Metatranscriptomic Comparison of Viromes in Endemic and Introduced Passerines in New Zealand
Abstract
1. Introduction
2. Materials and Methods
3. Results
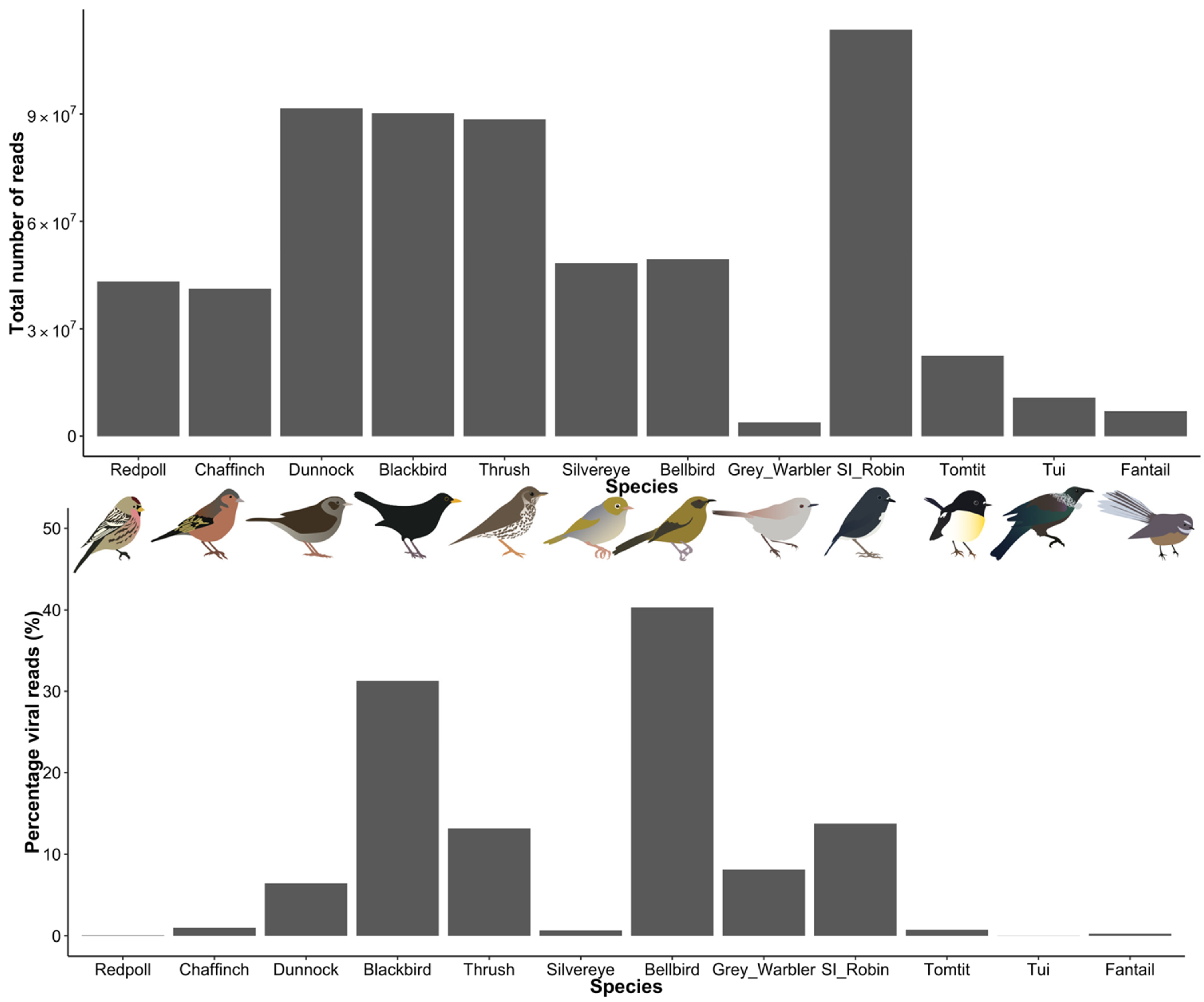
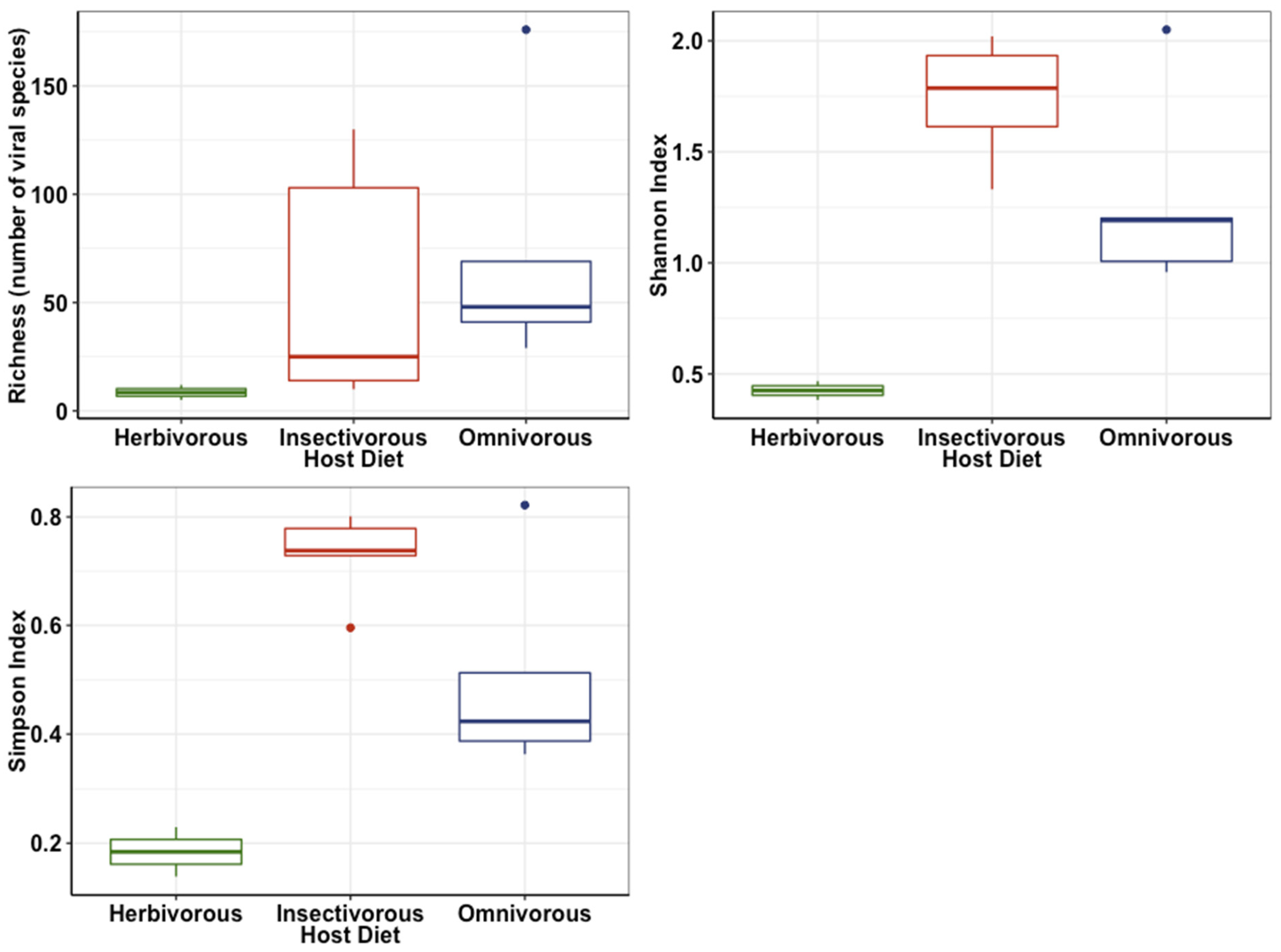
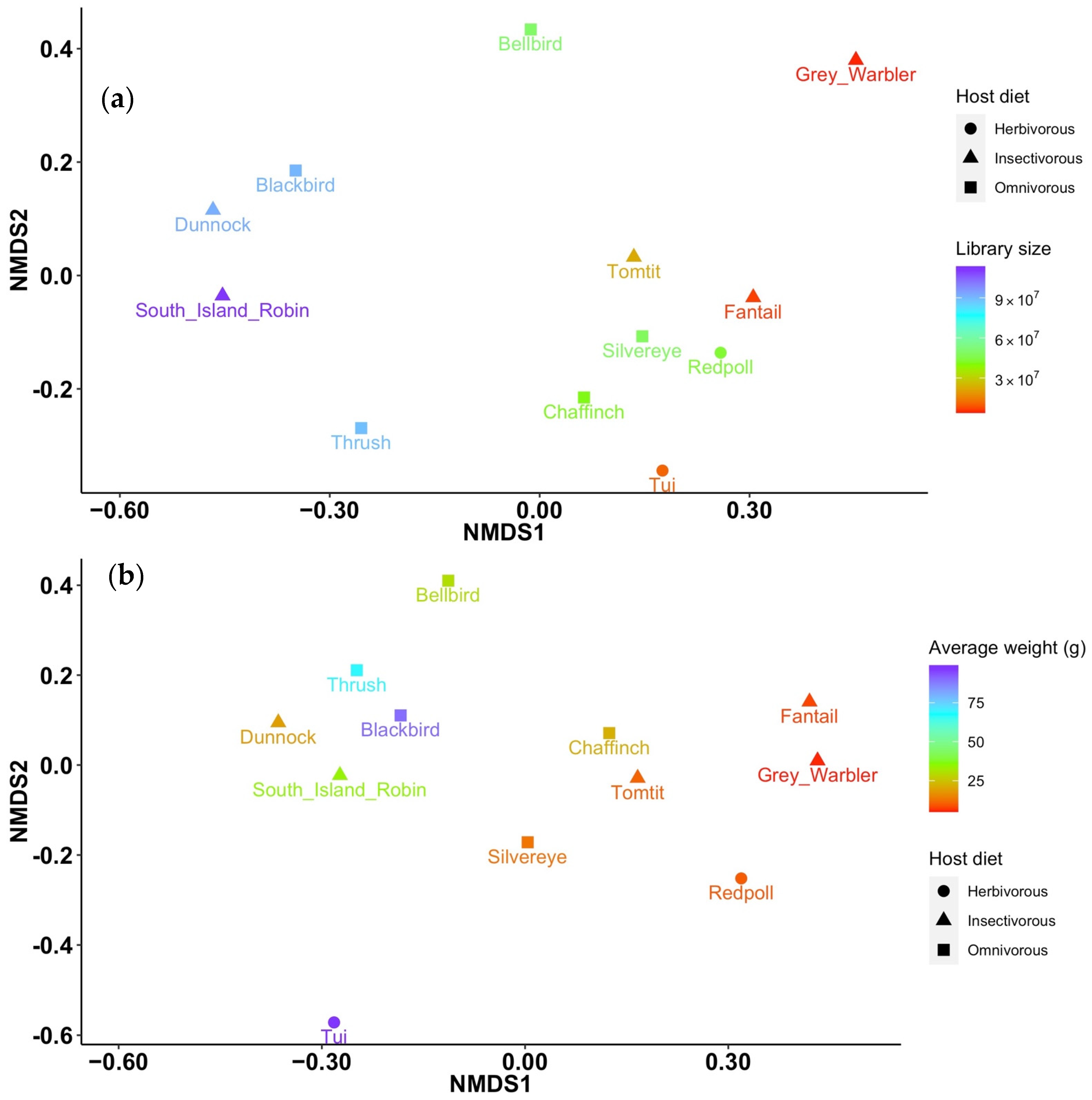
3.1. Viral Diversity
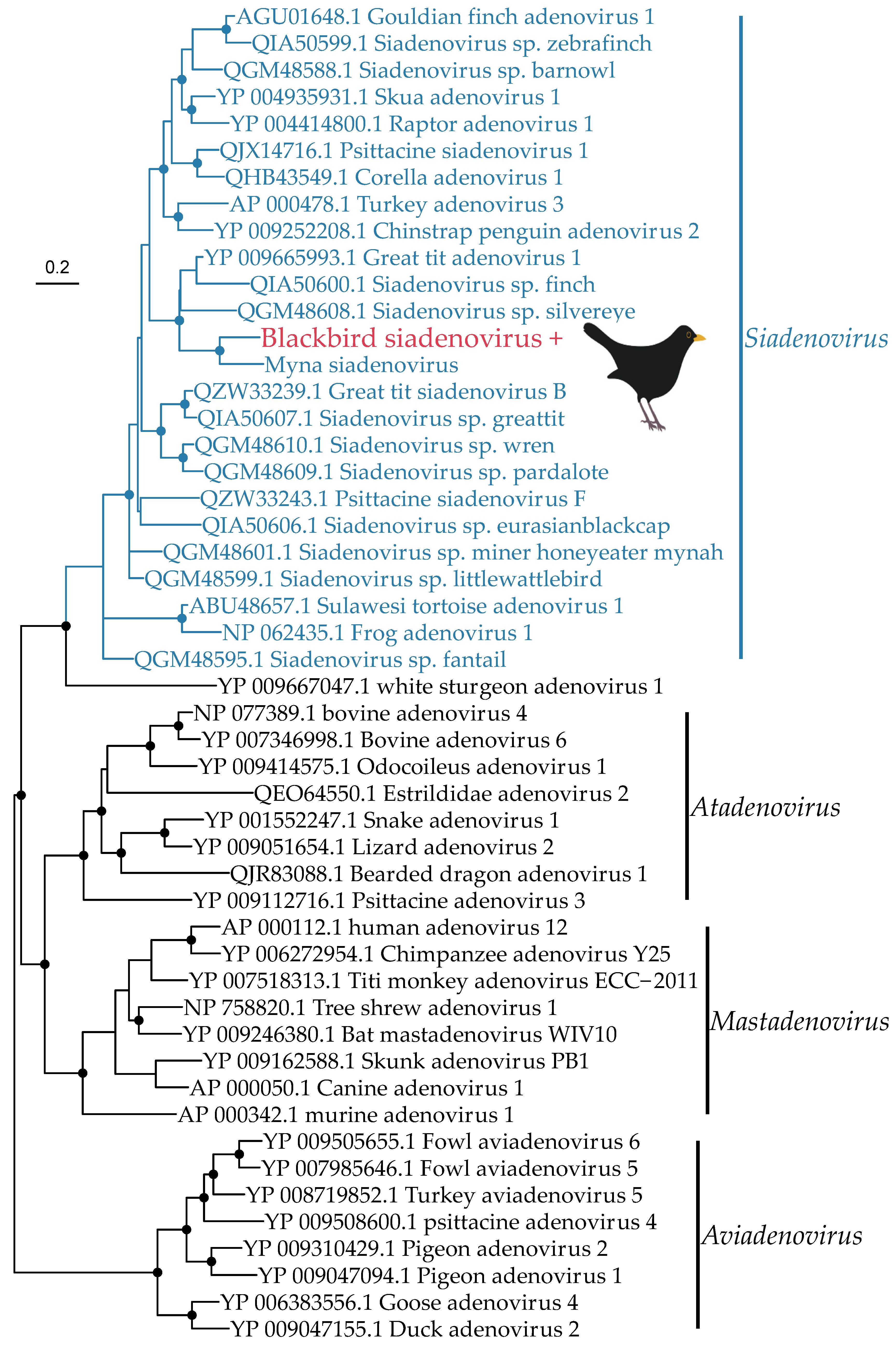
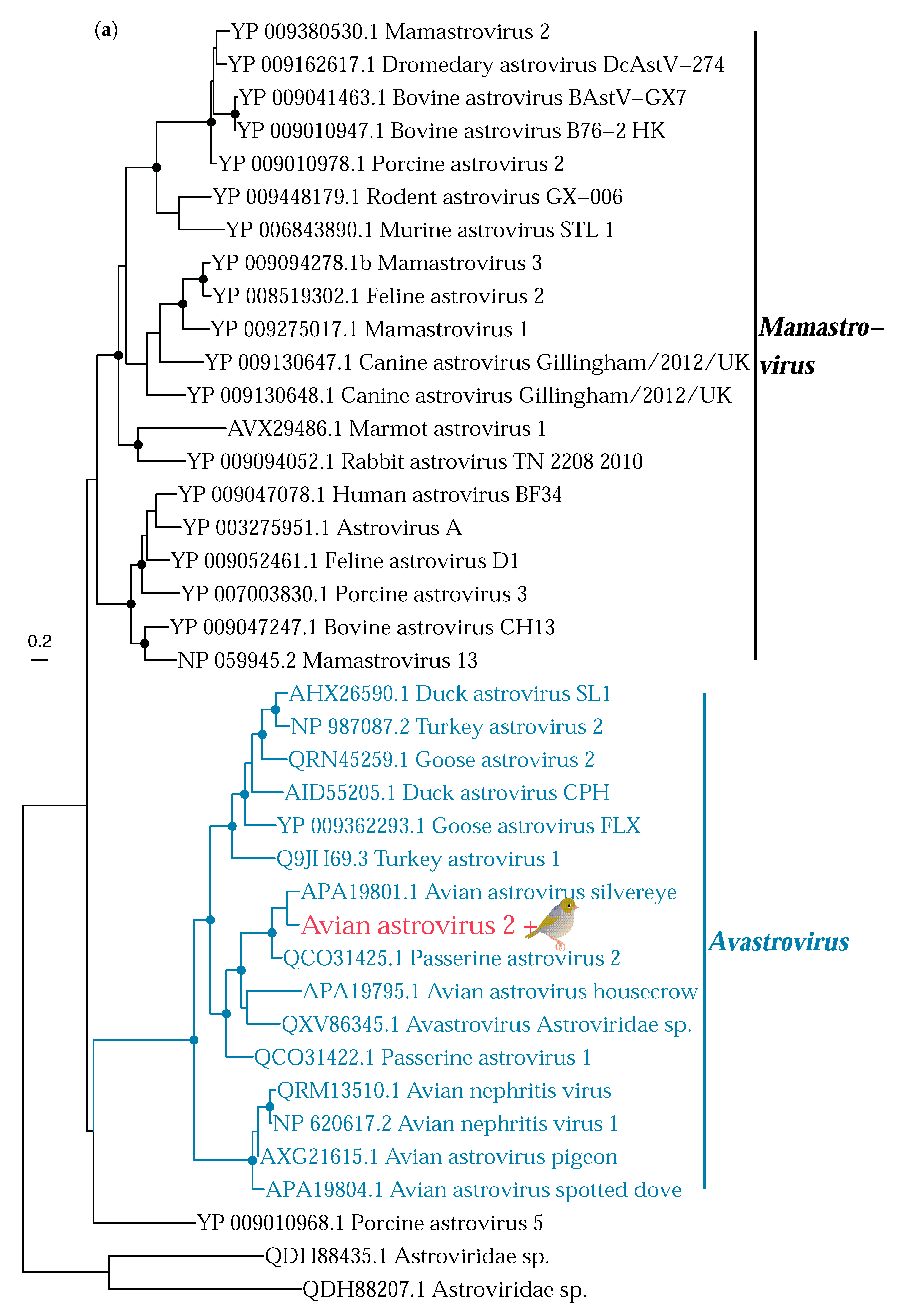
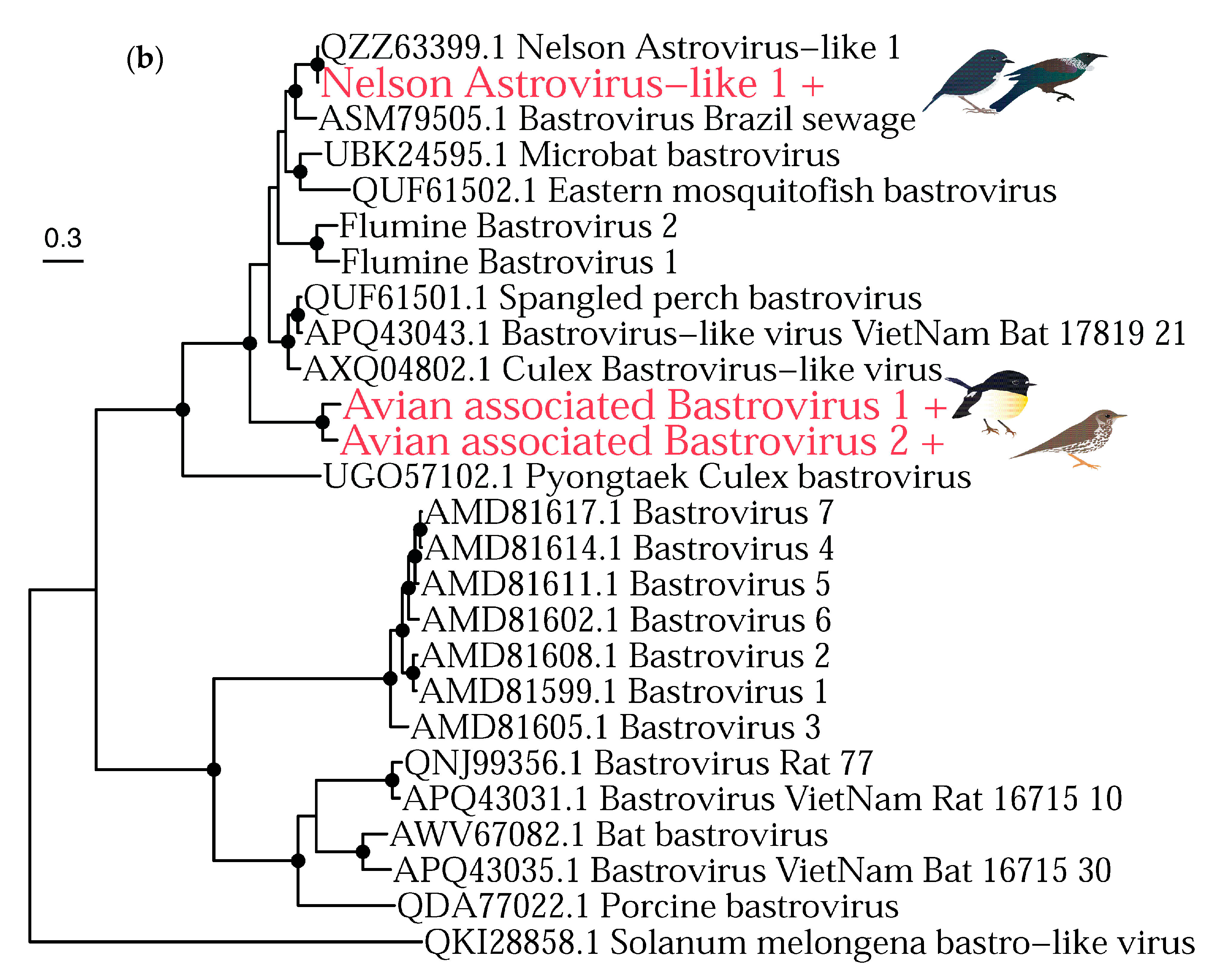
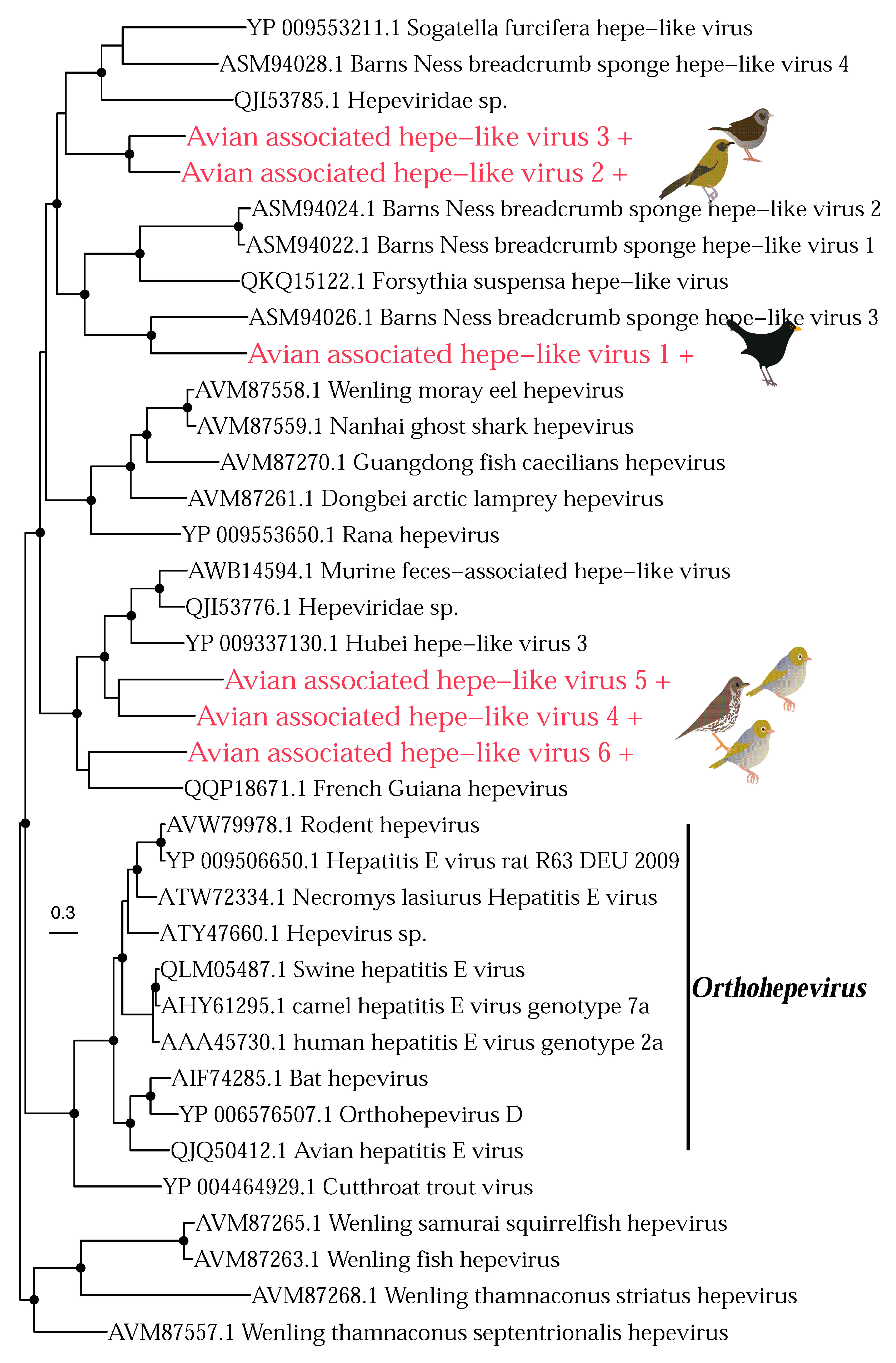
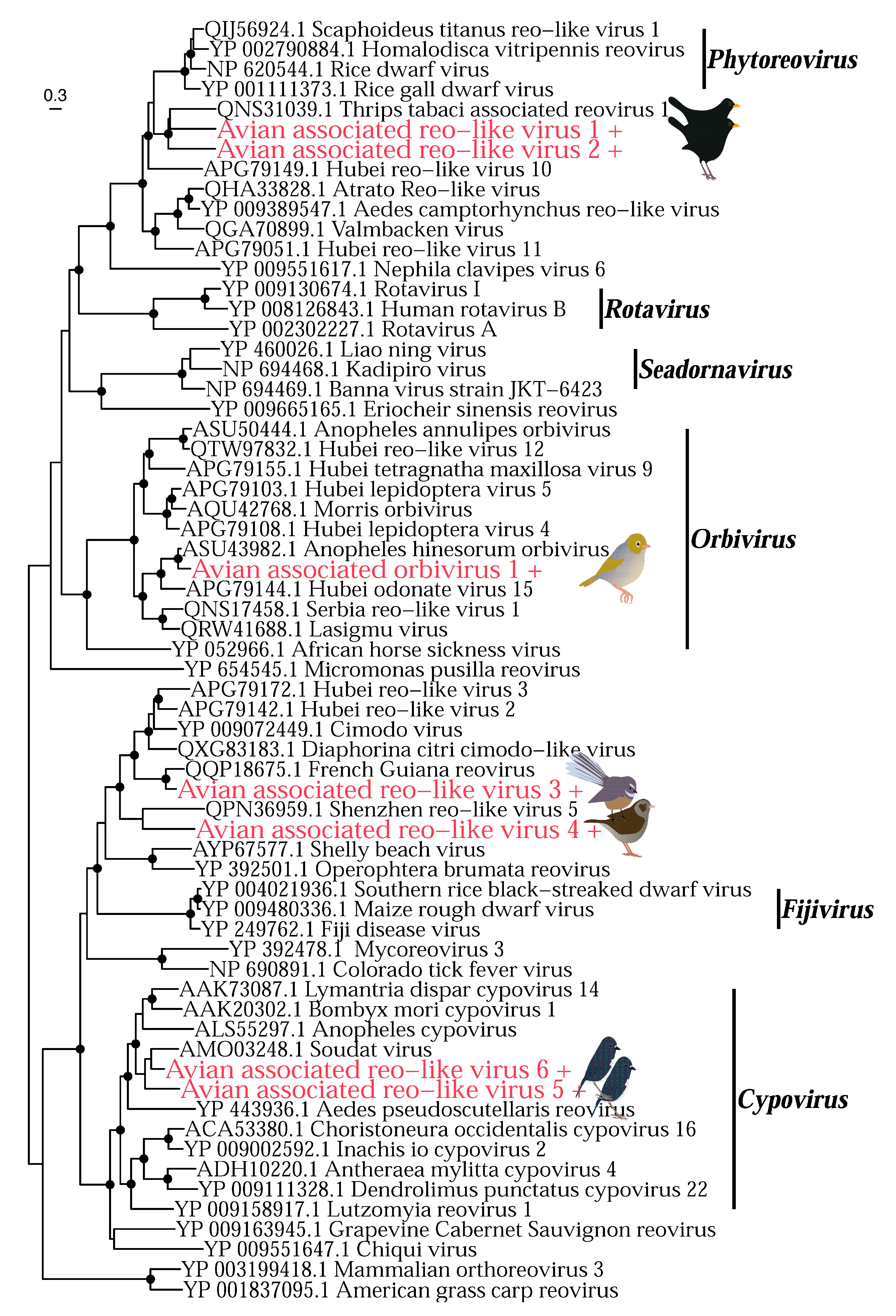
3.2. Phylogenetic Analysis of Putative Avian Viruses
3.2.1. DNA Viruses
3.2.2. RNA Viruses
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mitchell, K.J.; Llamas, B.; Soubrier, J.; Rawlence, N.J.; Worthy, T.H.; Wood, J.; Lee, M.S.; Cooper, A. Ancient DNA reveals elephant birds and kiwi are sister taxa and clarifies ratite bird evolution. Science 2014, 344, 898–900. [Google Scholar] [CrossRef] [PubMed]
- Gibb, G.C.; England, R.; Hartig, G.; McLenachan, P.A.; Taylor Smith, B.L.; McComish, B.J.; Cooper, A.; Penny, D. New Zealand passerines help clarify the diversification of major songbird lineages during the Oligocene. Genome Biol. Evol. 2015, 7, 2983–2995. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, K.J.; Wood, J.R.; Llamas, B.; McLenachan, P.A.; Kardailsky, O.; Scofield, R.P.; Worthy, T.H.; Cooper, A. Ancient mitochondrial genomes clarify the evolutionary history of New Zealand’s enigmatic acanthisittid wrens. Mol. Phy. Evol. 2016, 102, 295–304. [Google Scholar] [CrossRef] [PubMed]
