Occurrence, Genetic Variability of Tomato Yellow Ring Orthotospovirus Population and the Development of Reverse Transcription Loop-Mediated Isothermal Amplification Assay for Its Rapid Detection
Abstract
1. Introduction
2. Materials and Methods
2.1. Virus Source
2.2. Viruses Infecting Tomatoes in Poland
2.3. Analysis of the TYRV Polish Population
2.4. Phylogenetic-Trait Association
2.5. RT-LAMP
3. Results
3.1. Viruses Infecting Tomato in Poland
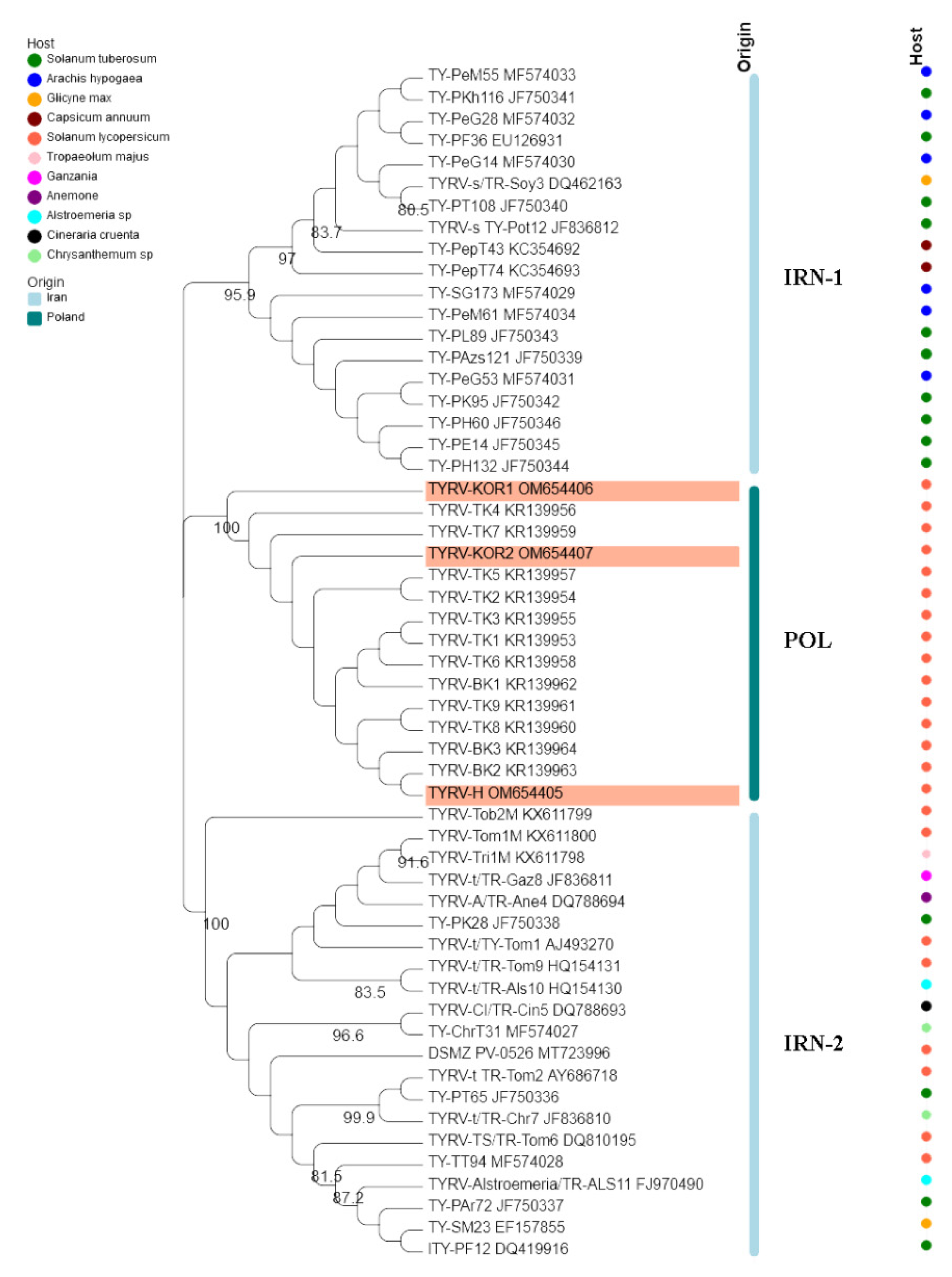
3.2. Analysis of TYRV Population
3.3. Host Driven Structure of the TYRV Population
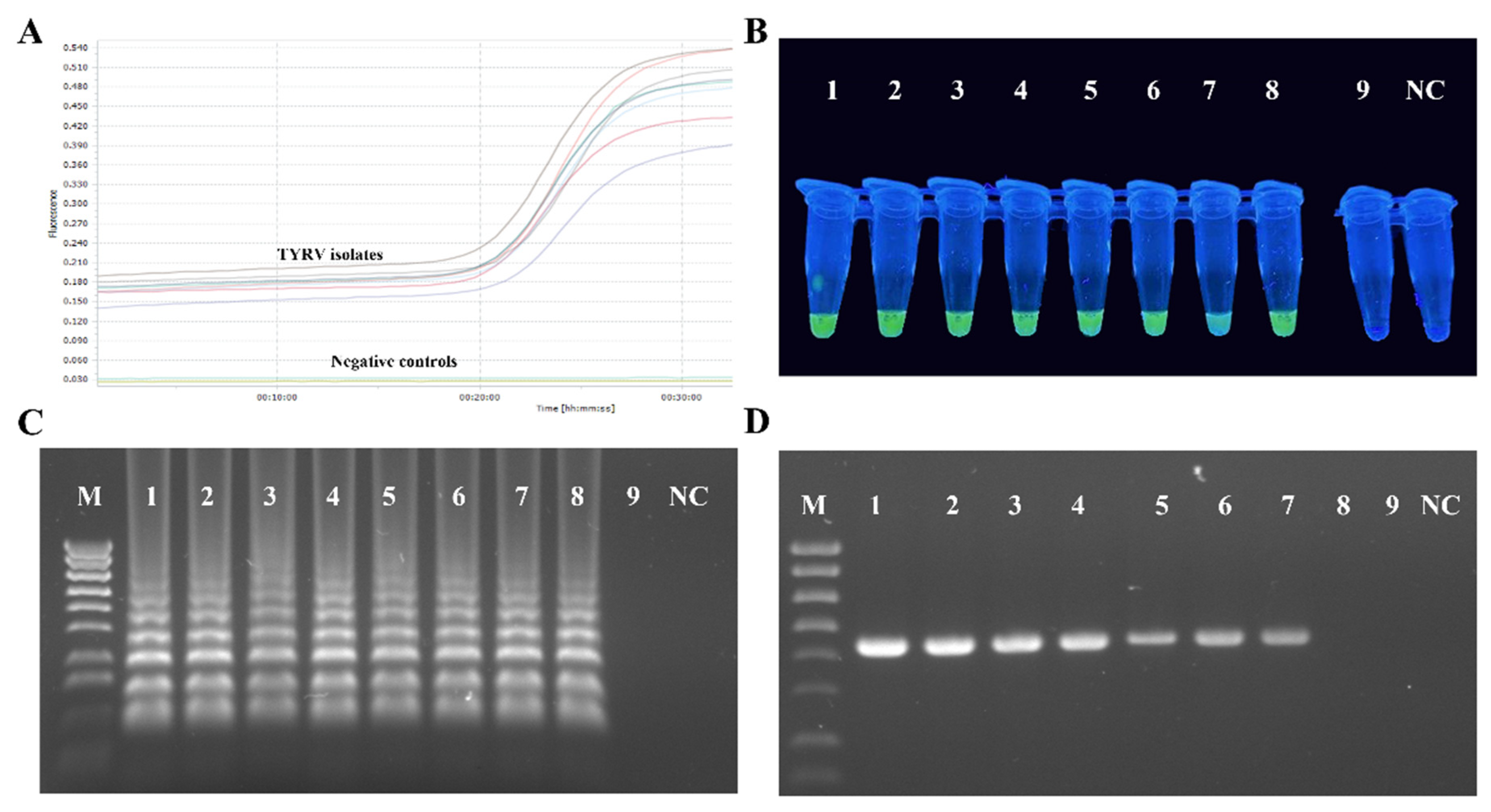
3.4. Optimisation of the RT-LAMP Conditions
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Food and Agriculture Organization of the United Nations. Available online: https://www.fao.org/home/en (accessed on 17 March 2022).
- Statistics|Eurostat. Available online: https://ec.europa.eu/eurostat/databrowser/view/tag00115/default/table?lang=en (accessed on 25 April 2022).
- Statistics Poland—Production and Foreign Trade of Agricultural Products in 2019. Available online: https://stat.gov.pl/obszary-tematyczne/rolnictwo-lesnictwo/uprawy-rolne-i-ogrodnicze/produkcja-i-handel-zagraniczny-produktami-rolnymi-w-2019-roku,1,16.html (accessed on 25 April 2022).
- Elena, S.F.; Fraile, A.; García-Arenal, F. Evolution and Emergence of Plant Viruses. Adv. Virus Res. 2014, 88, 161–191. [Google Scholar] [CrossRef] [PubMed]
- Rubio, L.; Guerri, J.; Moreno, P. Genetic Variability and Evolutionary Dynamics of Viruses of the Family Closteroviridae. Front. Microbiol. 2013, 4, 151. [Google Scholar] [CrossRef] [PubMed]
- Jończyk, M.; Le Gall, O.; Pałucha, A.; Borodynko, N.; Pospieszny, H. Cloning and Sequencing of Full-Length CDNAs of RNA1 and RNA2 of a Tomato Black Ring Virus Isolate from Poland. Arch. Virol. 2004, 149, 799–807. [Google Scholar] [CrossRef] [PubMed]
- Pospieszny, H.; Borodynko, N.; Obrępalska-Stęplowska, A.; Hasiów, B. The First Report of Tomato Torrado Virus in Poland. Plant Dis. 2007, 91, 1364. [Google Scholar] [CrossRef]
- Hasiów-Jaroszewska, B.; Borodynko, N.; Jackowiak, P.; Figlerowicz, M.; Pospieszny, H. Pepino Mosaic Virus—A Pathogen of Tomato Crops in Poland: Biology, Evolution and Diagnostics. J. Plant Prot. Res. 2010, 50, 470–476. [Google Scholar] [CrossRef]
- Laskowska, D. Characterization of Tomato Spotted Wilt Disease on Tomato and Vector Role in Transmission. Stud. Rep. Inst. Soil Sci. Plant Cultiv. 2008, 13, 43–50. [Google Scholar]
- Pospieszny, H. Nasilone wystepowanie chorob wirusowych pomidora szklarniowego w Wielkopolsce, w roku 2002. Prog. Plant Prot. 2003, 43, 321–324. [Google Scholar]
- Moycho, W.; Gubanski, M.; Fomaidis, B.; Lemanska, M.; Wajsbard, E. Występowanie wirusa mozaiki tytoniowej na plantacjach pomidorów miasta Łodzi i okolicy. Postępy Nauk Rolniczych 1960, 7, 79–82. [Google Scholar]
- Minicka, J.; Zarzyńska-Nowak, A.; Budzyńska, D.; Borodynko-Filas, N.; Hasiów-Jaroszewska, B. High-Throughput Sequencing Facilitates Discovery of New Plant Viruses in Poland. Plants 2020, 9, 820. [Google Scholar] [CrossRef]
