Gastrointestinal Involvement in SARS-CoV-2 Infection
Abstract
:1. Introduction
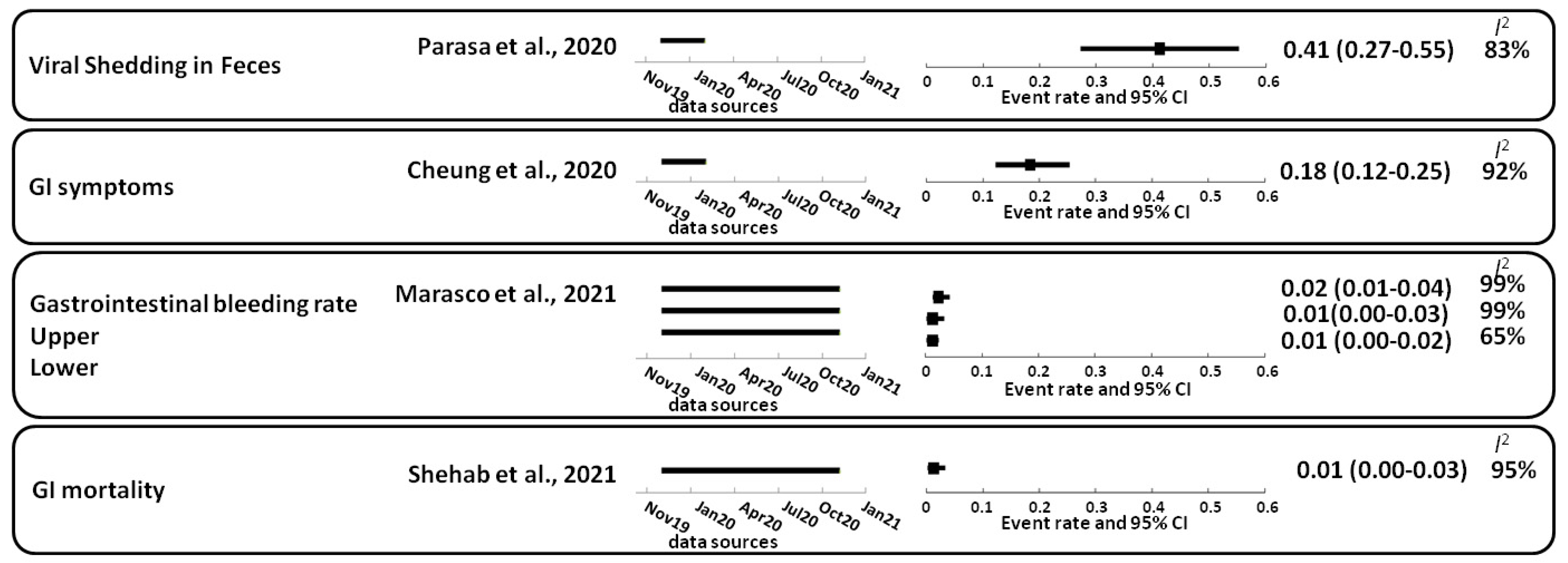
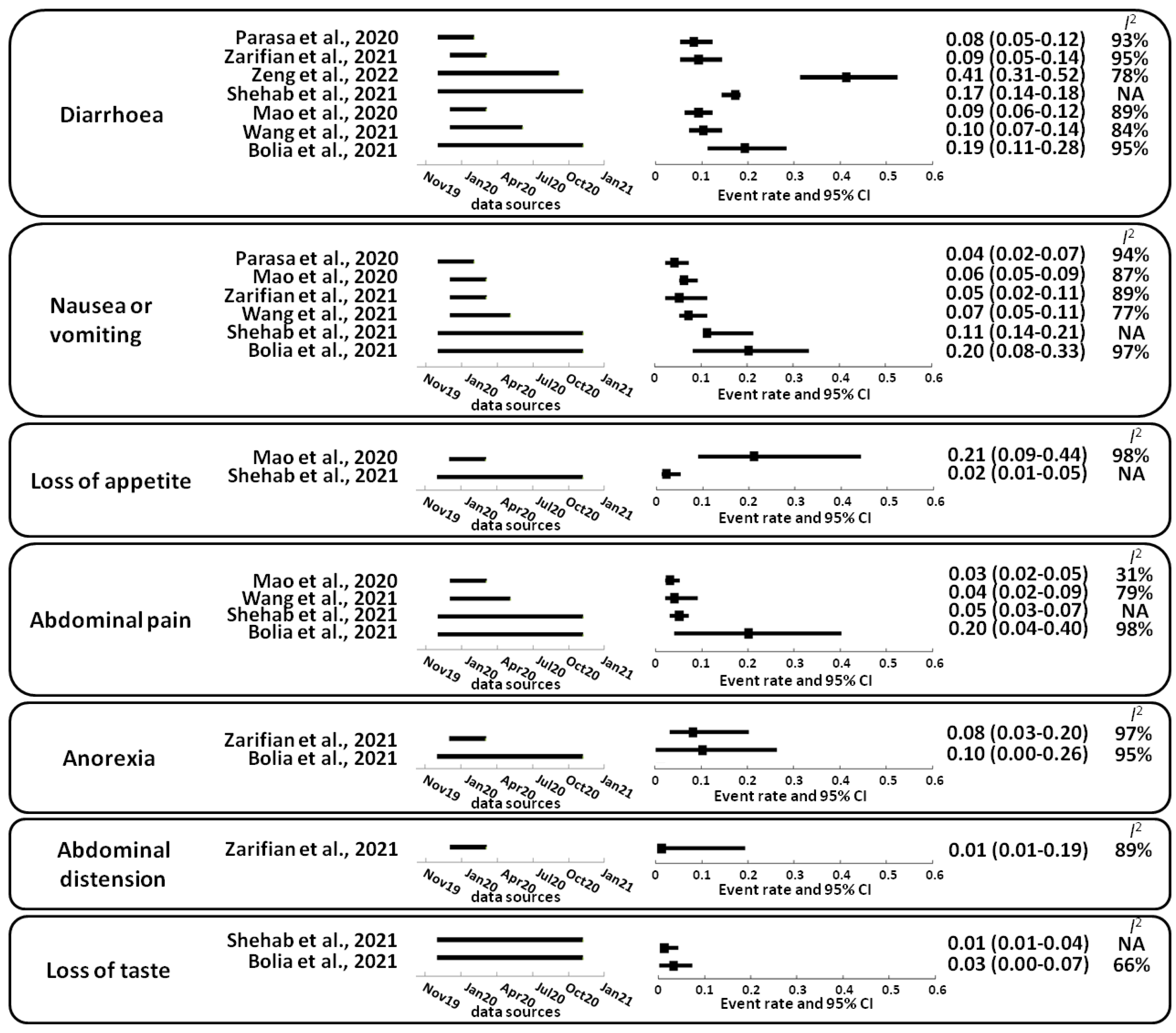
2. Gastrointestinal Complications Caused by SARS-CoV-2
3. SARS-CoV-2 Infection
3.1. Initial Steps of SARS-CoV-2 Infection
3.2. Angiotensin-Converting Enzyme 2 Expression and Genetic Variation
3.3. SARS-CoV-2 Infection in the Gastrointestinal Tract
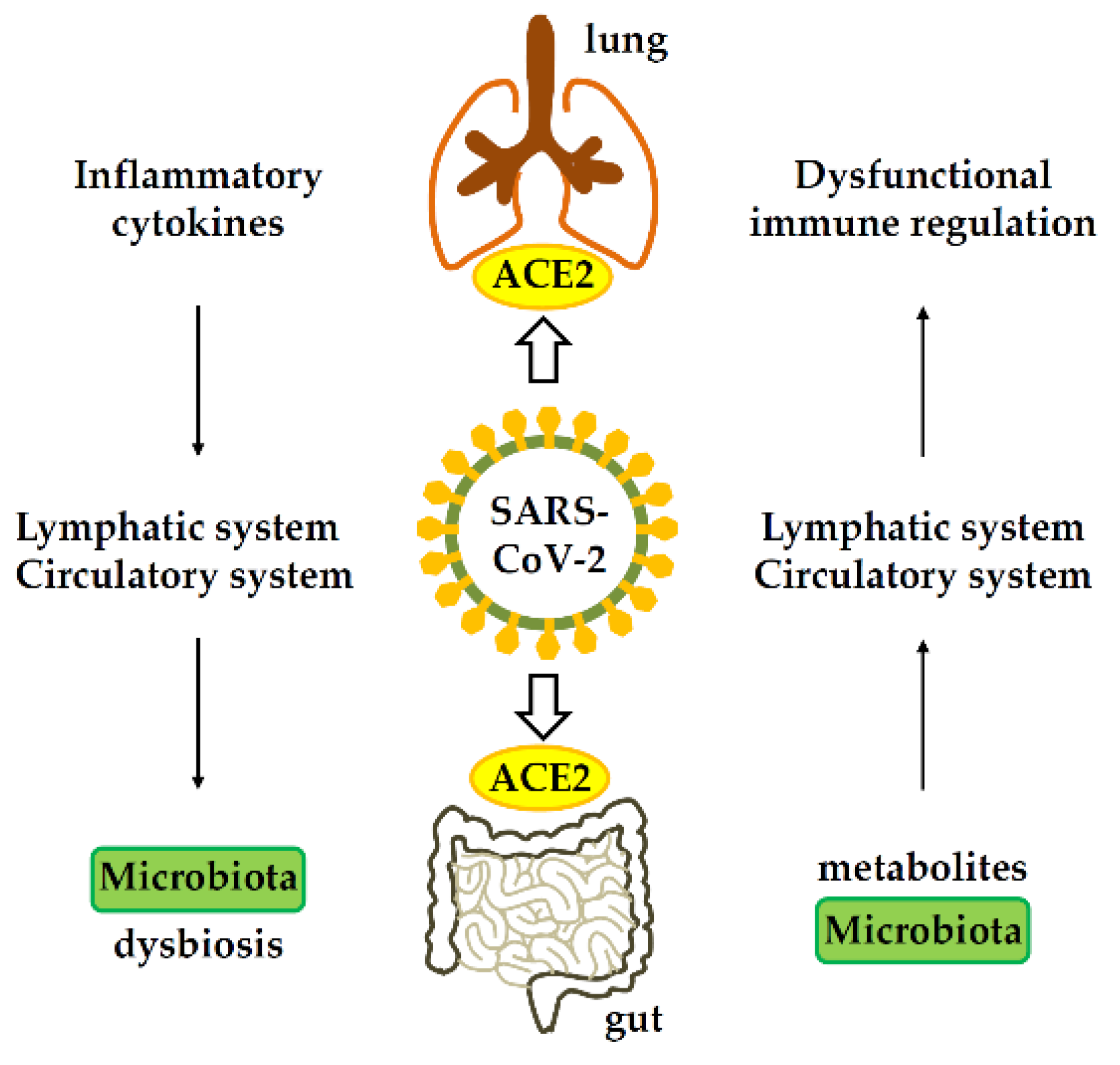
3.4. SARS-CoV-2 and Gut Microbiome
4. Host Immune Response Induced by SARS-CoV-2
4.1. Immune Response in the Gut Affects Gastrointestinal Tract
4.2. Immune Response in the Gastrointestinal Tract Affects Lung
4.3. The Mechanism Pathogenesis of COVID-19-Associated Gastrointestinal Manifestation
5. Immunization and Prevention via the Gastrointestinal
5.1. Oral Vaccine Candidates
5.2. Maintain Intestinal Microbiota Homeostasis
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Coronaviridae Study Group of the International Committee on Taxonomy of Viruses. The species Severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020, 5, 536–544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- WHO. Coronavirus Disease (COVID-19) Situation Reports. Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports (accessed on 16 February 2022).
- WHO. Statement on the Second Meeting of the International Health Regulations Emergency Committee Regarding the Outbreak of Novel Coronavirus (2019-nCoV). 2005. Available online: https://www.who.int/news-room/detail/30-01-2020-statement-on-the-second-meeting-of-the-international-health-regulations-(2005)-emergency-committee-regarding-the-outbreak-of-novel-coronavirus-(2019-ncov) (accessed on 16 February 2022).
