In-Depth Temporal Transcriptome Profiling of an Alphaherpesvirus Using Nanopore Sequencing
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells and Viruses
2.2. Cycloheximide Treatment
2.3. RNA Purification
2.4. Poly(A) RNA Isolation
2.5. Direct cDNA Sequencing
2.6. Pre-Processing and Data Analysis
3. Results
3.1. Time Course Analysis of BoHV-1 Transcriptome Using Nanopore Sequencing
3.2. BoHV-1 Expresses Four Immediate-Early Genes
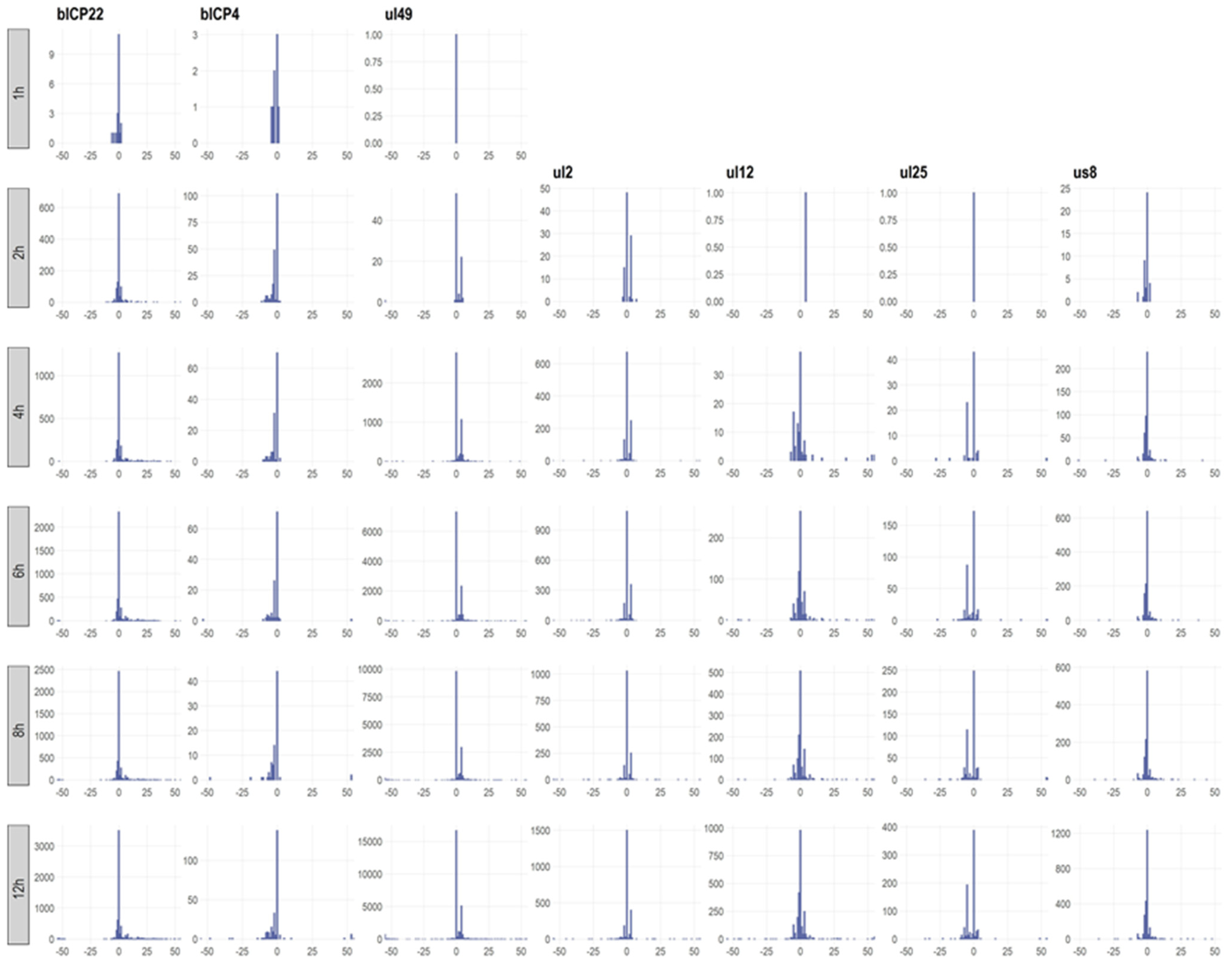
3.3. Transcription Start Sites
3.4. Transcription Start Site Clusters
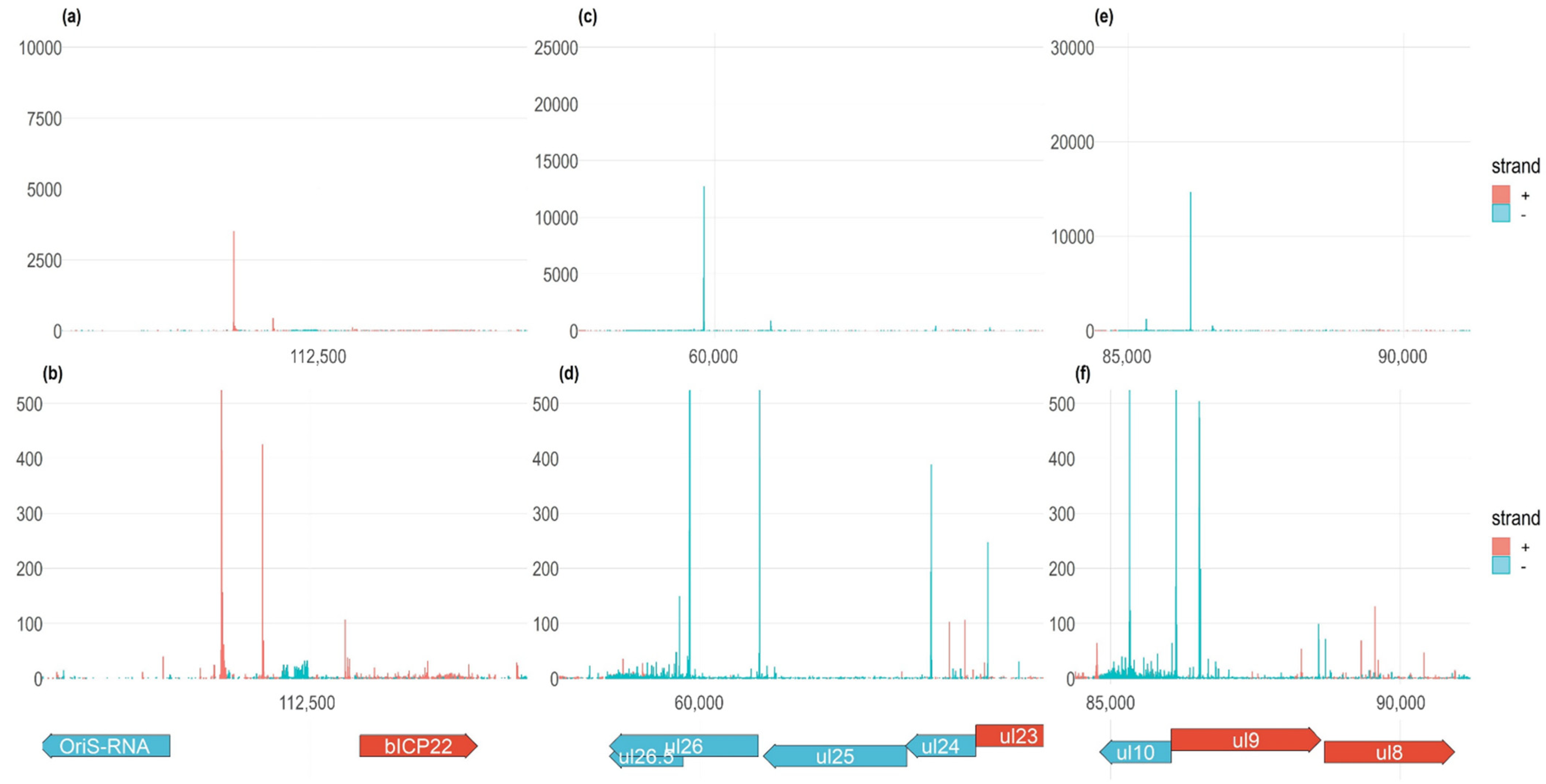
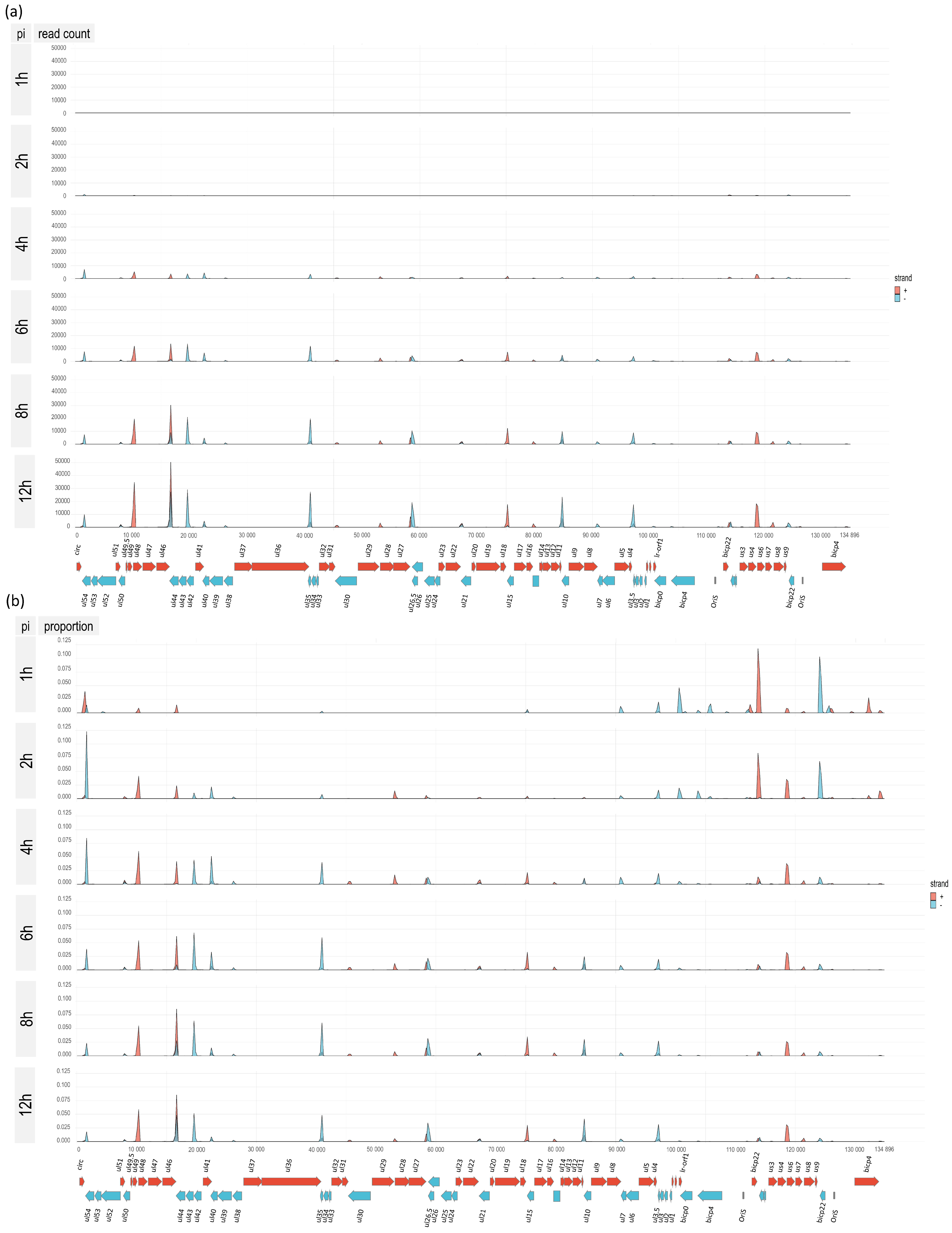
3.5. Time-Course Genome-Wide Expression of Transcription Start Sites
3.6. Transcription End Sites
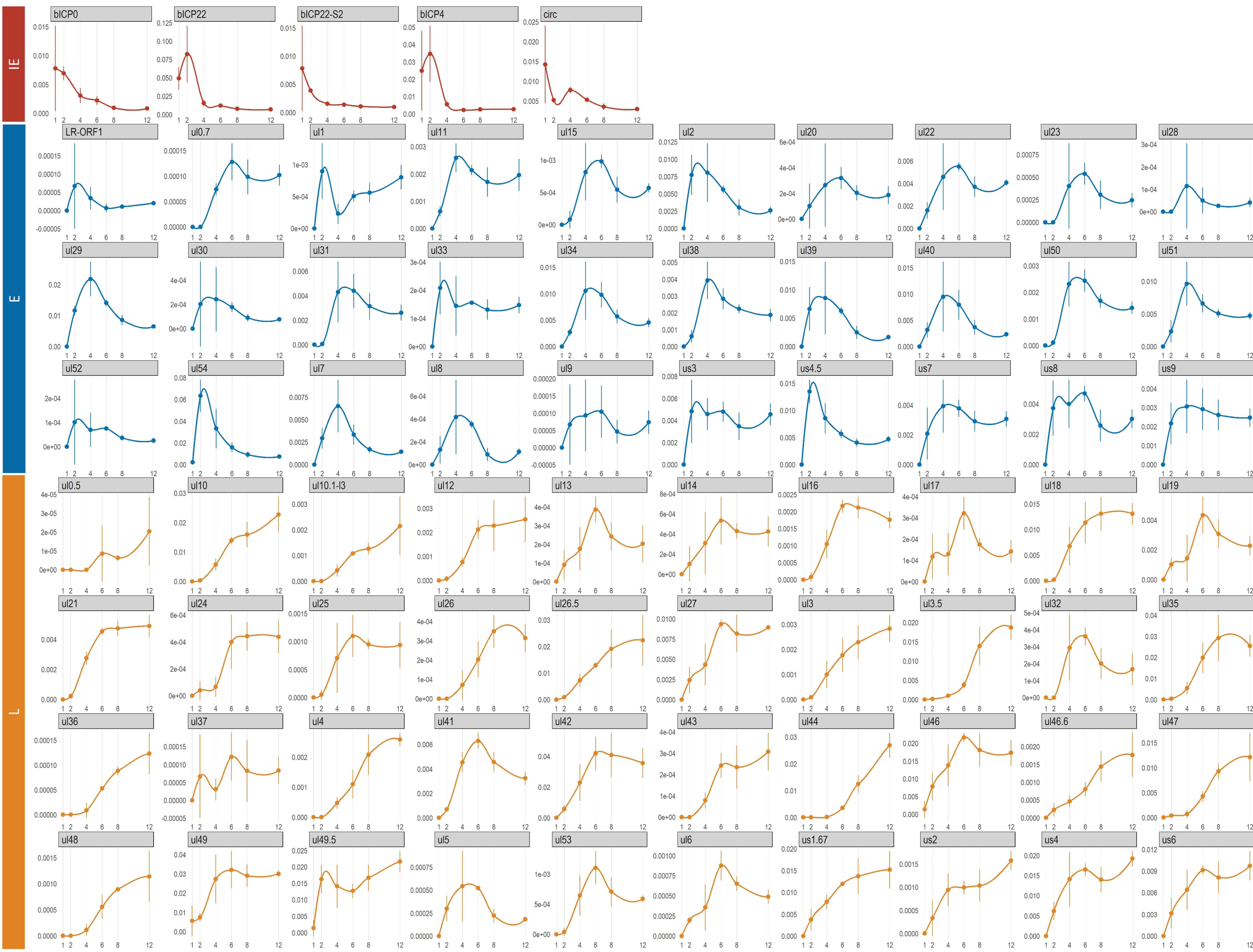
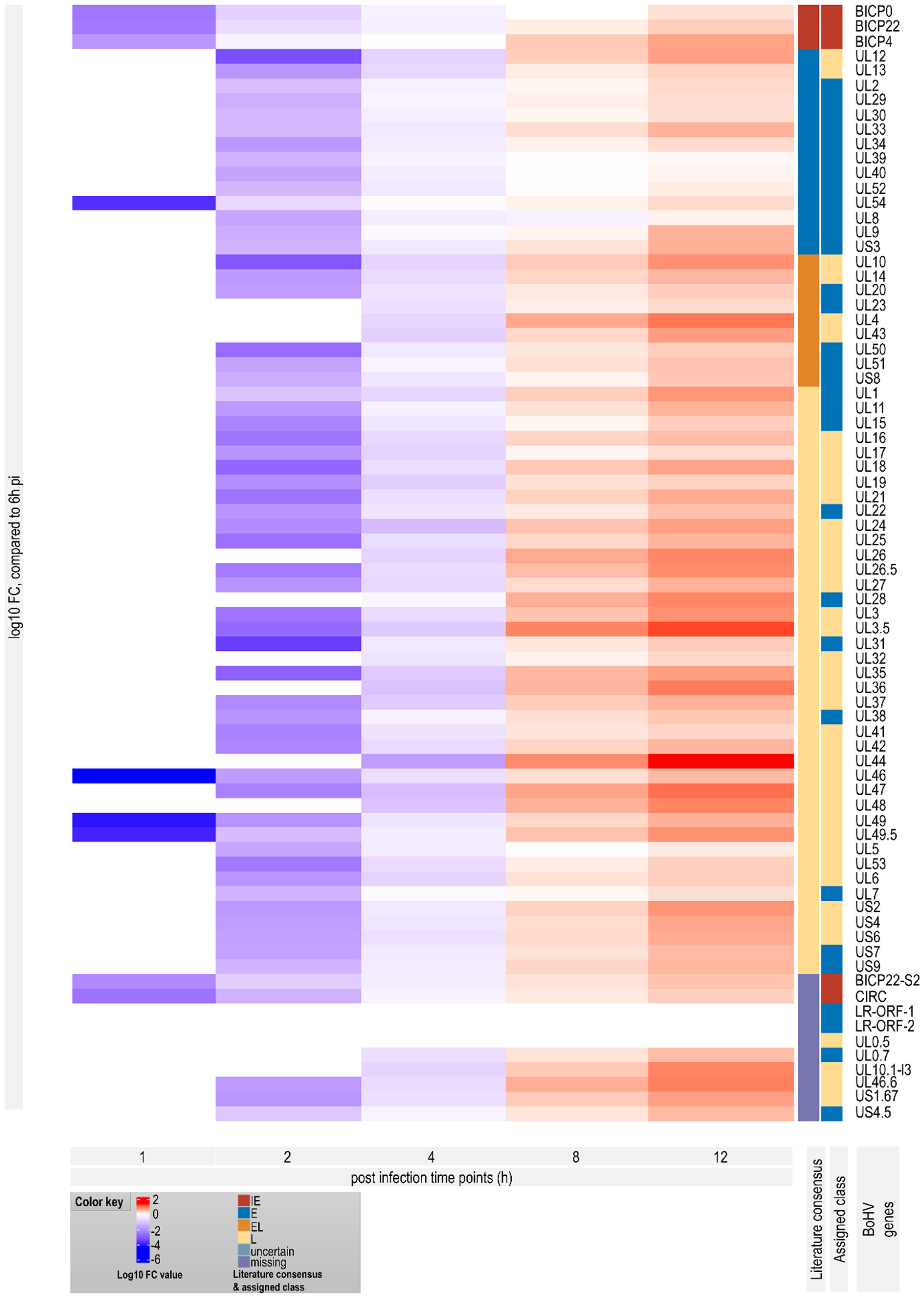
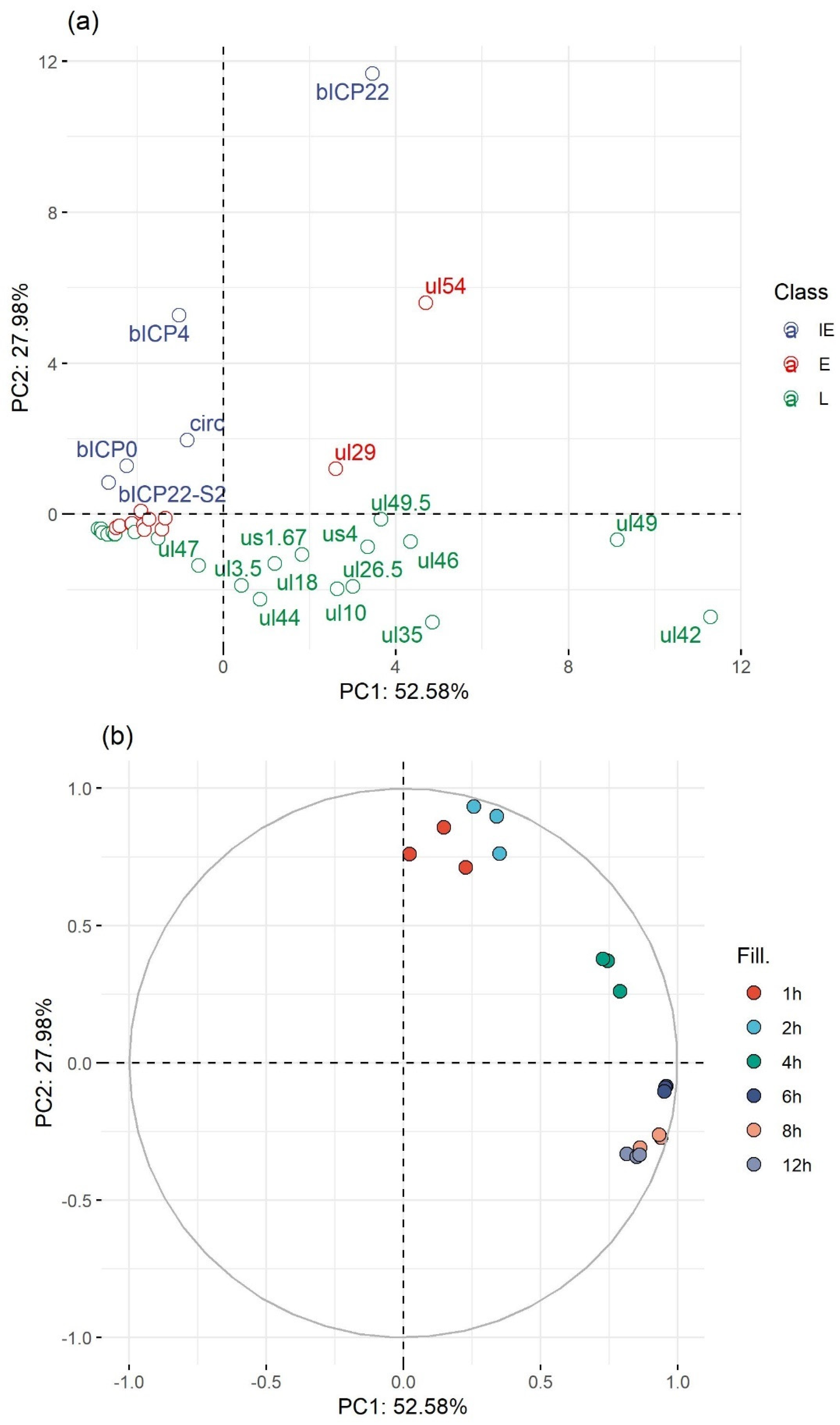
3.7. Genome-Wide Expression Dynamics of BoHV-1 Transcripts
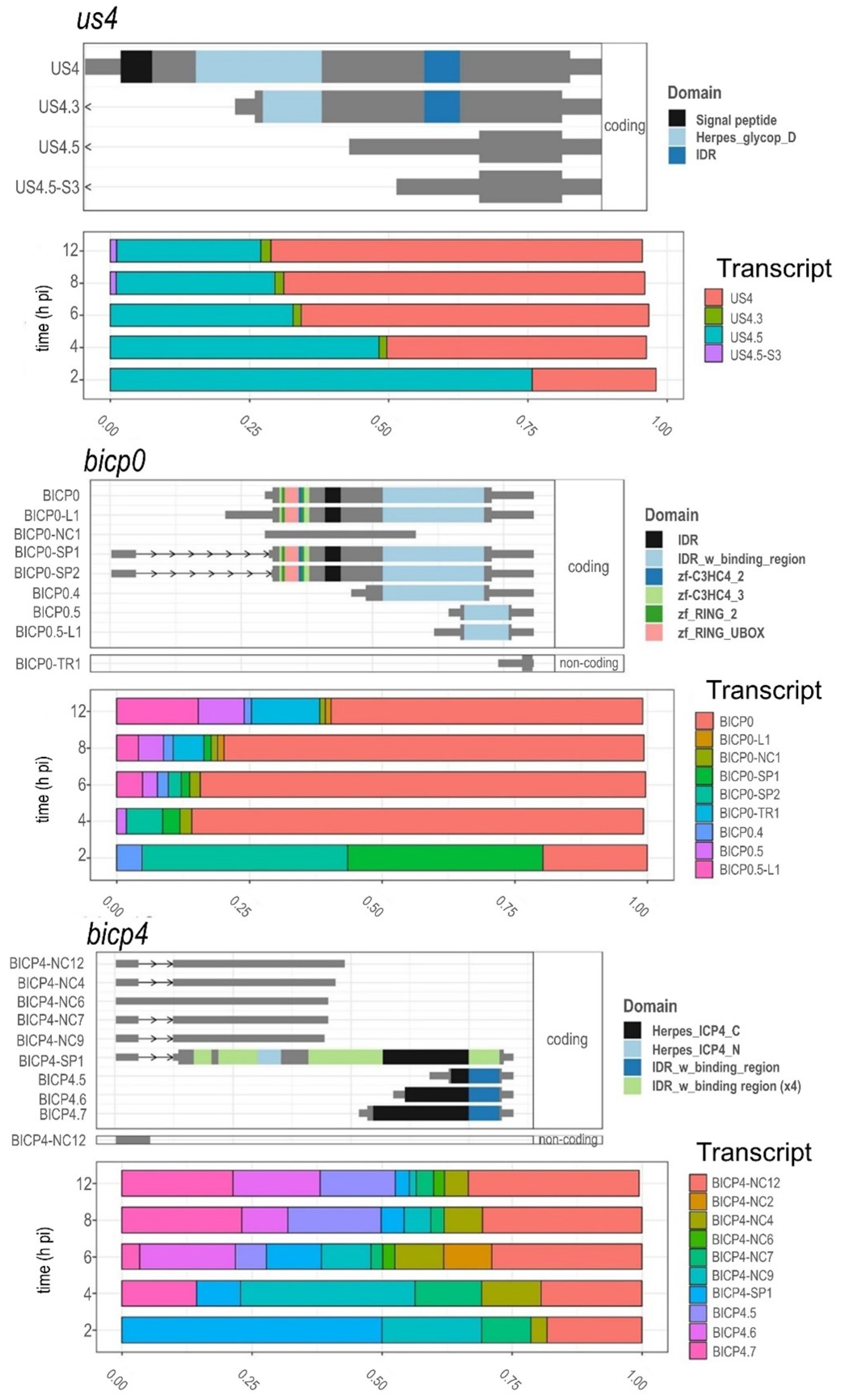
3.8. Time-Dependent Expression of Viral Gene Domains
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Van Oirschot, J.T. Bovine herpesvirus 1 in semen of bulls and the risk of transmission: A brief review. Vet. Q. 1995, 17, 29–33. [Google Scholar] [CrossRef] [PubMed]
- Khattar, S.K.; Van Drunen Littel-Van Den Hurk, S.; Babiuk, L.A.; Tikoo, S.K. Identification and transcriptional analysis of a 3′-coterminal gene cluster containing UL1, UL2, UL3, and UL3.5 open reading frames of bovine herpesvirus-1. Virology 1995, 213, 28–37. [Google Scholar] [CrossRef] [PubMed]
- Vlček, Č.; Benes, V.; Lu, Z.; Kutish, G.F.; Paces, V.; Rock, D.; Letchworth, G.J.; Schwyzer, M. Nucleotide sequence analysis of a 30-kb region of the bovine herpesvirus 1 genome which exhibits a colinear gene arrangement with the UL21 to UL4 genes of herpes simplex virus. Virology 1995, 210, 100–108. [Google Scholar] [CrossRef] [PubMed]
- d’Offay, J.M.; Fulton, R.W.; Eberle, R. Complete genome sequence of the NVSL BoHV-1.1 Cooper reference strain. Arch. Virol. 2013, 158, 1109–1113. [Google Scholar] [CrossRef] [PubMed]
- D’Offay, J.M.; Eberle, R.; Fulton, R.W.; Kirkland, P.D. Complete genomic sequence and comparative analysis of four genital and respiratory isolates of bovine herpesvirus subtype 1.2b (BoHV-1.2b), including the prototype virus strain K22. Arch. Virol. 2016, 161, 3269–3274. [Google Scholar] [CrossRef]
- Moldován, N.; Torma, G.; Gulyás, G.; Hornyák, Á.; Zádori, Z.; Jefferson, V.A.; Csabai, Z.; Boldogkői, M.; Tombácz, D.; Meyer, F.; et al. Time-course profiling of bovine herpesvirus type 1.1 transcriptome using multiplatform sequencing. Sci. Rep. 2020, 10, 20496. [Google Scholar] [CrossRef]