- Minicka, J.; Hasiów-Jaroszewska, B.; Borodynko-Filas, N.; Pospieszny, H.; Hanssen, I.M. Rapid Evolutionary Dynamics of the Pepino Mosaic Virus—Status and Future Perspectives. J. Plant Prot. Res. 2016, 56, 337–345. [Google Scholar] [CrossRef]
- Hasiów-Jaroszewska, B.; Stachecka, J.; Minicka, J.; Sowiński, M.; Borodynko, N. Variability of Potato Virus Y in Tomato Crops in Poland and Development of a Reverse-Transcription Loop-Mediated Isothermal Amplification Method for Virus Detection. Phytopathology 2015, 105, 1270–1276. [Google Scholar] [CrossRef] [PubMed]
- Gould, A.R.; Palukaitis, P.; Symons, R.H.; Mossop, D.W. Characterization of a Satellite RNA Associated with Cucumber Mosaic Virus. Virology 1978, 84, 443–455. [Google Scholar] [CrossRef]
- Aranda, M.A.; Fraile, A.; Garcia-Arenal, F. Genetic Variability and Evolution of the Satellite RNA of Cucumber Mosaic Virus during Natural Epidemics. J. Virol. 1993, 67, 5896–5901. [Google Scholar] [CrossRef] [PubMed]
- Murant, A.F.; Mayo, M.A.; Harrison, B.D.; Goold, R.A.Y. Evidence for Two Functional RNA Species and a ‘Satellite’ RNA in Tomato Black Ring Virus. J. Gen. Virol. 1973, 19, 275–278. [Google Scholar] [CrossRef]
- Oncino, C.; Hemmer, O.; Fritsch, C. Specificity in the Association of Tomato Black Ring Virus Satellite RNA with Helper Virus. Virology 1995, 213, 87–96. [Google Scholar] [CrossRef]
- Hemmer, O.; Meyer, M.; Greif, C.; Fritsch, C.Y. Comparison of the Nucleotide Sequences of Five Tomato Black Ring Virus Satellite RNAs. J. Gen. Virol. 1987, 68, 1823–1833. [Google Scholar] [CrossRef]
- Shimura, H.; Pantaleo, V.; Ishihara, T.; Myojo, N.; Inaba, J.; Sueda, K.; Burgyán, J.; Masuta, C. A Viral Satellite RNA Induces Yellow Symptoms on Tobacco by Targeting a Gene Involved in Chlorophyll Biosynthesis Using the RNA Silencing Machinery. PLoS Pathog. 2011, 7, e1002021. [Google Scholar] [CrossRef]
- Escriu, F.; Fraile, A.; García-Arenal, F. Evolution of Virulence in Natural Populations of the Satellite RNA of Cucumber Mosaic Virus. Phytopathology 2000, 90, 480–485. [Google Scholar] [CrossRef]
- Takanami, Y. A Striking Change in Symptoms on Cucumber Mosaic Virus-Infected Tobacco Plants Induced by a Satellite RNA. Virology 1981, 109, 120–126. [Google Scholar] [CrossRef]
- Xu, P.; Roossinck, M.J. Cucumber Mosaic Virus D Satellite RNA–Induced Programmed Cell Death in Tomato. Plant Cell 2000, 12, 1079–1092. [Google Scholar] [CrossRef]
- Zarzyńska-Nowak, A.; Rymelska, N.; Borodynko, N.; Hasiów-Jaroszewska, B. The Occurrence of Tomato Yellow Ring Virus on Tomato in Poland. Plant Dis. 2016, 100, 234. [Google Scholar] [CrossRef]
- Winter, S.; Koerbler, M.; Shahraeen, N.; Katul, L.; Lesemann, D.E. Characterization of a New Tospovirus Species Infecting Tomato in Iran. In Proceedings of the First Joint Conference of International Working Groups on Legume Viruses and Vegetable Viruses, Bonn, Germany, 4–9 August 2002. [Google Scholar]