- Innes, J.; Kelly, D.; Overton, J.M.; Gillies, C. Predation and other factors currently limiting New Zealand forest birds. NZ J. Ecol. 2010, 34, 86. [Google Scholar]
- Valente, L.; Etienne, R.S.; Garcia-R, J.C. Deep macroevolutionary impact of humans on New Zealand’s unique avifauna. Curr. Biol. 2019, 29, 2563–2569.e2564. [Google Scholar] [CrossRef]
- Barnagaud, J.-Y.; Barbaro, L.; Papaix, J.; Deconchat, M.; Brockerhoff, E.G. Habitat filtering by landscape and local forest composition in native and exotic New Zealand birds. Ecology 2014, 95, 78–87. [Google Scholar] [CrossRef]
- van Heezik, Y.; Smyth, A.; Mathieu, R. Diversity of native and exotic birds across an urban gradient in a New Zealand city. Landscape Urban Plan. 2008, 87, 223–232. [Google Scholar] [CrossRef]
- Thomson, G.M. The Naturalisation of Animals and Plants in New Zealand; Cambridge University Press: Cambridge, UK, 2011. [Google Scholar]
- Mees, G.F. A Systematic Review of the Indo-Australian Zosteropidae (Part III); EJ Brill: Leiden, The Netherlands, 1969. [Google Scholar]
- Hale, K.; Briskie, J. Decreased immunocompetence in a severely bottlenecked population of an endemic New Zealand bird. Anim. Conserv. 2007, 10, 2–10. [Google Scholar] [CrossRef]