- Wirth, U.V.; Gunkel, K.; Engels, M.; Schwyzer, M. Spatial and temporal distribution of bovine herpesvirus 1 transcripts. J. Virol. 1989, 63, 4882–4889. [Google Scholar] [CrossRef]
- Wirth, U.V.; Fraefel, C.; Vogt, B.; Vlcek, C.; Paces, V.; Schwyzer, M. Immediate-early RNA 2.9 and early RNA 2.6 of bovine herpesvirus 1 are 3′- coterminal and encode a putative zinc finger transactivator protein. J. Virol. 1992, 66, 2763–2772. [Google Scholar] [CrossRef]
- Misra, V.; Walker, S.; Hayes, S.; O’Hare, P. The bovine herpesvirus α gene trans-inducing factor activates transcription by mechanisms different from those of its herpes simplex virus type 1 counterpart VP16. J. Virol. 1995, 69, 5209–5216. [Google Scholar] [CrossRef]
- Guo, J.; Li, Q.; Jones, C. The bovine herpesvirus 1 regulatory proteins, bICP4 and bICP22, are expressed during the escape from latency. J. Neurovirol. 2019, 25, 42–49. [Google Scholar] [CrossRef]
- Köppel, R.; Vogt, B.; Schwyzer, M. Immediate-early protein BICP22 of bovine herpesvirus 1 trans-represses viral promoters of different kinetic classes and is itself regulated by BICP0 at transcriptional and posttranscriptional levels. Arch. Virol. 1997, 142, 2447–2464. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Wu, W.J.; Liu, L.D.; Wang, L.C.; Zhang, Y.; Wu, L.Q.; Guan, Y.; Li, Q.H. Herpes Simplex Virus 1 ICP22 Inhibits the Transcription of Viral Gene Promoters by Binding to and Blocking the Recruitment of P-TEFb. PLoS ONE 2012, 7, e45749. [Google Scholar] [CrossRef] [PubMed]
- Fraefel, C.; Wirth, U.V.; Vogt, B.; Schwyzer, M. Immediate-early transcription over covalently joined genome ends of bovine herpesvirus 1: The circ gene. J. Virol. 1993, 67, 1328–1333. [Google Scholar] [CrossRef] [PubMed]
- Kronstad, L.M.; Brulois, K.F.; Jung, J.U.; Glaunsinger, B.A. Dual short upstream open reading frames control translation of a herpesviral polycistronic mRNA. PLoS Pathog. 2013, 9, e1003156. [Google Scholar] [CrossRef] [PubMed]
- Jones, C. Herpes simplex virus type 1 and bovine herpesvirus 1 latency. Clin. Microbiol. Rev. 2003, 16, 79–95. [Google Scholar] [CrossRef]
- Oláh, P.; Tombácz, D.; Csabai, Z.; Póka, N.; Prazsák, I.; Boldogkői, Z. Characterization of pseudorabies virus transcriptome by Illumina sequencing. BMC Microbiol. 2015, 15, 130. [Google Scholar] [CrossRef]