- Hu, B.; Guo, H.; Zhou, P.; Shi, Z.L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2021, 19, 141–154. [Google Scholar] [CrossRef] [PubMed]
- National Center for Immunization and Respiratory Diseases (NCIRD), Division of Viral Diseases. SARS-CoV-2 Variant Classifications and Definitions. Last Updated 26 April 2022. Available online: https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-classifications.html (accessed on 25 May 2022).
- Gao, S.J.; Guo, H.; Luo, G. Omicron variant (B.1.1.529) of SARS-CoV-2, a global urgent public health alert! J. Med. Virol. 2022, 94, 1255–1256. [Google Scholar] [CrossRef] [PubMed]
- Parasa, S.; Desai, M.; Thoguluva Chandrasekar, V.; Patel, H.K.; Kennedy, K.F.; Roesch, T.; Spadaccini, M.; Colombo, M.; Gabbiadini, R.; Artifon, E.L.A.; et al. Prevalence of Gastrointestinal Symptoms and Fecal Viral Shedding in Patients With Coronavirus Disease 2019: A Systematic Review and Meta-analysis. JAMA Netw. Open 2020, 3, e2011335. [Google Scholar] [CrossRef]
- Holshue, M.L.; DeBolt, C.; Lindquist, S.; Lofy, K.H.; Wiesman, J.; Bruce, H.; Spitters, C.; Ericson, K.; Wilkerson, S.; Tural, A.; et al. First Case of 2019 Novel Coronavirus in the United States. N. Engl. J. Med. 2020, 382, 929–936. [Google Scholar] [CrossRef]
- Cheung, K.S.; Hung, I.F.N.; Chan, P.P.Y.; Lung, K.C.; Tso, E.; Liu, R.; Ng, Y.Y.; Chu, M.Y.; Chung, T.W.H.; Tam, A.R.; et al. Gastrointestinal Manifestations of SARS-CoV-2 Infection and Virus Load in Fecal Samples From a Hong Kong Cohort: Systematic Review and Meta-analysis. Gastroenterology 2020, 159, 81–95. [Google Scholar] [CrossRef]
- Ye, Q.; Wang, B.; Zhang, T.; Xu, J.; Shang, S. The mechanism and treatment of gastrointestinal symptoms in patients with COVID-19. Am. J. Physiol. Gastrointest. Liver Physiol. 2020, 319, G245–G252. [Google Scholar] [CrossRef]
- Marasco, G.; Maida, M.; Morreale, G.C.; Licata, M.; Renzulli, M.; Cremon, C.; Stanghellini, V.; Barbara, G. Gastrointestinal Bleeding in COVID-19 Patients: A Systematic Review with Meta-Analysis. Can. J. Gastroenterol. Hepatol. 2021, 2021, 2534975. [Google Scholar] [CrossRef]
- Shehab, M.; Alrashed, F.; Shuaibi, S.; Alajmi, D.; Barkun, A. Gastroenterological and hepatic manifestations of patients with COVID-19, prevalence, mortality by country, and intensive care admission rate: Systematic review and meta-analysis. BMJ Open Gastroenterol. 2021, 8, e000571. [Google Scholar] [CrossRef]
- Pan, L.; Mu, M.; Yang, P.; Sun, Y.; Wang, R.; Yan, J.; Li, P.; Hu, B.; Wang, J.; Hu, C.; et al. Clinical Characteristics of COVID-19 Patients With Digestive Symptoms in Hubei, China: A Descriptive, Cross-Sectional, Multicenter Study. Am. J. Gastroenterol. 2020, 115, 766–773. [Google Scholar] [CrossRef]
- Guan, W.J.; Ni, Z.Y.; Hu, Y.; Liang, W.H.; Ou, C.Q.; He, J.X.; Liu, L.; Shan, H.; Lei, C.L.; Hui, D.S.C.; et al. Clinical Characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 2020, 382, 1708–1720. [Google Scholar] [CrossRef] [PubMed]
