhang, Q.C.; Ding, Q. Identification of Functional Cis-Acting RNA Elements in the Hepatitis E Virus Genome Required for Viral Replication. PLoS Pathog. 2020, 16, e1008488. [Google Scholar] [CrossRef] [PubMed]
- Kanade, G.D.; Pingale, K.D.; Karpe, Y.A. Activities of Thrombin and Factor Xa Are Essential for Replication of Hepatitis E Virus and Are Possibly Implicated in the ORF1 Polyprotein Processing. J. Virol. 2018, 92, e01853-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karpe, Y.A.; Lole, K.S. Deubiquitination Activity Associated with Hepatitis E Virus Putative Papain-like Cysteine Protease. J. Gen. Virol. 2011, 92, 2088–2092. [Google Scholar] [CrossRef]
- Kumar, M.; Hooda, P.; Khanna, M.; Patel, U.; Sehgal, D. Development of BacMam Induced Hepatitis E Virus Replication Model in Hepatoma Cells to Study the Polyprotein Processing. Front. Microbiol. 2020, 11, 1347. [Google Scholar] [CrossRef]
- Paliwal, D.; Panda, S.K.; Kapur, N.; Varma, S.P.K.; Durgapal, H. Hepatitis E Virus (HEV) Protease: A Chymotrypsin-like Enzyme That Processes Both Non-Structural (PORF1) and Capsid (PORF2) Protein. J. Gen. Virol. 2014, 95, 1689–1700. [Google Scholar] [CrossRef]
- Panda, S.K.; Ansari, I.H.; Durgapal, H.; Agrawal, S.; Jameel, S. The in Vitro-Synthesized RNA from a CDNA Clone of Hepatitis E Virus Is Infectious. J. Virol. 2000, 74, 2430–2437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parvez, M.K. Molecular Characterization of Hepatitis E Virus ORF1 Gene Supports a Papain-like Cysteine Protease (PCP)-Domain Activity. Virus Res. 2013, 178, 553–556. [Google Scholar] [CrossRef]
- Perttilä, J.; Spuul, P.; Ahola, T. Early Secretory Pathway Localization and Lack of Processing for Hepatitis E Virus Replication Protein PORF1. J. Gen. Virol. 2013, 94, 807–816. [Google Scholar] [CrossRef]
- Ropp, S.L.; Tam, A.W.; Beames, B.; Purdy, M.; Frey, T.K. Expression of the Hepatitis E Virus ORF1. Arch. Virol. 2000, 145, 1321–1337. [Google Scholar] [CrossRef]
- Sehgal, D.; Thomas, S.; Chakraborty, M.; Jameel, S. Expression and Processing of the Hepatitis E Virus ORF1 Nonstructural Polyprotein. Virol. J. 2006, 3, 38. [Google Scholar] [CrossRef] [Green Version]
- Suppiah, S.; Zhou, Y.; Frey, T.K. Lack of Processing of the Expressed ORF1 Gene Product of Hepatitis E Virus. Virol. J. 2011, 8, 245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szkolnicka, D.; Pollán, A.; Da Silva, N.; Oechslin, N.; Gouttenoire, J.; Moradpour, D. Recombinant Hepatitis E Viruses Harboring Tags in the ORF1 Protein. J. Virol. 2019, 93, e00459-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jameel, S.; Zafrullah, M.; Ozdener, M.H.; Panda, S.K. Expression in Animal Cells and Characterization of the Hepatitis E Virus Structural Proteins. J. Virol. 1996, 70, 207–216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, Q.; Heller, B.; Capuccino, J.M.V.; Song, B.; Nimgaonkar, I.; Hrebikova, G.; Contreras, J.E.; Ploss, A. Hepatitis E Virus ORF3 Is a Functional Ion Channel Required for Release of Infectious Particles. Proc. Natl. Acad. Sci. USA 2017, 114, 1147–1152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koonin, E.V.; Dolja, V.V.; Krupovic, M. Origins and Evolution of Viruses of Eukaryotes: The Ultimate Modularity. Virology 2015, 479–480, 2–25. [Google Scholar] [CrossRef] [Green Version]
- Pietilä, M.K.; Hellström, K.; Ahola, T. Alphavirus Polymerase and RNA Replication. Virus Res. 2017, 234, 44–57. [Google Scholar] [CrossRef] [Green Version]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Tunyasuvunakool, K.; Adler, J.; Wu, Z.; Green, T.; Zielinski, M.; Žídek, A.; Bridgland, A.; Cowie, A.; Meyer, C.; Laydon, A.; et al. Highly Accurate Protein Structure Prediction for the Human Proteome. Nature 2021, 596, 590–596. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Applying and Improving AlphaFold at CASP14. Proteins Struct. Funct. Bioinforma. 2021, 89, 1711–1721. [Google Scholar] [CrossRef]