- Hassani-Mehraban, A.; Saaijer, J.; Peters, D.; Goldbach, R.W.; Kormelink, R. Molecular and Biological Comparison of Two Tomato Yellow Ring Virus (TYRV) Isolates: Challenging the Tospovirus Species Concept. Arch. Virol. 2007, 152, 85–96. [Google Scholar] [CrossRef] [PubMed]
- Rasoulpour, R.; Izadpanah, K. Characterisation of Cineraria Strain of Tomato Yellow Ring Virus from Iran. Australas. Plant Pathol. 2007, 36, 286–294. [Google Scholar] [CrossRef]
- Golnaraghi, A.R.; Pourrahim, R.; Ahoonmanesh, A.; Zamani-Zadeh, H.R.; Farzadfar, S. Detection and Characterization of a Distinct Isolate of Tomato Yellow Fruit Ring Virus from Potato. Plant Dis. 2008, 92, 1280–1287. [Google Scholar] [CrossRef] [PubMed]
- Beikzadeh, N.; Bayat, H.; Jafarpour, B.; Rohani, H.; Peters, D.; Hassani-Mehraban, A. Infection of Alstroemeria Plants with Tomato Yellow Ring Virus in Iran. J. Phytopathol. 2012, 160, 45–47. [Google Scholar] [CrossRef]
- Golnaraghi, A.; Shahraeen, N.; Nguyen, H.D. Characterization and Genetic Structure of a Tospovirus Causing Chlorotic Ring Spots and Chlorosis Disease on Peanut; Comparison with Iranian and Polish Populations of Tomato Yellow Fruit Ring Virus. Plant Dis. 2018, 102, 1509–1519. [Google Scholar] [CrossRef]
- Birithia, R.; Subramanian, S.; Villinger, J.; Muthomi, J.W.; Narla, R.D.; Pappu, H.R. First Report of Tomato Yellow Ring Virus (Tospovirus, Bunyaviridae) Infecting Tomato in Kenya. Plant Dis. 2012, 96, 1384. [Google Scholar] [CrossRef]
- Mortazavi, N.; Aleosfoor, M.; Minaei, K. Transmission of Cineraria Isolate of Tomato Yellow Ring Virus by Frankliniella occidentalis and Thrips tabaci (Thysanoptera, Thripidae). Linz. Biol. Beiträge J. 2014, 45, 2011–2018. [Google Scholar]
- Mortazavi, N.; Aleosfoor, M. Efficiency of Thrips tabaci and Frankliniella occidentalis Populations in Transmission of Tomato Yellow Ring Virus. Zool. Ecol. 2015, 25, 241–246. [Google Scholar] [CrossRef]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-Mediated Isothermal Amplification of DNA. Nucleic Acids Res. 2000, 28, e63. [Google Scholar] [CrossRef]
- Chaouch, M. Loop-Mediated Isothermal Amplification (LAMP): An Effective Molecular Point-of-Care Technique for the Rapid Diagnosis of Coronavirus SARS-CoV-2. Rev. Med. Virol. 2021, 31, e2215. [Google Scholar] [CrossRef] [PubMed]
- Nagamine, K.; Hase, T.; Notomi, T. Accelerated Reaction by Loop-Mediated Isothermal Amplification Using Loop Primers. Mol. Cell. Probes 2002, 16, 223–229. [Google Scholar] [CrossRef] [PubMed]
- Trzmiel, K.; Hasiów-Jaroszewska, B. Development of Loop-Mediated Isothermal Amplification Assay for Rapid Detection of Genetically Different Wheat Dwarf Virus Isolates. Mol. Biol. Rep. 2020, 47, 8325–8329. [Google Scholar] [CrossRef] [PubMed]
- Liang, R.; Liang, L.; Ren, X.; Jia, Y.; Han, K.; Zhao, J.; Song, C.; Cui, S. Development of a TaqMan Loop-Mediated Isothermal Amplification Assay for the Rapid Detection of Pigeon Paramyxovirus Type 1. Arch. Virol. 2021, 166, 1599–1605. [Google Scholar] [CrossRef]