- Redd, W.D.; Zhou, J.C.; Hathorn, K.E.; McCarty, T.R.; Bazarbashi, A.N.; Thompson, C.C.; Shen, L.; Chan, W.W. Prevalence and Characteristics of Gastrointestinal Symptoms in Patients With Severe Acute Respiratory Syndrome Coronavirus 2 Infection in the United States: A Multicenter Cohort Study. Gastroenterology 2020, 159, 765–767. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Jiang, X.; Zhang, Z.; Huang, S.; Zhang, Z.; Fang, Z.; Gu, Z.; Gao, L.; Shi, H.; Mai, L.; et al. Gastrointestinal symptoms of 95 cases with SARS-CoV-2 infection. Gut 2020, 69, 997–1001. [Google Scholar] [CrossRef] [PubMed]
- Remes-Troche, J.M.; Ramos-de-la-Medina, A.; Manriquez-Reyes, M.; Martinez-Perez-Maldonado, L.; Lara, E.L.; Solis-Gonzalez, M.A. Initial Gastrointestinal Manifestations in Patients With Severe Acute Respiratory Syndrome Coronavirus 2 Infection in 112 Patients From Veracruz in Southeastern Mexico. Gastroenterology 2020, 159, 1179–1181. [Google Scholar] [CrossRef] [PubMed]
- Boraschi, P.; Giugliano, L.; Mercogliano, G.; Donati, F.; Romano, S.; Neri, E. Abdominal and gastrointestinal manifestations in COVID-19 patients: Is imaging useful? World J. Gastroenterol. 2021, 27, 4143–4159. [Google Scholar] [CrossRef] [PubMed]
- Zarifian, A.; Zamiri Bidary, M.; Arekhi, S.; Rafiee, M.; Gholamalizadeh, H.; Amiriani, A.; Ghaderi, M.S.; Khadem-Rezaiyan, M.; Amini, M.; Ganji, A. Gastrointestinal and hepatic abnormalities in patients with confirmed COVID-19: A systematic review and meta-analysis. J. Med. Virol. 2021, 93, 336–350. [Google Scholar] [CrossRef]
- Zeng, W.; Qi, K.; Ye, M.; Zheng, L.; Liu, X.; Hu, S.; Zhang, W.; Tang, W.; Xu, J.; Yu, D.; et al. Gastrointestinal symptoms are associated with severity of coronavirus disease 2019: A systematic review and meta-analysis. Eur. J. Gastroenterol. Hepatol. 2022, 34, 168–176. [Google Scholar] [CrossRef]
- Mao, R.; Qiu, Y.; He, J.S.; Tan, J.Y.; Li, X.H.; Liang, J.; Shen, J.; Zhu, L.R.; Chen, Y.; Iacucci, M.; et al. Manifestations and prognosis of gastrointestinal and liver involvement in patients with COVID-19: A systematic review and meta-analysis. Lancet Gastroenterol. Hepatol. 2020, 5, 667–678. [Google Scholar] [CrossRef]
- Wang, J.; Yuan, X. Digestive system symptoms and function in children with COVID-19: A meta-analysis. Medicine 2021, 100, e24897. [Google Scholar] [CrossRef]
- Bolia, R.; Dhanesh Goel, A.; Badkur, M.; Jain, V. Gastrointestinal Manifestations of Pediatric Coronavirus Disease and Their Relationship with a Severe Clinical Course: A Systematic Review and Meta-analysis. J. Trop. Pediatr. 2021, 67, fmab051. [Google Scholar] [CrossRef]
- Das, S.; Jayaratne, R.; Barrett, K.E. The Role of Ion Transporters in the Pathophysiology of Infectious Diarrhea. Cell Mol. Gastroenterol. Hepatol. 2018, 6, 33–45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sueyoshi, R.; Ignatoski, K.M.; Daignault, S.; Okawada, M.; Teitelbaum, D.H. Angiotensin converting enzyme-inhibitor reduces colitis severity in an IL-10 knockout model. Dig. Dis. Sci. 2013, 58, 3165–3177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kariyawasam, J.C.; Jayarajah, U.; Riza, R.; Abeysuriya, V.; Seneviratne, S.L. Gastrointestinal manifestations in COVID-19. Trans. R. Soc. Trop. Med. Hyg. 2021, 115, 1362–1388. [Google Scholar] [CrossRef] [PubMed]
- Luo, S.; Zhang, X.; Xu, H. Don’t Overlook Digestive Symptoms in Patients With 2019 Novel Coronavirus Disease (COVID-19). Clin. Gastroenterol. Hepatol. 2020, 18, 1636–1637. [Google Scholar] [CrossRef]