- Wikelski, M.; Foufopoulos, J.; Vargas, H.; Snell, H. Galápagos birds and diseases: Invasive pathogens as threats for island species. Ecol. Soc. 2004, 9, 5. [Google Scholar] [CrossRef]
- Streicker, D.G.; Turmelle, A.S.; Vonhof, M.J.; Kuzmin, I.V.; McCracken, G.F.; Rupprecht, C.E. Host phylogeny constrains cross-species emergence and establishment of rabies virus in bats. Science 2010, 329, 676–679. [Google Scholar] [CrossRef]
- Miles, J. The ecology of Whataroa virus, an alphavirus, in South Westland, New Zealand. Epidem. Infect. 1973, 71, 701–713. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tompkins, D.; Paterson, R.; Massey, B.; Gleeson, D. Whataroa virus four decades on: Emerging, persisting, or fading out? J. R. Soc. NZ 2010, 40, 1–9. [Google Scholar] [CrossRef]
- Ha, H.J.; Howe, L.; Alley, M.; Gartrell, B. The phylogenetic analysis of avipoxvirus in New Zealand. Vet. Microbiol. 2011, 150, 80–87. [Google Scholar] [CrossRef] [PubMed]
- Gartrell, B.; Stone, M.; King, C.; Wang, J. An outbreak of disease caused by psittacinepoxvirus in rosellas. Surveillance 2003, 30, 11–13. [Google Scholar]
- Cracraft, J.; Barker, F. Passerine birds (Passeriformes). In The Timetree of Life; Oxford University Press: New York, NY, USA, 2009; pp. 423–431. [Google Scholar]