- Evans, R.; O’Neill, M.; Pritzel, A.; Antropova, N.; Senior, A.; Green, T.; Žídek, A.; Bates, R.; Blackwell, S.; Yim, J.; et al. Protein Complex Prediction with AlphaFold-Multimer. bioRxiv 2021. [Google Scholar] [CrossRef]
- Goulet, A.; Cambillau, C. Structure and Topology Prediction of Phage Adhesion Devices Using AlphaFold2: The Case of Two Oenococcus Oeni Phages. Microorganisms 2021, 9, 2151. [Google Scholar] [CrossRef] [PubMed]
- Goulet, A.; Cambillau, C. Present Impact of AlphaFold2 Revolution on Structural Biology, and an Illustration With the Structure Prediction of the Bacteriophage J-1 Host Adhesion Device. Front. Mol. Biosci. 2022, 9, 907452. [Google Scholar] [CrossRef] [PubMed]
- Proudfoot, A.; Hyrina, A.; Holdorf, M.; Frank, A.O.; Bussiere, D. First Crystal Structure of a Nonstructural Hepatitis E Viral Protein Identifies a Putative Novel Zinc-Binding Protein. J. Virol. 2019, 93, e00170-19. [Google Scholar] [CrossRef]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and Development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef] [Green Version]
- Holm, L. DALI and the Persistence of Protein Shape. Protein Sci. Publ. Protein Soc. 2020, 29, 128–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Meng, E.C.; Couch, G.S.; Croll, T.I.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Structure Visualization for Researchers, Educators, and Developers. Protein Sci. Publ. Protein Soc. 2021, 30, 70–82. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.; Bragagnolo, G.; Arranz, R.; Reguera, J. Capping Pores of Alphavirus NsP1 Gate Membranous Viral Replication Factories. Nature 2021, 589, 615–619. [Google Scholar] [CrossRef]
- Hammond, R.G.; Schormann, N.; McPherson, R.L.; Leung, A.K.L.; Deivanayagam, C.C.S.; Johnson, M.A. ADP-Ribose and Analogues Bound to the DeMARylating Macrodomain from the Bat Coronavirus HKU4. Proc. Natl. Acad. Sci. USA 2021, 118, e2004500118. [Google Scholar] [CrossRef]
- Law, Y.-S.; Utt, A.; Tan, Y.B.; Zheng, J.; Wang, S.; Chen, M.W.; Griffin, P.R.; Merits, A.; Luo, D. Structural Insights into RNA Recognition by the Chikungunya Virus NsP2 Helicase. Proc. Natl. Acad. Sci. USA 2019, 116, 9558–9567. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Wu, B.; Soca, W.A.; An, L. Crystal Structure of Classical Swine Fever Virus NS5B Reveals a Novel N-Terminal Domain. J. Virol. 2018, 92, e00324-18. [Google Scholar] [CrossRef] [Green Version]
- Holm, L.; Laakso, L.M. Dali Server Update. Nucleic Acids Res. 2016, 44, W351–W355. [Google Scholar] [CrossRef] [PubMed]
- Schneidman-Duhovny, D.; Inbar, Y.; Nussinov, R.; Wolfson, H.J. PatchDock and SymmDock: Servers for Rigid and Symmetric Docking. Nucleic Acids Res. 2005, 33, W363–W367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, C.; Debing, Y.; Jankevicius, G.; Neyts, J.; Ahel, I.; Coutard, B.; Canard, B. Viral Macro Domains Reverse Protein ADP-Ribosylation. J. Virol. 2016, 90, 8478–8486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poch, O.; Sauvaget, I.; Delarue, M.; Tordo, N. Identification of Four Conserved Motifs among the RNA-Dependent Polymerase Encoding Elements. EMBO J. 1989, 8, 3867–3874. [Google Scholar] [CrossRef]
- Wang, M.; Li, R.; Shu, B.; Jing, X.; Ye, H.-Q.; Gong, P. Stringent Control of the RNA-Dependent RNA Polymerase Translocation Revealed by Multiple Intermediate Structures. Nat. Commun. 2020, 11, 2605. [Google Scholar] [CrossRef] [PubMed]