- Jin, X.; Lian, J.S.; Hu, J.H.; Gao, J.; Zheng, L.; Zhang, Y.M.; Hao, S.R.; Jia, H.Y.; Cai, H.; Zhang, X.L.; et al. Epidemiological, clinical and virological characteristics of 74 cases of coronavirus-infected disease 2019 (COVID-19) with gastrointestinal symptoms. Gut 2020, 69, 1002–1009. [Google Scholar] [CrossRef] [Green Version]
- Lee, I.C.; Huo, T.I.; Huang, Y.H. Gastrointestinal and liver manifestations in patients with COVID-19. J. Chin. Med. Assoc. 2020, 83, 521–523. [Google Scholar] [CrossRef]
- Wang, H.; Qiu, P.; Liu, J.; Wang, F.; Zhao, Q. The liver injury and gastrointestinal symptoms in patients with Coronavirus Disease 19: A systematic review and meta-analysis. Clin. Res. Hepatol. Gastroenterol. 2020, 44, 653–661. [Google Scholar] [CrossRef]
- Tian, Y.; Rong, L.; Nian, W.; He, Y. Review article: Gastrointestinal features in COVID-19 and the possibility of faecal transmission. Aliment. Pharmacol. Ther. 2020, 51, 843–851. [Google Scholar] [CrossRef]
- El Moheb, M.; Naar, L.; Christensen, M.A.; Kapoen, C.; Maurer, L.R.; Farhat, M.; Kaafarani, H.M.A. Gastrointestinal Complications in Critically Ill Patients With and Without COVID-19. JAMA 2020, 324, 1899–1901. [Google Scholar] [CrossRef]
- Nobel, Y.R.; Phipps, M.; Zucker, J.; Lebwohl, B.; Wang, T.C.; Sobieszczyk, M.E.; Freedberg, D.E. Gastrointestinal Symptoms and Coronavirus Disease 2019: A Case-Control Study From the United States. Gastroenterology 2020, 159, 373–375. [Google Scholar] [CrossRef]
- Aghemo, A.; Piovani, D.; Parigi, T.L.; Brunetta, E.; Pugliese, N.; Vespa, E.; Omodei, P.D.; Preatoni, P.; Lleo, A.; Repici, A.; et al. COVID-19 Digestive System Involvement and Clinical Outcomes in a Large Academic Hospital in Milan, Italy. Clin. Gastroenterol. Hepatol. 2020, 18, 2366–2368. [Google Scholar] [CrossRef] [PubMed]
- Livanos, A.E.; Jha, D.; Cossarini, F.; Gonzalez-Reiche, A.S.; Tokuyama, M.; Aydillo, T.; Parigi, T.L.; Ladinsky, M.S.; Ramos, I.; Dunleavy, K.; et al. Intestinal Host Response to SARS-CoV-2 Infection and COVID-19 Outcomes in Patients with Gastrointestinal Symptoms. Gastroenterology 2021, 160, 2435–2450. [Google Scholar] [CrossRef] [PubMed]
- Klok, F.A.; Kruip, M.; van der Meer, N.J.M.; Arbous, M.S.; Gommers, D.; Kant, K.M.; Kaptein, F.H.J.; van Paassen, J.; Stals, M.A.M.; Huisman, M.V.; et al. Incidence of thrombotic complications in critically ill ICU patients with COVID-19. Thromb. Res. 2020, 191, 145–147. [Google Scholar] [CrossRef]
- Connors, J.M.; Levy, J.H. COVID-19 and its implications for thrombosis and anticoagulation. Blood 2020, 135, 2033–2040. [Google Scholar] [CrossRef] [PubMed]
- Wichmann, D.; Sperhake, J.P.; Lutgehetmann, M.; Steurer, S.; Edler, C.; Heinemann, A.; Heinrich, F.; Mushumba, H.; Kniep, I.; Schroder, A.S.; et al. Autopsy Findings and Venous Thromboembolism in Patients With COVID-19: A Prospective Cohort Study. Ann. Intern. Med. 2020, 173, 268–277. [Google Scholar] [CrossRef]
- Salehi, S.; Gholamrezanezhad, A. Reply to “Segmental Pulmonary Vascular Changes in COVID-19 Pneumonia”. AJR Am. J. Roentgenol. 2020, 215, W34. [Google Scholar] [CrossRef]