- Lundström, J.O.; Lindström, K.M.; Olsen, B.; Dufva, R.; Krakower, D.S. Prevalence of Sindbis virus neutralizing antibodies among Swedish passerines indicates that thrushes are the main amplifying hosts. J. Med. Entomol. 2001, 38, 289–297. [Google Scholar] [CrossRef]
- Roiz, D.; Vázquez, A.; Ruiz, S.; Tenorio, A.; Soriguer, R.; Figuerola, J. Evidence that passerine birds act as amplifying hosts for usutu virus circulation. Ecohealth 2019, 16, 734–742. [Google Scholar] [CrossRef]
- Nemeth, N.M.; Oesterle, P.T.; Bowen, R.A. Passive immunity to West Nile virus provides limited protection in a common passerine species. Am. J. Trop. Med. Hyg. 2008, 79, 283–290. [Google Scholar] [CrossRef]
- Fernández-Correa, I.; Truchado, D.A.; Gomez-Lucia, E.; Doménech, A.; Pérez-Tris, J.; Schmidt-Chanasit, J.; Cadar, D.; Benítez, L. A novel group of avian astroviruses from Neotropical passerine birds broaden the diversity and host range of Astroviridae. Sci. Rep. 2019, 9, 9513. [Google Scholar] [CrossRef]
- Truchado, D.A.; Diaz-Piqueras, J.M.; Gomez-Lucia, E.; Doménech, A.; Milá, B.; Pérez-Tris, J.; Schmidt-Chanasit, J.; Cadar, D.; Benítez, L. A novel and divergent gyrovirus with unusual genomic features detected in wild passerine birds from a remote rainforest in French Guiana. Viruses 2019, 11, 1148. [Google Scholar] [CrossRef]