- Wade, J.T.; Grainger, D.C. Pervasive transcription: Illuminating the dark matter of bacterial transcriptomes. Nat. Rev. Microbiol. 2014, 12, 647–653. [Google Scholar] [CrossRef]
- Shiraki, T.; Kondo, S.; Katayama, S.; Waki, K.; Kasukawa, T.; Kawaji, H.; Kodzius, R.; Watahiki, A.; Nakamura, M.; Arakawa, T.; et al. Cap analysis gene expression for high-throughput analysis of transcriptional starting point and identification of promoter usage. Proc. Natl. Acad. Sci. USA 2003, 100, 15776–15781. [Google Scholar] [CrossRef]
- Batut, P.; Dobin, A.; Plessy, C.; Carninci, P.; Gingeras, T.R. High-fidelity promoter profiling reveals wide- spread alternative promoter usage and transposon-driven developmental gene expression. Genome Res. 2013, 23, 169–180. [Google Scholar] [CrossRef]
- Kawaji, H.; Kasukawa, T.; Forrest, A.; Carninci, P.; Hayashizaki, Y. The FANTOM5 collection, a data series underpinning mammalian transcriptome atlases in diverse cell types. Sci. Data 2017, 4, 170113. [Google Scholar] [CrossRef]
- Gupta, I.; Collier, P.G.; Haase, B.; Mahfouz, A.; Joglekar, A.; Floyd, T.; Koopmans, F.; Barres, B.; Smit, A.B.; Sloan, S.A.; et al. Single-cell isoform RNA sequencing characterizes isoforms in thousands of cerebellar cells. Nat. Biotechnol. 2018, 36, 1197–1202. [Google Scholar] [CrossRef] [PubMed]
- Hardwick, S.A.; Hu, W.; Joglekar, A.; Fan, L.; Collier, P.G.; Foord, C.; Balacco, J.; Lanjewar, S.; Sampson, M.M.; Koopmans, F.; et al. Single-nuclei isoform RNA sequencing unlocks barcoded exon connectivity in frozen brain tissue. Nat. Biotechnol. 2022. [Google Scholar] [CrossRef] [PubMed]
- Glazov, E.A.; Horwood, P.F.; Assavalapsakul, W.; Kongsuwan, K.; Mitchell, R.W.; Mitter, N.; Mahony, T.J. Characterization of microRNAs encoded by the bovine herpesvirus 1 genome. J. Gen. Virol. 2010, 91, 32–41. [Google Scholar] [CrossRef]
- Byrne, A.; Beaudin, A.E.; Olsen, H.E.; Jain, M.; Cole, C.; Palmer, T. Nanopore long-read RNAseq reveals widespread transcriptional variation among the surface receptors of individual B cells. Nat. Commun. 2017, 8, 16027. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.-Y.; Deng, F.; Jia, X.; Li, C.; Lai, S.-J. A transcriptome atlas of rabbit revealed by PacBio single-molecule long-read sequencing. Sci. Rep. 2017, 7, 7648. [Google Scholar] [CrossRef] [PubMed]
- Kakuk, B.; Kiss, A.A.; Torma, G.; Csabai, Z.; Prazsák, I.; Mizik, M.; Megyeri, K.; Tombácz, D.; Boldogkői, Z. Nanopore Assay Reveals Cell-Type-Dependent Gene Expression of Vesicular Stomatitis Indiana Virus and Differential Host Cell Response. Pathogens 2021, 10, 1196. [Google Scholar] [CrossRef]