- Salehi, S.; Abedi, A.; Gholamrezanezhad, A. Reply to “Vascular Changes Detected With Thoracic CT in Coronavirus Disease (COVID-19) Might Be Significant Determinants for Accurate Diagnosis and Optimal Patient Management”. AJR Am. J. Roentgenol. 2020, 215, W16. [Google Scholar] [CrossRef]
- Eslambolchi, A.; Aghaghazvini, L.; Gholamrezanezhad, A.; Kavosi, H.; Radmard, A.R. Coronavirus disease 2019 (COVID-19) in patients with systemic autoimmune diseases or vasculitis: Radiologic presentation. J. Thromb. Thrombolysis 2021, 51, 339–348. [Google Scholar] [CrossRef]
- Kooraki, S.; Hosseiny, M.; Myers, L.; Gholamrezanezhad, A. Coronavirus (COVID-19) Outbreak: What the Department of Radiology Should Know. J. Am. Coll. Radiol. 2020, 17, 447–451. [Google Scholar] [CrossRef]
- Keshavarz, P.; Rafiee, F.; Kavandi, H.; Goudarzi, S.; Heidari, F.; Gholamrezanezhad, A. Ischemic gastrointestinal complications of COVID-19: A systematic review on imaging presentation. Clin. Imaging 2021, 73, 86–95. [Google Scholar] [CrossRef]
- Subissi, L.; Posthuma, C.C.; Collet, A.; Zevenhoven-Dobbe, J.C.; Gorbalenya, A.E.; Decroly, E.; Snijder, E.J.; Canard, B.; Imbert, I. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc. Natl. Acad. Sci. USA 2014, 111, E3900–E3909. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rota, P.A.; Oberste, M.S.; Monroe, S.S.; Nix, W.A.; Campagnoli, R.; Icenogle, J.P.; Penaranda, S.; Bankamp, B.; Maher, K.; Chen, M.H.; et al. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 2003, 300, 1394–1399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- V’Kovski, P.; Kratzel, A.; Steiner, S.; Stalder, H.; Thiel, V. Coronavirus biology and replication: Implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021, 19, 155–170. [Google Scholar] [CrossRef] [PubMed]
- Brian, D.A.; Baric, R.S. Coronavirus genome structure and replication. Curr. Top Microbiol. Immunol. 2005, 287, 369. [Google Scholar]
- Henry, B.M.; de Oliveira, M.H.S.; Benoit, S.; Plebani, M.; Lippi, G. Hematologic, biochemical and immune biomarker abnormalities associated with severe illness and mortality in coronavirus disease 2019 (COVID-19): A meta-analysis. Clin. Chem. Lab. Med. 2020, 58, 1021–1028. [Google Scholar] [CrossRef] [Green Version]
- Choudhry, N.; Zhao, X.; Xu, D.; Zanin, M.; Chen, W.; Yang, Z.; Chen, J. Chinese Therapeutic Strategy for Fighting COVID-19 and Potential Small-Molecule Inhibitors against Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). J. Med. Chem. 2020, 63, 13205–13227. [Google Scholar] [CrossRef]
- Jackson, C.B.; Farzan, M.; Chen, B.; Choe, H. Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Biol. 2022, 23, 3–20. [Google Scholar] [CrossRef]
- Yan, R.; Zhang, Y.; Li, Y.; Xia, L.; Guo, Y.; Zhou, Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 2020, 367, 1444–1448. [Google Scholar] [CrossRef] [Green Version]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef] [Green Version]
- Parmar, M.S. TMPRSS2: An Equally Important Protease as ACE2 in the Pathogenicity of SARS-CoV-2 Infection. Mayo Clin. Proc. 2021, 96, 2748–2752. [Google Scholar] [CrossRef]