- Truchado, D.A.; Llanos-Garrido, A.; Oropesa-Olmedo, D.A.; Cerrada, B.; Cea, P.; Moens, M.A.; Gomez-Lucia, E.; Doménech, A.; Milá, B.; Pérez-Tris, J. Comparative metagenomics of Palearctic and Neotropical avian cloacal viromes reveal geographic bias in virus discovery. Microorganisms 2020, 8, 1869. [Google Scholar] [CrossRef]
- Shan, T.; Yang, S.; Wang, H.; Wang, H.; Zhang, J.; Gong, G.; Xiao, Y.; Yang, J.; Wang, X.; Lu, J. Virome in the cloaca of wild and breeding birds revealed a diversity of significant viruses. Microbiome 2022, 10, 60. [Google Scholar] [CrossRef]
- Custer, J.M.; White, R.; Taylor, H.; Schmidlin, K.; Fontenele, R.S.; Stainton, D.; Kraberger, S.; Briskie, J.V.; Varsani, A. Diverse single-stranded DNA viruses identified in New Zealand (Aotearoa) South Island robin (Petroica australis) fecal samples. Virology 2022, 565, 38–51. [Google Scholar] [CrossRef] [PubMed]
- French, R.K.; Holmes, E.C. An ecosystems perspective on virus evolution and emergence. Trends Microbiol. 2020, 28, 165–175. [Google Scholar] [CrossRef] [PubMed]
- Wille, M.; Shi, M.; Hurt, A.C.; Klaassen, M.; Holmes, E.C. RNA virome abundance and diversity is associated with host age in a bird species. Virology 2021, 561, 98–106. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Bushnell, B. BBMap Short Read Aligner; University of California: Berkeley, CA, USA, 2016; Available online: https://sourceforge.net/projects/bbmap (accessed on 1 January 2022).