- Moldován, N.; Balázs, Z.; Tombácz, D.; Csabai, Z.; Szűcs, A.; Snyder, M.; Boldogkői, Z. Multi-platform analysis reveals a complex transcriptome architecture of a circovirus. Virus Res. 2017, 237, 37–46. [Google Scholar] [CrossRef][Green Version]
- Nudelman, G.; Frasca, A.; Kent, B.; Sadler, K.C.; Sealfon, S.C.; Walsh, M.J.; Zaslavsky, E. High resolution annotation of zebrafish transcriptome using long-read sequencing. Genome Res. 2018, 28, 1415–1425. [Google Scholar] [CrossRef]
- Tombácz, D.; Prazsák, I.; Szucs, A.; Dénes, B.; Snyder, M.; Boldogkoi, Z. Dynamic transcriptome profiling dataset of vaccinia virus obtained from long-read sequencing techniques. Gigascience 2018, 7, giy139. [Google Scholar] [CrossRef]
- Li, Y.; Fang, C.; Fu, Y.; Hu, A.; Li, C.; Zou, C.; Li, X.; Zhao, S.; Zhang, C.; Li, C. A survey of transcriptome complexity in Sus scrofa using single-molecule long-read sequencing. DNA Res. 2018, 25, 421–437. [Google Scholar] [CrossRef]
- Moldován, N.; Tombácz, D.; Szűcs, A.; Csabai, Z.; Balázs, Z.; Kis, E.; Molnár, J.; Boldogkői, Z. Third-generation sequencing reveals extensive polycistronism and transcriptional overlapping in a baculovirus. Sci. Rep. 2018, 8, 8604. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Zhang, H.; Kohnen, M.V.; Prasad, K.V.S.K.; Gu, L.; Reddy, A.S.N. Analysis of transcriptome and epitranscriptome in plants using PacBio iso-seq and nanopore-based direct RNA sequencing. Front. Genet. 2019, 10, 253. [Google Scholar] [CrossRef] [PubMed]
- Boldogkői, Z.; Moldován, N.; Balázs, Z.; Snyder, M.; Tombácz, D. Long-read sequencing—A powerful tool in viral transcriptome research. Trends Microbiol. 2019, 27, 578–592. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; Zhang, J.; Liu, Q.; Liu, X.; Wang, H.; He, J.; Kang, L. Long-read direct RNA sequencing by 5′-Cap capturing reveals the impact of Piwi on the widespread exonization of transposable elements in locusts. RNA Biol. 2019, 16, 950–959. [Google Scholar] [CrossRef]
- Zhang, B.; Liu, J.; Wang, X.; Wei, Z. Full-length RNA sequencing reveals unique transcriptome composition in bermudagrass. Plant Physiol. Biochem. 2018, 132, 95–103. [Google Scholar] [CrossRef]
- Tombácz, D.; Csabai, Z.; Oláh, P.; Havelda, Z.; Sharon, D.; Snyder, M.; Boldogkői, Z. Characterization of novel transcripts in pseudorabies virus. Viruses 2015, 7, 2727–2744. [Google Scholar] [CrossRef]