- Jang, W.S.; Lim, D.H.; Yoon, J.; Kim, A.; Lim, M.; Nam, J.; Yanagihara, R.; Ryu, S.-W.; Jung, B.K.; Ryoo, N.-H.; et al. Development of a Multiplex Loop-Mediated Isothermal Amplification (LAMP) Assay for on-Site Diagnosis of SARS CoV-2. PLoS ONE 2021, 16, e0248042. [Google Scholar] [CrossRef]
- Zarzyńska-Nowak, A.; Hasiów-Jaroszewska, B.; Jeżewska, M. Molecular Analysis of Barley Stripe Mosaic Virus Isolates Differing in Their Biological Properties and the Development of Reverse Transcription Loop-Mediated Isothermal Amplification Assays for Their Detection. Arch. Virol. 2018, 163, 1163–1170. [Google Scholar] [CrossRef]
- Hasiów-Jaroszewska, B.; Borodynko, N. Detection of Pepino Mosaic Virus Isolates from Tomato by One-Step Reverse Transcription Loop-Mediated Isothermal Amplification. Arch. Virol. 2013, 158, 2153–2156. [Google Scholar] [CrossRef]
- Hasiów-Jaroszewska, B.; Kasprowicz, M.; Borodynko-Filas, N. Rapid Detection of Cucumber Mosaic Virus Isolates Representing Distinct Phylogenetic Subgroups by Reverse Transcription, Loop-Mediated Isothermal Amplification. J. Plant Dis. Prot. 2018, 125, 227–230. [Google Scholar] [CrossRef]
- Bashir, N.S.; Kalhor, M.R.; Zarghani, S.N. Detection, Differentiation and Phylogenetic Analysis of Cucumber Mosaic Virus Isolates from Cucurbits in the Northwest Region of Iran. Virus Genes 2006, 32, 277–288. [Google Scholar] [CrossRef]
- Škorić, D.; Krajačić, M.; Perica, M.Ć.; Halupecki, E.; Topić, J.; Igrc-Barčić, J. Cucumber Mosaic Virus (Cucumovirus) and Associated SatRNA in Weed Species under the Natural Epidemic Conditions of Tomato Lethal Necrosis in Croatia/Gurken Mosaik Virus (Cucumovirus) Und Verwandte SatRNA, Die in Kroatien Unter Natürlichen Bedingungen in Unkrautarten Gefunden Wurden, Führten Zu Letaler Tomatennekrose. Z. Pflanzenkrankh. Pflanzenschutz/J. Plant Dis. Prot. 2000, 107, 304–309. [Google Scholar]
- Rigotti, S.; Gugerli, P. Rapid Identification of Potato Virus Y Strains by One-Step Triplex RT-PCR. J. Virol. Methods 2007, 140, 90–94. [Google Scholar] [CrossRef] [PubMed]
- Zarzyńska-Nowak, A.; Hasiów-Jaroszewska, B.; Budzyńska, D.; Trzmiel, K. Genetic Variability of Polish Tomato Black Ring Virus Isolates and Their Satellite RNAs. Plant Pathol. 2020, 69, 1034–1041. [Google Scholar] [CrossRef]
- PM 7/146 (1); Tomato Brown Rugose Fruit Virus. EPPO: Luxembourg, 2021; Volume 51, pp. 178–197. [CrossRef]
- Segev, L.; Wintermantel, W.M.; Polston, J.E.; Lapidot, M. First Report of Tomato Chlorosis Virus in Israel. Plant Dis. 2004, 88, 1160. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Udaya Shankar, A.C.; Nayaka, S.C.; Lund, O.S.; Prakash, H.S. Detection of Tobacco Mosaic Virus and Tomato Mosaic Virus in Pepper and Tomato by Multiplex RT–PCR. Lett. Appl. Microbiol. 2011, 53, 359–363. [Google Scholar] [CrossRef]
- Budziszewska, M.; Obrepalska-Steplowska, A.; Wieczorek, P.; Pospieszny, H. The Nucleotide Sequence of a Polish Isolate of Tomato Torrado Virus. Virus Genes 2008, 37, 400–406. [Google Scholar] [CrossRef]