- Piccininni, S.; Varaklioti, A.; Nardelli, M.; Dave, B.; Raney, K.D.; McCarthy, J.E.G. Modulation of the Hepatitis C Virus RNA-Dependent RNA Polymerase Activity by the Non-Structural (NS) 3 Helicase and the NS4B Membrane Protein. J. Biol. Chem. 2002, 277, 45670–45679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, Z.; Yan, L.; Ren, Z.; Wu, L.; Wang, J.; Guo, J.; Zheng, L.; Ming, Z.; Zhang, L.; Lou, Z.; et al. Delicate Structural Coordination of the Severe Acute Respiratory Syndrome Coronavirus Nsp13 upon ATP Hydrolysis. Nucleic Acids Res. 2019, 47, 6538–6550. [Google Scholar] [CrossRef] [Green Version]
- Grange, Z.L.; Goldstein, T.; Johnson, C.K.; Anthony, S.; Gilardi, K.; Daszak, P.; Olival, K.J.; O’Rourke, T.; Murray, S.; Olson, S.H.; et al. Ranking the Risk of Animal-to-Human Spillover for Newly Discovered Viruses. Proc. Natl. Acad. Sci. USA 2021, 118, e2002324118. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.; Charpilienne, A.; Parent, A.; Boussac, A.; D’Autreaux, B.; Poupon, J.; Poncet, D. The Rotavirus Nonstructural Protein NSP5 Coordinates a [2Fe-2S] Iron-Sulfur Cluster That Modulates Interaction to RNA. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2013, 27, 1074–1083. [Google Scholar] [CrossRef]
- Maio, N.; Lafont, B.A.P.; Sil, D.; Li, Y.; Bollinger, J.M.; Krebs, C.; Pierson, T.C.; Linehan, W.M.; Rouault, T.A. Fe-S Cofactors in the SARS-CoV-2 RNA-Dependent RNA Polymerase Are Potential Antiviral Targets. Science 2021, 373, 236–241. [Google Scholar] [CrossRef]
- Krietsch, J.; Rouleau, M.; Pic, É.; Ethier, C.; Dawson, T.M.; Dawson, V.L.; Masson, J.-Y.; Poirier, G.G.; Gagné, J.-P. Reprogramming Cellular Events by Poly(ADP-Ribose)-Binding Proteins. Mol. Aspects Med. 2013, 34, 1066–1087. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Malone, B.; Llewellyn, E.; Grasso, M.; Shelton, P.M.M.; Olinares, P.D.B.; Maruthi, K.; Eng, E.T.; Vatandaslar, H.; Chait, B.T.; et al. Structural Basis for Helicase-Polymerase Coupling in the SARS-CoV-2 Replication-Transcription Complex. Cell 2020, 182, 1560–1573.e13. [Google Scholar] [CrossRef] [PubMed]
- De Clercq, E.; Li, G. Approved Antiviral Drugs over the Past 50 Years. Clin. Microbiol. Rev. 2016, 29, 695–747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| nsP | Boundaries | PDB-ID | Z-Dali | Rmsd Å | Aligned Residues | Function | Organism | Reference |
|---|---|---|---|---|---|---|---|---|
| 1 | 9–459 | 6z0v | 20.2 | 3.8 | 347/452 | capping pore | CHIKV | [37] |
| 2 | 516–689 | 6nu9 | 29.6 | 1.1 | 166/168 | ZBD | HEV | [33] |
| 3 | 793–941 | 6mea | 18.4 | 1.9 | 138/162 | macro domain | bat CoV-HKU4 | [38] |
| 4 | 944–1223 | 6jim | 21.4 | 3.2 | 262/455 | helicase | CHIKV | [39] |
| 5 | 1242–1700 | 5y6r | 20.7 | 3.5 | 363/667 | RdRp | CSFV | [40] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Goulet, A.; Cambillau, C.; Roussel, A.; Imbert, I. Structure Prediction and Analysis of Hepatitis E Virus Non-Structural Proteins from the Replication and Transcription Machinery by AlphaFold2. Viruses 2022, 14, 1537. https://doi.org/10.3390/v14071537
Goulet A, Cambillau C, Roussel A, Imbert I. Structure Prediction and Analysis of Hepatitis E Virus Non-Structural Proteins from the Replication and Transcription Machinery by AlphaFold2. Viruses. 2022; 14(7):1537. https://doi.org/10.3390/v14071537
Chicago/Turabian StyleGoulet, Adeline, Christian Cambillau, Alain Roussel, and Isabelle Imbert. 2022. "Structure Prediction and Analysis of Hepatitis E Virus Non-Structural Proteins from the Replication and Transcription Machinery by AlphaFold2" Viruses 14, no. 7: 1537. https://doi.org/10.3390/v14071537
APA StyleGoulet, A., Cambillau, C., Roussel, A., & Imbert, I. (2022). Structure Prediction and Analysis of Hepatitis E Virus Non-Structural Proteins from the Replication and Transcription Machinery by AlphaFold2. Viruses, 14(7), 1537. https://doi.org/10.3390/v14071537