- O’Grady, T.; Wang, X.; Höner Zu Bentrup, K.; Baddoo, M.; Concha, M.; Flemington, E.K. Global transcript structure resolution of high gene density genomes through multi-platform data integration. Nucleic Acids Res. 2016, 44, e145. [Google Scholar] [CrossRef]
- Tombácz, D.; Csabai, Z.; Oláh, P.; Balázs, Z.; Likó, I.; Zsigmond, L.; Sharon, D.; Snyder, M.; Boldogkői, Z. Full-Length Isoform Sequencing Reveals Novel Transcripts and Substantial Transcriptional Overlaps in a Herpesvirus. PLoS ONE 2016, 11, e0162868. [Google Scholar] [CrossRef]
- Moldován, N.; Tombácz, D.; Szűcs, A.; Csabai, Z.; Snyder, M.; Boldogkői, Z. Multi-Platform Sequencing Approach Reveals a Novel Transcriptome Profile in Pseudorabies Virus. Front. Microbiol. 2018, 8, 2708. [Google Scholar] [CrossRef]
- Balázs, Z.; Tombácz, D.; Csabai, Z.; Szűcs, A.; Megyeri, K.; Petrov, A.N.; Snyder, M.; Boldogkői, Z. Long-Read Sequencing of Human Cytomegalovirus Transcriptome Reveals RNA Isoforms Carrying Distinct Coding Potentials. Sci. Rep. 2017, 7, 15989. [Google Scholar] [CrossRef]
- Tombácz, D.; Balázs, Z.; Gulyás, G.; Csabai, Z.; Boldogkői, M.; Snyder, M.; Boldogkői, Z. Multiple Long-read Sequencing Survey of Herpes Simplex Virus Lytic Transcriptome. Front. Genet. 2019, 10, 834. [Google Scholar] [CrossRef] [PubMed]
- Depledge, D.P.; Srinivas, K.P.; Sadaoka, T.; Bready, D.; Mori, Y.; Placantonakis, D.G.; Mohr, I.; Wilson, A.C. Direct RNA sequencing on nanopore arrays redefines the transcriptional complexity of a viral pathogen. Nat. Commun. 2019, 10, 754. [Google Scholar] [CrossRef] [PubMed]
- Tombácz, D.; Torma, G.; Gulyás, G.; Moldován, N.; Snyder, M.; Boldogkői, Z. Meta-analytic approach for transcriptome profiling of herpes simplex virus type 1. Sci. Data 2020, 7, 223. [Google Scholar] [CrossRef] [PubMed]
- Haberle, V.; Stark, A. Eukaryotic core promoters and the functional basis of transcription initiation. Nat. Rev. Mol. Cell. Biol. 2018, 19, 621–637. [Google Scholar] [CrossRef] [PubMed]
- Tombácz, D.; Prazsák, I.; Csabai, Z.; Moldován, N.; Balázs, Z.; Dénes, B.; Snyder, M.; Boldogkői, Z. Long-read Assays Shed New Light on the Transcriptome Complexity of a Viral Pathogen. Sci. Rep. 2020, 10, 13822. [Google Scholar] [CrossRef]
- Kakuk, B.; Tombácz, D.; Balázs, Z.; Moldován, N.; Csabai, Z.; Torma, G.; Megyeri, K.; Snyder, M.; Boldogkői, Z. Combined Nanopore and Single-Molecule Real-Time Sequencing Survey of Human Betaherpesvirus 5 Transcriptome. Sci. Rep. 2021, 11, 14487. [Google Scholar] [CrossRef]