- Zarzyńska-Nowak, A.; Hasiów-Jaroszewska, B.; Korbecka-Glinka, G.; Przybyś, M.; Borodynko-Filas, N. A Multiplex RT-PCR Assay for Simultaneous Detection of Tomato Spotted Wilt Virus and Tomato Yellow Ring Virus in Tomato Plants. Can. J. Plant Pathol. 2018, 40, 580–586. [Google Scholar] [CrossRef]
- Hall, T. BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Edgar, R.C. MUSCLE: A Multiple Sequence Alignment Method with Reduced Time and Space Complexity. BMC Bioinform. 2004, 5, 113. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Muhire, B.M.; Varsani, A.; Martin, D.P. SDT: A Virus Classification Tool Based on Pairwise Sequence Alignment and Identity Calculation. PLoS ONE 2014, 9, e108277. [Google Scholar] [CrossRef]
- Kosakovsky Pond, S.L.; Posada, D.; Gravenor, M.B.; Woelk, C.H.; Frost, S.D.W. Automated Phylogenetic Detection of Recombination Using a Genetic Algorithm. Mol. Biol. Evol. 2006, 23, 1891–1901. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, B.; Gao, S.; Lercher, M.J.; Hu, S.; Chen, W.-H. Evolview v3: A Webserver for Visualization, Annotation, and Management of Phylogenetic Trees. Nucleic Acids Res. 2019, 47, W270–W275. [Google Scholar] [CrossRef] [PubMed]
- Weaver, S.; Shank, S.D.; Spielman, S.J.; Li, M.; Muse, S.V.; Kosakovsky Pond, S.L. Datamonkey 2.0: A Modern Web Application for Characterizing Selective and Other Evolutionary Processes. Mol. Biol. Evol. 2018, 35, 773–777. [Google Scholar] [CrossRef] [PubMed]
- Bouckaert, R.; Vaughan, T.G.; Barido-Sottani, J.; Duchêne, S.; Fourment, M.; Gavryushkina, A.; Heled, J.; Jones, G.; Kühnert, D.; Maio, N.D.; et al. BEAST 2.5: An Advanced Software Platform for Bayesian Evolutionary Analysis. PLoS Comput. Biol. 2019, 15, e1006650. [Google Scholar] [CrossRef] [PubMed]
- Posada, D. JModelTest: Phylogenetic Model Averaging. Mol. Biol. Evol. 2008, 25, 1253–1256. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef]
- Parker, J.; Rambaut, A.; Pybus, O.G. Correlating Viral Phenotypes with Phylogeny: Accounting for Phylogenetic Uncertainty. Infect. Genet. Evol. 2008, 8, 239–246. [Google Scholar] [CrossRef]
- Hanssen, I.M.; Lapidot, M.; Thomma, B.P.H.J. Emerging Viral Diseases of Tomato Crops. Mol. Plant Microbe Interact. 2010, 23, 539–548. [Google Scholar] [CrossRef]
- Oladokun, J.O.; Halabi, M.H.; Barua, P.; Nath, P.D. Tomato Brown Rugose Fruit Disease: Current Distribution, Knowledge and Future Prospects. Plant Pathol. 2019, 68, 1579–1586. [Google Scholar] [CrossRef]
- Maree, H.J.; Fox, A.; Al Rwahnih, M.; Boonham, N.; Candresse, T. Application of HTS for Routine Plant Virus Diagnostics: State of the Art and Challenges. Front. Plant Sci. 2018, 9, 1082. [Google Scholar] [CrossRef]
- Hasiów-Jaroszewska, B.; Boezen, D.; Zwart, M.P. Metagenomic Studies of Viruses in Weeds and Wild Plants: A Powerful Approach to Characterise Variable Virus Communities. Viruses 2021, 13, 1939. [Google Scholar] [CrossRef] [PubMed]