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q. Trinity: Reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat. Biotechnol. 2011, 29, 644. [Google Scholar] [CrossRef] [PubMed]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinformat. 2009, 10, 421. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Clausen, P.T.; Aarestrup, F.M.; Lund, O. Rapid and precise alignment of raw reads against redundant databases with KMA. BMC Bioinformat. 2018, 19, 307. [Google Scholar] [CrossRef]
- Marcelino, V.R.; Clausen, P.T.; Buchmann, J.P.; Wille, M.; Iredell, J.R.; Meyer, W.; Lund, O.; Sorrell, T.C.; Holmes, E.C. CCMetagen: Comprehensive and accurate identification of eukaryotes and prokaryotes in metagenomic data. Genome Biol. 2020, 21, 103. [Google Scholar] [CrossRef] [PubMed]
- Nagendra, H. Opposite trends in response for the Shannon and Simpson indices of landscape diversity. Appl. Geogr. 2002, 22, 175–186. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Paradis, E.; Schliep, K. ape 5.0: An environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 2019, 35, 526–528. [Google Scholar] [CrossRef]
- Yu, G.; Smith, D.K.; Zhu, H.; Guan, Y.; Lam, T.T.Y. ggtree: An R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 2017, 8, 28–36. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2. Wiley Interdiscip. Rev. Comput. Stat. 2011, 3, 180–185. [Google Scholar] [CrossRef]
- Pebesma, E.J. Simple features for R: Standardized support for spatial vector data. R J. 2018, 10, 439. [Google Scholar] [CrossRef]
- Dunnington, D. ggspatial: Spatial Data Framework for ggplot2; R package version 1.1.5; 2021. Available online: https://cloud.r-project.org/web/packages/ggspatial/index.html (accessed on 20 March 2022).
- South, A. Rnaturalearth: World Map Data from Natural Earth; R package version 0.1.0; 2017. Available online: https://cran.r-project.org/web/packages/rnaturalearth/README.html (accessed on 20 March 2022).
- Porter, A.F.; Cobbin, J.; Li, C.-X.; Eden, J.-S.; Holmes, E.C. Metagenomic identification of viral sequences in laboratory reagents. Viruses 2021, 13, 2122. [Google Scholar] [CrossRef] [PubMed]
- Wille, M.; Eden, J.S.; Shi, M.; Klaassen, M.; Hurt, A.C.; Holmes, E.C. Virus–virus interactions and host ecology are associated with RNA virome structure in wild birds. Mol. Ecol. 2018, 27, 5263–5278. [Google Scholar] [CrossRef] [PubMed]
- Wille, M.; Harvey, E.; Shi, M.; Gonzalez-Acuña, D.; Holmes, E.C.; Hurt, A.C. Sustained RNA virome diversity in Antarctic penguins and their ticks. ISME J. 2020, 14, 1768–1782. [Google Scholar] [CrossRef] [PubMed]
- Lukashov, V.V.; Goudsmit, J. Evolutionary relationships among Astroviridae. J. Gen. Virol. 2002, 83, 1397–1405. [Google Scholar] [CrossRef]
- Remnant, E.J.; Baty, J.W.; Bulgarella, M.; Dobelmann, J.; Quinn, O.; Gruber, M.A.; Lester, P.J. A diverse viral community from predatory wasps in their native and invaded range, with a new virus infectious to honey bees. Viruses 2021, 13, 1431. [Google Scholar] [CrossRef]
- Roberts, J.M.; Simbiken, N.; Dale, C.; Armstrong, J.; Anderson, D.L. Tolerance of honey bees to Varroa mite in the absence of deformed wing virus. Viruses 2020, 12, 575. [Google Scholar] [CrossRef]
- Sherman, J.; Unwin, S.; Travis, D.A.; Oram, F.; Wich, S.A.; Jaya, R.L.; Voigt, M.; Santika, T.; Massingham, E.; Seaman, D.J. Disease risk and conservation implications of orangutan translocations. Front. Vet. Sci. 2021, 8, 749547. [Google Scholar] [CrossRef]
- Shi, M.; Lin, X.-D.; Tian, J.-H.; Chen, L.-J.; Chen, X.; Li, C.-X.; Qin, X.-C.; Li, J.; Cao, J.-P.; Eden, J.-S. Redefining the invertebrate RNA virosphere. Nature 2016, 540, 539–543. [Google Scholar] [CrossRef]
- Thureen, D.R.; Keeler, C.L., Jr. Psittacid herpesvirus 1 and infectious laryngotracheitis virus: Comparative genome sequence analysis of two avian alphaherpesviruses. J. Virol. 2006, 80, 7863–7872. [Google Scholar] [CrossRef]
- Wellehan, J.F.; Gagea, M.; Smith, D.A.; Taylor, W.M.; Berhane, Y.; Bienzle, D. Characterization of a herpesvirus associated with tracheitis in Gouldian finches (Erythrura [Chloebia] gouldiae). J. Clin. Microbiol. 2003, 41, 4054–4057. [Google Scholar] [CrossRef]
- Azab, W.; Dayaram, A.; Greenwood, A.D.; Osterrieder, N. How host specific are herpesviruses? Lessons from herpesviruses infecting wild and endangered mammals. Annu. Rev. Virol. 2018, 5, 53–68. [Google Scholar] [CrossRef] [PubMed]
- Woźniakowski, G.J.; Samorek-Salamonowicz, E.; Szymański, P.; Wencel, P.; Houszka, M. Phylogenetic analysis of Columbid herpesvirus-1 in rock pigeons, birds of prey and non-raptorial birds in Poland. BMC Vet. Res. 2013, 9, 52. [Google Scholar] [CrossRef] [PubMed]