- Torma, G.; Tombácz, D.; Csabai, Z.; Moldován, N.; Mészáros, I.; Zádori, Z.; Boldogkői, Z. Combined Short and Long-read Sequencing Reveals a Complex Transcriptomic Architecture of African Swine Fever Virus. Viruses 2021, 13, 579. [Google Scholar] [CrossRef]
- Fülöp, Á.; Torma, G.; Moldován, N.; Szenthe, K.; Bánáti, F.; Almsarrhad, I.A.; Csabai, Z.; Tombácz, D.; Minárovits, J.; Boldogkői, Z. Integrative Profiling of Epstein-Barr Virus Transcriptome Using a Multiplatform Approach. Virol. J. 2022, 19, 7. [Google Scholar] [CrossRef]
- Hampsey, M. Molecular genetics of the RNA polymerase II general transcriptional machinery. Microbiol. Mol. Biol. Rev. 1998, 62, 465–503. [Google Scholar] [CrossRef]
- Carninci, P.; Sandelin, A.; Lenhard, B.; Katayama, S.; Shimokawa, K.; Ponjavic, J.; Semple, C.A.; Taylor, M.S.; Engström, P.G.; Frith, M.C.; et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet. 2006, 38, 626–635. [Google Scholar] [CrossRef]
- Ni, T.; Corcoran, D.L.; Rach, E.A.; Song, S.; Spana, E.P.; Gao, Y.; Ohler, U.; Zhu, J. A paired-end sequencing strategy to map the complex landscape of transcription initiation. Nat. Methods. 2010, 7, 521–527. [Google Scholar] [CrossRef] [PubMed]
- FANTOM Consortium and the RIKEN PMI and CLST (DGT); Forrest, A.R.R.; Kawaji, H.; Rehli, M.; Baillie, J.K.; de Hoon, M.J.L.; Haberle, V.; Lassmann, T.; Kulakovskiy, I.V.; Lizio, M.; et al. A promoter-level mammalian expression atlas. Nature 2014, 507, 462–470. [Google Scholar] [CrossRef] [PubMed]
- Tombácz, D.; Balázs, Z.; Csabai, Z.; Moldován, N.; Szűcs, A.; Sharon, D.; Snyder, M.; Boldogkői, Z. Characterization of the Dynamic Transcriptome of a Herpesvirus with Long-read Single Molecule Real-Time Sequencing. Sci. Rep. 2017, 7, 43751. [Google Scholar] [CrossRef] [PubMed]
- Balázs, Z.; Tombácz, D.; Csabai, Z.; Moldován, N.; Snyder, M.; Boldogkői, Z. Template-switching artifacts resemble alternative polyadenylation. BMC Genom. 2019, 20, 824. [Google Scholar] [CrossRef]
- Sessegolo, C.; Cruaud, C.; Da Silva, C.; Cologne, A.; Dubarry, M.; Derrien, T.; Lacroix, V.; Aury, J.M. Transcriptome profiling of mouse samples using nanopore sequencing of cDNA and RNA molecules. Sci. Rep. 2019, 9, 14908. [Google Scholar] [CrossRef]
- Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef]
- Tombácz, D.; Moldován, N.; Torma, G.; Nagy, T.; Hornyák, Á.; Csabai, Z.; Gulyás, G.; Boldogkői, M.; Jefferson, V.A.; Zádori, Z.; et al. Dynamic Transcriptome Sequencing of Bovine Alphaherpesvirus Type 1 and Host Cells Carried Out by a Multi-Technique Approach. Front. Genet. 2021, 7, 619056. [Google Scholar] [CrossRef]