- Hasiów-Jaroszewska, B.; Pospieszny, H.; Borodynko, N. New Necrotic Isolates of Pepino Mosaic Virus Representing the Ch2 Genotype. J. Phytopathol. 2009, 157, 494–496. [Google Scholar] [CrossRef]
- Minicka, J.; Rymelska, N.; Elena, S.F.; Czerwoniec, A.; Hasiów-Jaroszewska, B. Molecular Evolution of Pepino Mosaic Virus during Long-Term Passaging in Different Hosts and Its Impact on Virus Virulence. Ann. Appl. Biol. 2015, 166, 389–401. [Google Scholar] [CrossRef]
- Ghotbi, T.; Shahraeen, N.; Winter, S. Occurrence of Tospoviruses in Ornamental and Weed Species in Markazi and Tehran Provinces in Iran. Plant Dis. 2005, 89, 425–429. [Google Scholar] [CrossRef]
- Jacquemond, M. Cucumber Mosaic Virus. In Advances in Virus Research; Loebenstein, G., Lecoq, H., Eds.; Viruses and Virus Diseases of Vegetables in the Mediterranean Basin; Academic Press: London, UK, 2012; Chapter 13; Volume 84, pp. 439–504. [Google Scholar]
- Hasiów-Jaroszewska, B.; Budzyńska, D.; Rymelska, N.; Korpys, P.; Borodynko-Filas, N. Phylogenetic Evidence of Natural Reassortants in the Cucumber Mosaic Virus Population in Poland. Can. J. Plant Pathol. 2018, 40, 587–593. [Google Scholar] [CrossRef]
- Hasiów-Jaroszewska, B.; Chrzanowski, M.; Budzyńska, D.; Rymelska, N.; Borodynko-Filas, N. Genetic Diversity, Distant Phylogenetic Relationships and the Occurrence of Recombination Events among Cucumber Mosaic Virus Isolates from Zucchini in Poland. Arch. Virol. 2017, 162, 1751–1756. [Google Scholar] [CrossRef] [PubMed]
- Kaper, J.M.; Waterworth, H.E. Cucumber Mosaic Virus Associated RNA 5: Causal Agent for Tomato Necrosis. Science 1977, 196, 429–431. [Google Scholar] [CrossRef] [PubMed]
- Gallitelli, D.; Di Franco, A.; Vovlas, C.; Kaper, J. Infezioni Miste del Virus del Mosaico del Cetriolo (CMV) e di Potyvirus in Colture Ortive di Puglia e Basilicata. Inf. Fitopatol. 1988, 38, 57–64. [Google Scholar]
- Jorda, C.; Alfaro, A.; Aranda, A.; Moriones, E.; Garcia, F. Epidemic of Cucumber Mosaic Virus plus Satellite RNA in Tomatoes of Eastern Spain. Plant Dis. 1992, 76, 363–366. [Google Scholar] [CrossRef]
- Giakountis, A.; Tsarmpopoulos, I.; Chatzivassiliou, E.K. Cucumber Mosaic Virus Isolates from Greek Legumes Are Associated with Satellite RNAs That Are Necrogenic for Tomato. Plant Dis. 2018, 102, 2268–2276. [Google Scholar] [CrossRef]
- Pospieszny, H.; Borodynko, N.; Hasiow-Jaroszewska, B. Wirus Y ziemniaka (Potato Virus Y, PVY) na pomidorze szklarniowym. Prog. Plant Prot. 2009, 49, 1327–1330. [Google Scholar]
- Jeżewska, M.; Trzmiel, K.; Zarzyńska-Nowak, A. Detection of Infectious Tobamoviruses in Irrigation and Drainage Canals in Greater Poland. J. Plant Prot. Res. 2018, 58, 202–205. [Google Scholar] [CrossRef]
- Salem, N.; Mansour, A.; Ciuffo, M.; Falk, B.W.; Turina, M. A New Tobamovirus Infecting Tomato Crops in Jordan. Arch. Virol. 2016, 161, 503–506. [Google Scholar] [CrossRef] [PubMed]
- EPPO Tomato Brown Rugose Fruit Virus (TOBRFV) [World Distribution]. EPPO Global Database. Available online: https://gd.eppo.int/taxon/TOBRFV/distribution (accessed on 15 March 2022).