- Tomaszewski, E.K.; Kaleta, E.F.; Phalen, D.N. Molecular phylogeny of the psittacid herpesviruses causing Pacheco’s disease: Correlation of genotype with phenotypic expression. J. Virol. 2003, 77, 11260–11267. [Google Scholar] [CrossRef] [PubMed]
- Chang, W.-S.; Rose, K.; Holmes, E.C. Meta-transcriptomic analysis of the virome and microbiome of the invasive Indian myna (Acridotheres tristis) in Australia. One Health 2021, 13, 100360. [Google Scholar] [CrossRef] [PubMed]
- Rinder, M.; Schmitz, A.; Baas, N.; Korbel, R. Molecular identification of novel and genetically diverse adenoviruses in Passeriform birds. Virus Genes 2020, 56, 316–324. [Google Scholar] [CrossRef]
- Vaz, F.F.; Raso, T.F.; Agius, J.E.; Hunt, T.; Leishman, A.; Eden, J.-S.; Phalen, D.N. Opportunistic sampling of wild native and invasive birds reveals a rich diversity of adenoviruses in Australia. Virus Evol. 2020, 6, veaa024. [Google Scholar] [CrossRef]
- Beach, N.M.; Duncan, R.B.; Larsen, C.T.; Meng, X.-J.; Sriranganathan, N.; Pierson, F.W. Comparison of 12 turkey hemorrhagic enteritis virus isolates allows prediction of genetic factors affecting virulence. J. Gen. Virol. 2009, 90, 1978–1985. [Google Scholar] [CrossRef]
- Kovács, E.R.; Benkő, M. Confirmation of a novel siadenovirus species detected in raptors: Partial sequence and phylogenetic analysis. Virus Res. 2009, 140, 64–70. [Google Scholar] [CrossRef]
- Kovács, E.R.; Jánoska, M.; Dán, Á.; Harrach, B.; Benkő, M. Recognition and partial genome characterization by non-specific DNA amplification and PCR of a new siadenovirus species in a sample originating from Parus major, a great tit. J. Virol. Methods 2010, 163, 262–268. [Google Scholar] [CrossRef]
- Park, Y.M.; Kim, J.-H.; Gu, S.H.; Lee, S.Y.; Lee, M.-G.; Kang, Y.K.; Kang, S.-H.; Kim, H.J.; Song, J.-W. Full genome analysis of a novel adenovirus from the South Polar skua (Catharacta maccormicki) in Antarctica. Virology 2012, 422, 144–150. [Google Scholar] [CrossRef]
- Wellehan, J.F., Jr.; Greenacre, C.B.; Fleming, G.J.; Stetter, M.D.; Childress, A.L.; Terrell, S.P. Siadenovirus infection in two psittacine bird species. Avian Pathol. 2009, 38, 413–417. [Google Scholar] [CrossRef] [PubMed]
- Cimsit, B.; Tichy, E.M.; Patel, S.B.; Rosencrantz, R.; Emre, S. Treatment of adenovirus hepatitis with cidofovir in a pediatric liver transplant recipient. Pediatric Transplant. 2012, 16, E90–E93. [Google Scholar] [CrossRef] [PubMed]
- Pintore, M.D.; Corbellini, D.; Chieppa, M.N.; Vallino Costassa, E.; Florio, C.L.; Varello, K.; Bozzetta, E.; Adriano, D.; Decaro, N.; Casalone, C. Canine adenovirus type 1 and Pasteurella pneumotropica co-infection in a puppy. Vet. Ital. 2016, 52, 57. [Google Scholar] [PubMed]
- Zöller, M.; Mätz-Rensing, K.; Kaup, F.J. Adenoviral hepatitis in a SIV-infected rhesus monkey (Macaca mulatta). J. Med. Primatol. 2008, 37, 184–187. [Google Scholar] [CrossRef] [PubMed]
- Waldron, F.M.; Stone, G.N.; Obbard, D.J. Metagenomic sequencing suggests a diversity of RNA interference-like responses to viruses across multicellular eukaryotes. PLoS Genet. 2018, 14, e1007533. [Google Scholar] [CrossRef]
- Filion, A.; Niebuhr, C.; Deschamps, L.; Poulin, R. Anthropogenic landscape alteration promotes higher disease risk in wild New Zealand avian communities. PLoS ONE 2022, 17, e0265568. [Google Scholar] [CrossRef]
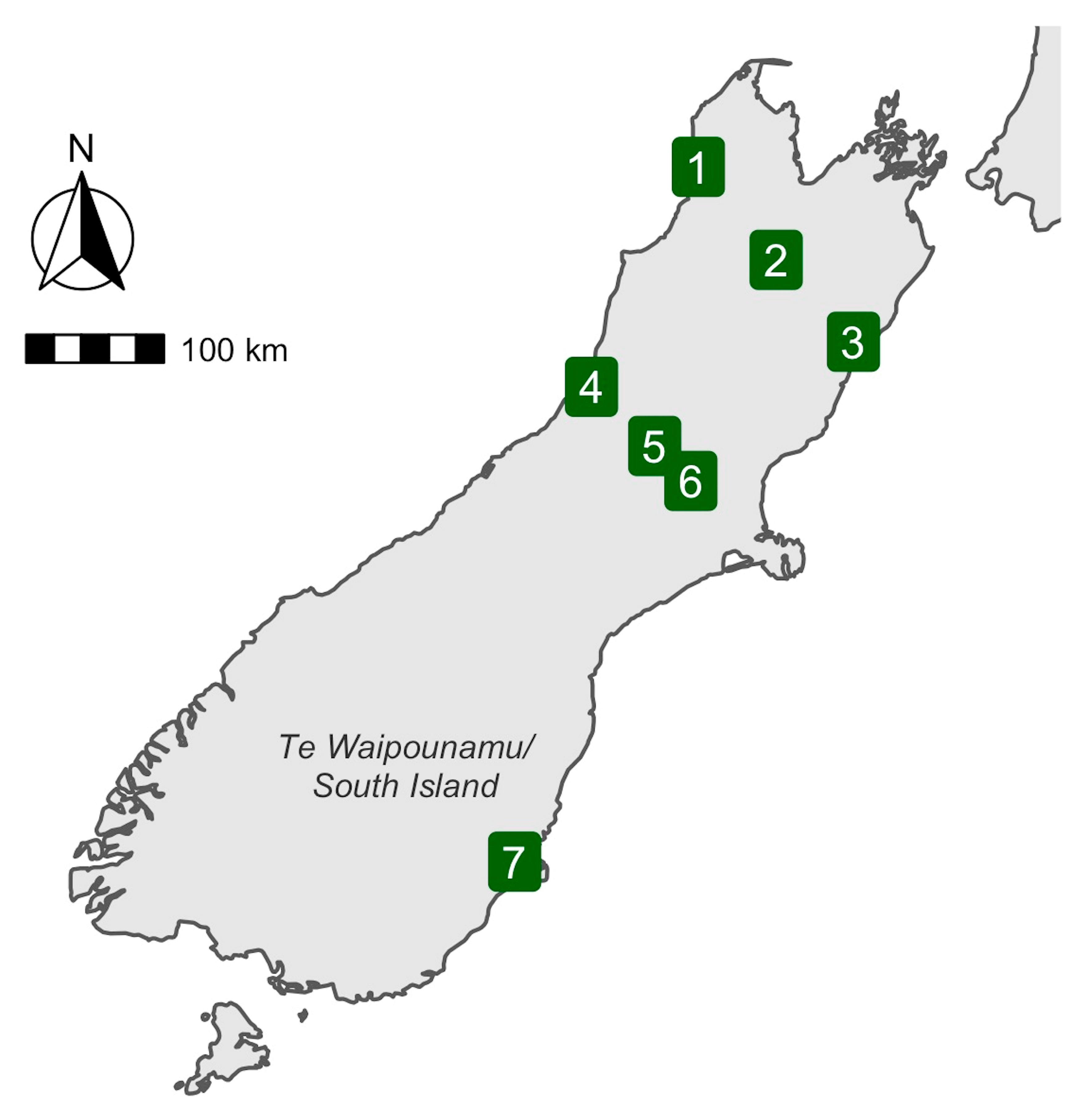



| Common Name(s) | Scientific Name | Species Origin | Number Sampled | Predominant Diet |
|---|---|---|---|---|
| Common redpoll | Carduelis flammea | Introduced | 4 | Herbivorous |
| Chaffinch | Fringilla coelebs | Introduced | 5 | Omnivorous |
| Dunnock | Prunella modularis | Introduced | 4 | Insectivorous |
| Eurasian blackbird | Turdus merula | Introduced | 4 | Omnivorous |
| Song thrush | Turdus philomelos | Introduced | 3 | Omnivorous |
| Silvereye/tauhou | Zosterops lateralis | Introduced | 11 | Omnivorous |
| Bellbird/korimako | Anthornis melanura | Endemic | 12 | Omnivorous |
| Grey warbler/riroriro | Gerygone igata | Endemic | 2 | Insectivorous |
| South Island robin/kakaruai | Petroica australis | Endemic | 4 | Insectivorous |
| Tomtit/miromiro | Petroica macrocephala | Endemic | 4 | Insectivorous |
| Tūī | Prosthemadera novaeseelandiae | Endemic | 3 | Herbivorous |
| Fantail/pīwakawaka | Rhipidura fuliginosa | Endemic | 3 | Insectivorous |
| Species/Library | Richness | Shannon | Simpson | Predominant Diet |
|---|---|---|---|---|
| Bellbird | 48 | 0.95839607 | 0.36300495 | Omnivorous |
| Common redpoll | 12 | 0.38264421 | 0.13870225 | Herbivorous |