- Morgan, M.; Pagès, H.; Obenchain, V.; Hayden, N. Rsamtools: Binary alignment (BAM), FASTA, variant call (BCF), and tabix file import. R Package Version 2.10.0 2021, 1, 677–689. [Google Scholar]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; ISBN 978-3-319-24277-4. [Google Scholar]
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef]
- Anders, S.; Reyes, A.; Huber, W. Detecting differential usage of exons from RNA-seq data. Genome Res. 2012, 22, 4025. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef] [PubMed]
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef] [PubMed]
- Mészáros, B.; Erdős, G.; Dosztányi, Z. IUPred2A: Context-dependent prediction of protein disorder as a function of redox state and protein binding. Nucleic Acids Res. 2018, 46, W329–W337. [Google Scholar] [CrossRef] [PubMed]
- Wirth, U.V.; Vogt, B.; Schwyzer, M. The three major immediate-early transcripts of bovine herpesvirus 1 arise from two divergent and spliced transcription units. J. Virol. 1991, 65, 195–205. [Google Scholar] [CrossRef] [PubMed]
- Fan, D.; Wang, M.; Cheng, A.; Jia, R.; Yang, Q.; Wu, Y.; Zhu, D.; Zhao, X.; Chen, S.; Liu, M.; et al. The Role of VP16 in the Life Cycle of Alphaherpesviruses. Front. Microbiol. 2020, 11, 1910. [Google Scholar] [CrossRef] [PubMed]
- Some Genetic Functions Encoded by Herpes Simplex Virus Type 1. Available online: http://darwin.bio.uci.edu/~faculty/wagner/table.html (accessed on 3 October 2003).
- The Genetic and Transcription Map of the HSV-1 Genome. Available online: http://darwin.bio.uci.edu/~faculty/wagner/hsvimg04z.jpg (accessed on 3 October 2003).
- Roizman, B. The function of herpes simplex virus genes: A primer for genetic engineering of novel vectors. Proc. Nat. Acad. Sci. USA 1996, 93, 11307–11312. [Google Scholar] [CrossRef]
- Roizman, B.; Campadelli-Fiume, G. Alphaherpes viral genes and their functions. In Human Herpesviruses-Biology, Therapy and Immunoprophylaxis; Arvin, A., Campadelli-Fiume, G., Mocarski, E., Moore, P.S., Roizman, B., Whitley, R., Yamanishi, K., Eds.; Cambridge University Press: Cambridge, UK, 2007; pp. 70–92. [Google Scholar]
- Pseudorabies Virus Gene Annotations. Available online: https://www.fli.de/en/institutes/institute-of-molecular-virology-and-cell-biology-imvz/laboratories/laboratory-for-virus-host-cell-interactions/fig-7-gene-and-transcript-arrangement-in-the-prv-genome/ (accessed on 1 September 2005).