- Wintermantel, W.M.; Wisler, G.C. Vector Specificity, Host Range, and Genetic Diversity of Tomato Chlorosis Virus. Plant Dis. 2006, 90, 814–819. [Google Scholar] [CrossRef]
- Pourrahim, R.; Golnaraghi, A.; Farzadfar, S.; Ohshima, K. Partial Biological and Molecular Characterization of Tomato Yellow Fruit Ring Virus Isolates from Potato. Plant Pathol. J. 2012, 28, 390–400. [Google Scholar] [CrossRef][Green Version]
- Golnaraghi, A.R.; Pourrahim, R.; Farzadfar, S.; Ahoonmanesh, A. Identification and Partial Characterization of a Tospovirus Causing Leaf and Stem Necrosis on Potato. Plant Pathol. J. 2007, 6, 227–234. [Google Scholar]
- Worobey, M.; Holmes, E.C. Evolutionary Aspects of Recombination in RNA Viruses. J. Gen. Virol. 1999, 80, 2535–2543. [Google Scholar] [CrossRef]
- Rubio, L.; Galipienso, L.; Ferriol, I. Detection of Plant Viruses and Disease Management: Relevance of Genetic Diversity and Evolution. Front. Plant Sci. 2020, 11, 1092. [Google Scholar] [CrossRef]
- Beikzadeh, N.; Jafarpour, B.; Rouhani, H.; Peters, D.; Hassani Mehraban, A. Molecular Diagnosis of Tomato Yellow Ring Virus (Tyrv) On Alstroemeria in Khorasan Razavi Province, Iran. Iran. Plant Prot. Res. J. Plant Prot. 2011, 25, 313–324. [Google Scholar]
- Bhat, A.I.; Aman, R.; Mahfouz, M. Onsite Detection of Plant Viruses Using Isothermal Amplification Assays. Plant Biotechnol. J. 2022. [Google Scholar] [CrossRef]
- Tomlinson, J.; Boonham, N. Potential of LAMP for Detection of Plant Pathogens. CAB Rev. Perspect. Agric. Vet. Sci. Nutr. Nat. Resour. 2008, 3, 1–7. [Google Scholar] [CrossRef]


| Virus Name | Primer Name | Primer Sequence 5′-3′ | Annealing Temp. | Reference | |
|---|---|---|---|---|---|
| cucumber mosaic virus | CMV | CMVCPf | GCTTCTCCGCGAG | 50 °C | [43] |
| CMVCPr | GCCGTAAGCTGGATGGAC | ||||
| cucumber mosaic virus | CMVsat-fwd | AAGGATCCGGGTCCTGBDDDGGAATG | 55 °C | [44] | |
| satRNA | CMVsat-rev | AAGGATCCGTTTTGTTTGWTRGAGAAT TGCGYRGAG | |||
| pepino mosaic virus | PepMV | TGB3F | GGTGGACAATATCAAGACGG | 51 °C | [41] |
| TGB3R | CTGTATTGGGATTTGAGAAGTC | ||||
| potato virus Y | PVY | PVYc3F | CAACGCAAAAACACTCAyAAAmGC | 57 °C | [45] |
| PVYfR | TAAGTGrACAGACCCTCTyTTCTC | ||||
| PVY3F | TGTAACGAAAGGGACTAGTGCAAAG | ||||
| PVY3R | CCGCTATGAGTAAGTCCTGCACA | ||||
| PVYCP2F | CCAGTCAAACCCGAACAAAGG | ||||
| PVYCP1R | GGCATAGCGTGCTAAACCCA | ||||
| tomato black ring virus | TBRV | TBRVR1-P1-KRF | GGTAAAAGTTCTGGGTGCT | 53 °C | [46] |
| TBRVR1-P1-KRR | GCAAATCCACCTCCTTATCC | ||||
| tomato black ring virus | CH_SAT_F1 | TAATTTTGAAAGTCTCTGA | 47 °C | [46] | |
| satRNA | CH_SAT_R2 | GGACAGCTCGTTGGTTCTTAGA | |||
| tomato brown rugose | ToBRFV | CaTa28 Fw | GGTGGTGTCAGTGTCTGTTT | 60 °C | [47] |
| fruit virus | CaTa28 Rv | GCGTCCTTGGTAGTGATGTT | |||
| CaTa28 Pr | 6FAM-AGAGAATGGAGAGAGCGGACGAGG-BHQ’1 | ||||
| CSP1325 Fw | CATTTGAAAGTGCATCCGGTT T | ||||
| CSP1325 Rv | GTACCACGTGTGTTTGCAGACA | ||||
| CSP1325 Pr | VIC-ATGGTCCTCTGCACCTGCATCTTGAGA-BHQ’1 | ||||
| tomato chlorosis virus | ToCV | ToCVCPF | ATGGAGAACAGTGCCGTTGC | 58 °C | [48] |
| ToCVCPR | TTAGCAACCAGTTATCGATGC | ||||
| tomato mosaic virus | ToMV | ToMV F | CGAGAGGGGCAACAAACAT | 66 °C | [49] |
| ToMV R | ACCTGTCTCCATCTCTTTGG | ||||
| tomato torrado virus | ToTV | 2TT5 | GATGAGAAAGGAAAGAAGCAG | 55 °C | [50] |
| 2TT6 | CATATCACCCAAATGCTTCTC | ||||
| tobacco mosaic virus | TMV | TMV F | CGACATCAGCCGATGCAGC | 66 °C | [49] |
| TMV R | ACCGTTTTCGAACCGAGACT | ||||
| tomato spotted wilt | TSWV | TS1-F | GCCTATGGATTACCTCTTG | 45 °C | [51] |
| orthotospovirus | TS1-R | GTTTCACTGTAATGTTCCA | |||
| tomato yellow ring | TYRV | TY2-F | CTAACAAAGCCATGAAGA | 45 °C | [51] |
| orthotospovirus | TY2-R | GAAGACCCAGCACCA |