| Chaffinch | 29 | 1.00701147 | 0.38721558 | Omnivorous |
| Grey warbler | 14 | 1.78694747 | 0.80119441 | Insectivorous |
| South Island robin | 103 | 1.33109767 | 0.59564466 | Insectivorous |
| Tomtit | 25 | 1.93339985 | 0.72866524 | Insectivorous |
| Dunnock | 130 | 2.02011625 | 0.77871149 | Insectivorous |
| Tūī | 5 | 0.46803031 | 0.2296815 | Herbivorous |
| Fantail | 10 | 1.61362017 | 0.73783657 | Insectivorous |
| Eurasian blackbird | 176 | 1.1909211 | 0.51274955 | Omnivorous |
| Song thrush | 69 | 2.04993336 | 0.82176403 | Omnivorous |
| Silvereye | 41 | 1.20118728 | 0.42349987 | Omnivorous |
| Virus | Viral Family | Host Species | Abundance (RPM) | Gene | Length (Amino Acids) |
|---|---|---|---|---|---|
| Blackbird siadenovirus | Adenoviridae | Blackbird | 21.45 | DNA polymerase | 130 |
| Avian astrovirus 2 | Astroviridae | Silvereye | 6.6 | RdRp | 211 |
| Nelson astrovirus-like 1 | Astroviridae | Robin | 40115.8 | RdRp | 1402 |
| Tūī | 0.93 | RdRp | 104 | ||
| Avian associated bastrovirus 1 | Astroviridae | Tomtit | 314.9 | RdRp | 134 |
| Avian associated bastrovirus 2 | Astroviridae | Thrush | 8956.92 | RdRp | 834 |
| Avian associated hepe-like virus 1 | Hepeviridae | Blackbird | 2.93 | Replicase | 205 |
| Avian associated hepe-like virus 2 | Hepeviridae | Bellbird | 20.33 | Replicase | 105 |
| Avian associated hepe-like virus 3 | Hepeviridae | Dunnock | 228.58 | Replicase | 1350 |
| Avian associated hepe-like virus 4 | Hepeviridae | Thrush | 170.6 | Replicase | 1760 |
| Avian associated hepe-like virus 5 | Hepeviridae | Silvereye | 26.99 | Replicase | 126 |
| Avian associated hepe-like virus 6 | Hepeviridae | Silvereye | 14.62 | Replicase | 109 |
| Turdid alphaherpesvirus 1 | Herpesviridae | Thrush | 1207.78 | Major capsid protein | 1459 |
| Avian associated calicivirus 1 | Caliciviridae | Dunnock | 417.46 | RdRp | 791 |
| Avian associated calicivirus 2 | Caliciviridae | Blackbird | 0.75 | RdRp | 145 |
| Avian associated calicivirus 3 | Caliciviridae | Blackbird | 0.77 | RdRp | 142 |
| Avian associated calicivirus 4 | Caliciviridae | Blackbird | 3.12 | RdRp | 148 |
| Avian associated picorna-like virus 1 | Picornaviridae | Bellbird | 16.34 | RdRp | 141 |
| Avian associated picorna-like virus 2 | Picornaviridae | Robin | 5.04 | RdRp | 193 |
| Avian associated picorna-like virus 3 | Picornaviridae | Thrush | 30.64 | RdRp | 281 |
| Avian associated picorna-like virus 4 | Picornaviridae | Thrush | 54.1 | RdRp | 360 |
| Avian associated picorna-like virus 5 | Picornaviridae | Thrush | 27.29 | RdRp | 407 |
| Avian associated picorna-like virus 6 | Picornaviridae | Blackbird | 1.03 | RdRp | 101 |
| Avian associated picorna-like virus 7 | Picornaviridae | Dunnock | 17.32 | RdRp | 186 |
| Avian associated picorna-like virus 8 | Picornaviridae | Blackbird | 1.73 | RdRp | 173 |
| Thrush | 250.73 | RdRp | 722 | ||
| Avian associated picorna-like virus 9 | Picornaviridae | Thrush | 6.96 | RdRp | 181 |
| Avian associated picorna-like virus 10 | Picornaviridae | Blackbird | 829.51 | RdRp | 1708 |
| Avian associated picorna-like virus 11 | Picornaviridae | Blackbird | 0.43 | RdRp | 130 |
| Avian associated orbivirus 1 | Reoviridae | Silvereye | 19.36 | RdRp | 142 |
| Avian associated reo-like virus 1 | Reoviridae | Blackbird | 5.37 | RdRp | 304 |
| Avian associated reo-like virus 2 | Reoviridae | Blackbird | 16.82 | RdRp | 897 |
| Avian associated reo-like virus 3 | Reoviridae | Fantail | 90.34 | RdRp | 113 |
| Avian associated reo-like virus 4 | Reoviridae | Dunnock | 5.84 | RdRp | 73 |
| Avian associated reo-like virus 5 | Reoviridae | Robin | 2.33 | RdRp | 161 |
| Avian associated reo-like virus 6 | Reoviridae | Robin | 1.48 | RdRp | 85 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
French, R.K.; Filion, A.; Niebuhr, C.N.; Holmes, E.C. Metatranscriptomic Comparison of Viromes in Endemic and Introduced Passerines in New Zealand. Viruses 2022, 14, 1364. https://doi.org/10.3390/v14071364
French RK, Filion A, Niebuhr CN, Holmes EC. Metatranscriptomic Comparison of Viromes in Endemic and Introduced Passerines in New Zealand. Viruses. 2022; 14(7):1364. https://doi.org/10.3390/v14071364
Chicago/Turabian StyleFrench, Rebecca K., Antoine Filion, Chris N. Niebuhr, and Edward C. Holmes. 2022. "Metatranscriptomic Comparison of Viromes in Endemic and Introduced Passerines in New Zealand" Viruses 14, no. 7: 1364. https://doi.org/10.3390/v14071364
APA StyleFrench, R. K., Filion, A., Niebuhr, C. N., & Holmes, E. C. (2022). Metatranscriptomic Comparison of Viromes in Endemic and Introduced Passerines in New Zealand. Viruses, 14(7), 1364. https://doi.org/10.3390/v14071364