- Vitting-Seerup, K.; Sandelin, A. The landscape of isoform switches in human cancers. Mol. Cancer Res. 2017, 15, 1206–1220. [Google Scholar] [CrossRef]
- Prazsák, I.; Moldován, N.; Balázs, Z.; Tombácz, D.; Megyeri, K.; Szűcs, A.; Csabai, Z.; Boldogkői, Z. Long-read sequencing uncovers a complex transcriptome topology in varicella zoster virus. BMC Genom. 2018, 19, 873. [Google Scholar] [CrossRef]
- Mathieu-Daudé, F.; Welsh, J.; Vogt, T.; McClelland, M. DNA rehybridization during PCR: The ‘Cot Effect’ and its consequences. Nucleic Acids Res. 1996, 24, 2080–2086. [Google Scholar] [CrossRef]
- Polz, M.F.; Cavanaugh, C.M. Bias in template-to-product ratios in multitemplate PCR. Appl. Environ. Microbiol. 1998, 64, 3724–3730. [Google Scholar] [CrossRef]
- Suzuki, Y.; Taira, H.; Tsunoda, T.; Mizushima-Sugano, J.; Sese, J.; Hata, H.; Ota, T.; Isogai, T.; Tanaka, T.; Morishita, S.; et al. Diverse transcriptional initiation revealed by fine, large-scale mapping of mRNA start sites. EMBO Rep. 2001, 2, 388–393. [Google Scholar] [CrossRef]
- Smale, S.T.; Kadonaga, J.T. The RNA polymerase II core promoter. Annu. Rev. Biochem. 2003, 72, 449–479. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Park, J.K.; Zhang, J.Z. Evidence that alternative transcriptional initiation is largely nonadaptive. PLoS Biol. 2019, 17, e3000197. [Google Scholar] [CrossRef] [PubMed]
- Kawaji, H.; Frith, M.C.; Katayama, S.; Sandelin, A.; Kai, C.; Kawai, J.; Carninci, P.; Hayashizaki, Y. Dynamic usage of transcription start sites within core promoters. Genome Biol. 2006, 7, R118. [Google Scholar] [CrossRef]
- Barber, K.A.; Daugherty, H.C.; Ander, S.E.; Jefferson, V.A.; Shack, L.A.; Pechan, T.; Nanduri, B.; Meyer, F. Protein Composition of the Bovine Herpesvirus 1.1 Virion. Vet Sci. 2017, 4, 11. [Google Scholar] [CrossRef] [PubMed]
- Pokhriyal, M.; Ratta, B.; Yadav, B.S.; Kumar, A.; Saxena, M.; Verma, O.P.; Sharma, B. Three newly identified Immediate Early Genes of Bovine herpesvirus 1 lack the characteristic Octamer binding motif-1. Sci. Rep. 2018, 8, 11441. [Google Scholar] [CrossRef] [PubMed]
- Boldogkői, Z.; Balázs, Z.; Moldován, N.; Prazsák, I.; Tombácz, D. Novel classes of replication-associated transcripts discovered in viruses. RNA Biol. 2019, 16, 166–175. [Google Scholar] [CrossRef] [PubMed]
- Boldogkői, Z.; Tombácz, D.; Balázs, Z. Interactions between the Transcription and Replication Machineries Regulate the RNA and DNA synthesis in the Herpesviruses. Virus Genes 2019, 55, 274–279. [Google Scholar] [CrossRef] [PubMed]
- Boldogkői, Z. Transcriptional interference networks coordinate the expression of functionally-related genes clustered in the same genomic loci. Front. Genet. 2012, 3, 122. [Google Scholar] [CrossRef]



| Time Point | Barcode | Barcode Sequence | |
|---|---|---|---|
| 1st replicate | 1 h | A1(BC01) | AAGAAAGTTGTCGGTGTCTTTGTG |
| 2 h | A2(BC02) | TCGATTCCGTTTGTAGTCGTCTGT | |
| 4 h | A3(BC03) | GAGTCTTGTGTCCCAGTTACCAGG | |
| 6 h | A4(BC04) | TTCGGATTCTATCGTGTTTCCCTA | |
| 8 h | A5(BC05) | CTTGTCCAGGGTTTGTGTAACCTT | |
| 12 h | A6(BC06) | TTCTCGCAAAGGCAGAAAGTAGTC | |
| MOCK | A7(BC07) | GTGTTACCGTGGGAATGAATCCTT | |
| 2nd replicate | 1 h | A8(BC08) | TTCAGGGAACAAACCAAGTTACGT |
| 2 h | A9(BC09) | AACTAGGCACAGCGAGTCTTGGTT | |
| 4 h | A10(BC10) | AAGCGTTGAAACCTTTGTCCTCTC | |
| 6 h | A11(BC11) | GTTTCATCTATCGGAGGGAATGGA | |
| 8 h | A12(BC24) | GCATAGTTCTGCATGATGGGTTAG | |
| 12 h | A1(BC01) | AAGAAAGTTGTCGGTGTCTTTGTG | |
| MOCK | A2(BC02) | TCGATTCCGTTTGTAGTCGTCTGT | |
| 3rd replicate | 1 h | A3(BC03) | GAGTCTTGTGTCCCAGTTACCAGG |
| 2 h | A4(BC04) | TTCGGATTCTATCGTGTTTCCCTA | |
| 4 h | A5(BC05) | CTTGTCCAGGGTTTGTGTAACCTT | |
| 6 h | A6(BC06) | TTCTCGCAAAGGCAGAAAGTAGTC | |
| 8 h | A7(BC07) | GTGTTACCGTGGGAATGAATCCTT | |
| 12 h | A8(BC08) | TTCAGGGAACAAACCAAGTTACGT | |
| MOCK | A9(BC09) | AACTAGGCACAGCGAGTCTTGGTT |
| Gene | Sequence Names | Start | End | 6 h 20 mg | 8 h 20 mg | 6 h 100 mg | 8 h 100 mg |
|---|---|---|---|---|---|---|---|
| bicp22 | JX898220.1 | 112,888 | 113,790 | 132,263 | 165,542 | 138,780 | 127,408 |
| bicp4 | JX898220.1 | 103,907 | 107,941 | 44,292 | 57,067 | 36,249 | 43,288 |
| circ | JX898220.1 | 487 | 1227 | 5781 | 8667 | 6137 | 5398 |
| bicp0 | JX898220.1 | 100,898 | 102,949 | 4459 | 5270 | 4529 | 4234 |
| ul54 | JX898220.1 | 1648 | 2850 | 13,953 | 62,679 | 4601 | 4841 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tombácz, D.; Kakuk, B.; Torma, G.; Csabai, Z.; Gulyás, G.; Tamás, V.; Zádori, Z.; Jefferson, V.A.; Meyer, F.; Boldogkői, Z. In-Depth Temporal Transcriptome Profiling of an Alphaherpesvirus Using Nanopore Sequencing. Viruses 2022, 14, 1289. https://doi.org/10.3390/v14061289
Tombácz D, Kakuk B, Torma G, Csabai Z, Gulyás G, Tamás V, Zádori Z, Jefferson VA, Meyer F, Boldogkői Z. In-Depth Temporal Transcriptome Profiling of an Alphaherpesvirus Using Nanopore Sequencing. Viruses. 2022; 14(6):1289. https://doi.org/10.3390/v14061289
Chicago/Turabian StyleTombácz, Dóra, Balázs Kakuk, Gábor Torma, Zsolt Csabai, Gábor Gulyás, Vivien Tamás, Zoltán Zádori, Victoria A. Jefferson, Florencia Meyer, and Zsolt Boldogkői. 2022. "In-Depth Temporal Transcriptome Profiling of an Alphaherpesvirus Using Nanopore Sequencing" Viruses 14, no. 6: 1289. https://doi.org/10.3390/v14061289
APA StyleTombácz, D., Kakuk, B., Torma, G., Csabai, Z., Gulyás, G., Tamás, V., Zádori, Z., Jefferson, V. A., Meyer, F., & Boldogkői, Z. (2022). In-Depth Temporal Transcriptome Profiling of an Alphaherpesvirus Using Nanopore Sequencing. Viruses, 14(6), 1289. https://doi.org/10.3390/v14061289