| Primers Name | Primer Sequence 5′-3′ |
|---|---|
| TYRV FIP | CACAGTAGAGCTAGGAACAACAATA-AAAATGGTTAAAGCAGGGC |
| TYRV BIP | GGTCAAGATGATTGGACATTCCGA-TGCATTTTCCACAGCAATG |
| TYRV F3 | GAGAAACAGAGCAGGGATT |
| TYRV B3 | TCATACATTTTCTGTTTCTCAGT |
| Year | No. of Collected Samples | No. of Infected Samples | Single Infection | Mixed Infection | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PepMV | TYRV | TSWV | CMV | PVY | ToMV | TBRV | TMV | ToTV | ToBRV * | ToCV * | TSWV+ TYRV | CMV+ PepMV | TSWV+ PepMV | CMV+ TYRV | |||
| 2014 | 54 | 25 | 7 | 8 | 6 | - | - | - | - | - | - | x | x | 4 | - | - | - |
| 2015 | 41 | 11 | 11 | - | - | - | - | - | - | - | - | x | x | - | - | - | - |
| 2016 | 37 | 11 | 10 | - | - | 1 | - | - | - | - | - | x | x | - | - | - | - |
| 2017 | 22 | 6 | 5 | 1 | - | - | - | - | - | - | - | x | x | - | - | - | - |
| 2018 | 24 | 6 | 6 | - | - | - | - | - | - | - | - | x | x | - | - | - | - |
| 2019 | 20 | 5 | 3 | - | - | 2 | - | - | - | - | - | x | - | - | - | - | |
| 2020 | 18 | 11 | 7 | - | - | - | 2 | - | - | - | - | - | - | - | 2 | - | - |
| 2021 | 18 | 14 | 7 | 1 | 1 | 1 | - | 1 | 1 | - | - | - | - | - | - | 1 | 1 |
| Total | 234 | 89 | 56 | 10 | 7 | 4 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 4 | 2 | 1 | 1 |
| Isolate Origin (Host Plant) | Observed Value | Null Value | p-Value | ||||
|---|---|---|---|---|---|---|---|
| Mean | Lower HPD | Upper HPD | Mean | Lower HPD | Upper HPD | ||
| 95% CI | 95% Cl | 95% CI | 95% CI | ||||
| AI | 2.96 | 2.45 | 3.47 | 4.00 | 3.49 | 4.59 | 0.00 |
| PS | 18.21 | 17.00 | 19.00 | 24.93 | 22.74 | 26.82 | 0.00 |
| MC (SL) | 15.00 | 15.00 | 15.00 | 2.70 | 2.00 | 3.66 | 0.01 |
| MC (AL) | 1.00 | 1.00 | 1.00 | 1.01 | 1.00 | 1.09 | 1.00 |
| MC (CH) | 1.00 | 1.00 | 1.00 | 1.01 | 1.00 | 1.06 | 1.00 |
| MC (GL) | 1.00 | 1.00 | 1.00 | 1.01 | 1.00 | 1.07 | 1.00 |
| MC (ST) | 3.11 | 2.00 | 5.00 | 1.89 | 1.28 | 2.52 | 0.04 |
| MC (AR) | 1.08 | 1.00 | 2.00 | 1.21 | 1.00 | 2.00 | 1.00 |
| MC (CA) | 1.00 | 1.00 | 1.00 | 1.02 | 1.00 | 1.04 | 1.00 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zarzyńska-Nowak, A.; Budzyńska, D.; Taberska, A.; Jędrzejczak, N.; Minicka, J.; Borodynko-Filas, N.; Hasiów-Jaroszewska, B. Occurrence, Genetic Variability of Tomato Yellow Ring Orthotospovirus Population and the Development of Reverse Transcription Loop-Mediated Isothermal Amplification Assay for Its Rapid Detection. Viruses 2022, 14, 1405. https://doi.org/10.3390/v14071405
Zarzyńska-Nowak A, Budzyńska D, Taberska A, Jędrzejczak N, Minicka J, Borodynko-Filas N, Hasiów-Jaroszewska B. Occurrence, Genetic Variability of Tomato Yellow Ring Orthotospovirus Population and the Development of Reverse Transcription Loop-Mediated Isothermal Amplification Assay for Its Rapid Detection. Viruses. 2022; 14(7):1405. https://doi.org/10.3390/v14071405
Chicago/Turabian StyleZarzyńska-Nowak, Aleksandra, Daria Budzyńska, Agnieszka Taberska, Norbert Jędrzejczak, Julia Minicka, Natasza Borodynko-Filas, and Beata Hasiów-Jaroszewska. 2022. "Occurrence, Genetic Variability of Tomato Yellow Ring Orthotospovirus Population and the Development of Reverse Transcription Loop-Mediated Isothermal Amplification Assay for Its Rapid Detection" Viruses 14, no. 7: 1405. https://doi.org/10.3390/v14071405
APA StyleZarzyńska-Nowak, A., Budzyńska, D., Taberska, A., Jędrzejczak, N., Minicka, J., Borodynko-Filas, N., & Hasiów-Jaroszewska, B. (2022). Occurrence, Genetic Variability of Tomato Yellow Ring Orthotospovirus Population and the Development of Reverse Transcription Loop-Mediated Isothermal Amplification Assay for Its Rapid Detection. Viruses, 14(7), 1405. https://doi.org/10.3390/v14071405