- Wu, L.; Zhou, L.; Mo, M.; Liu, T.; Wu, C.; Gong, C.; Lu, K.; Gong, L.; Zhu, W.; Xu, Z. SARS-CoV-2 Omicron RBD shows weaker binding affinity than the currently dominant Delta variant to human ACE2. Signal Transduct. Target. Ther. 2022, 7, 8. [Google Scholar] [CrossRef] [PubMed]
- Zou, X.; Chen, K.; Zou, J.; Han, P.; Hao, J.; Han, Z. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front. Med. 2020, 14, 185–192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.; Zhao, Z.; Wang, Y.; Zhou, Y.; Ma, Y.; Zuo, W. Single-Cell RNA Expression Profiling of ACE2, the Receptor of SARS-CoV-2. Am. J. Respir. Crit. Care Med. 2020, 202, 756–759. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Zhong, L.; Deng, J.; Peng, J.; Dan, H.; Zeng, X.; Li, T.; Chen, Q. High expression of ACE2 receptor of 2019-nCoV on the epithelial cells of oral mucosa. Int. J. Oral Sci. 2020, 12, 8. [Google Scholar] [CrossRef]
- Liang, W.; Feng, Z.; Rao, S.; Xiao, C.; Xue, X.; Lin, Z.; Zhang, Q.; Qi, W. Diarrhoea may be underestimated: A missing link in 2019 novel coronavirus. Gut 2020, 69, 1141–1143. [Google Scholar] [CrossRef] [Green Version]
- He, J.; Tao, H.; Yan, Y.; Huang, S.Y.; Xiao, Y. Molecular Mechanism of Evolution and Human Infection with SARS-CoV-2. Viruses 2020, 12, 428. [Google Scholar] [CrossRef] [Green Version]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Kruger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280. [Google Scholar] [CrossRef]
- Lamers, M.M.; Beumer, J.; van der Vaart, J.; Knoops, K.; Puschhof, J.; Breugem, T.I.; Ravelli, R.B.G.; Paul van Schayck, J.; Mykytyn, A.Z.; Duimel, H.Q.; et al. SARS-CoV-2 productively infects human gut enterocytes. Science 2020, 369, 50–54. [Google Scholar] [CrossRef]
- Sungnak, W.; Huang, N.; Becavin, C.; Berg, M.; Queen, R.; Litvinukova, M.; Talavera-Lopez, C.; Maatz, H.; Reichart, D.; Sampaziotis, F.; et al. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 2020, 26, 681–687. [Google Scholar] [CrossRef] [Green Version]
- Berni Canani, R.; Comegna, M.; Paparo, L.; Cernera, G.; Bruno, C.; Strisciuglio, C.; Zollo, I.; Gravina, A.G.; Miele, E.; Cantone, E.; et al. Age-Related Differences in the Expression of Most Relevant Mediators of SARS-CoV-2 Infection in Human Respiratory and Gastrointestinal Tract. Front. Pediatr. 2021, 9, 697390. [Google Scholar] [CrossRef]
- Gu, J.; Han, B.; Wang, J. COVID-19: Gastrointestinal Manifestations and Potential Fecal-Oral Transmission. Gastroenterology 2020, 158, 1518–1519. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Moore, M.J.; Vasilieva, N.; Sui, J.; Wong, S.K.; Berne, M.A.; Somasundaran, M.; Sullivan, J.L.; Luzuriaga, K.; Greenough, T.C.; et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 2003, 426, 450–454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hashimoto, T.; Perlot, T.; Rehman, A.; Trichereau, J.; Ishiguro, H.; Paolino, M.; Sigl, V.; Hanada, T.; Hanada, R.; Lipinski, S.; et al. ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature 2012, 487, 477–481. [Google Scholar] [CrossRef] [PubMed]
- Kuba, K.; Imai, Y.; Rao, S.; Gao, H.; Guo, F.; Guan, B.; Huan, Y.; Yang, P.; Zhang, Y.; Deng, W.; et al. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat. Med. 2005, 11, 875–879. [Google Scholar] [CrossRef]
- Cao, Y.; Li, L.; Feng, Z.; Wan, S.; Huang, P.; Sun, X.; Wen, F.; Huang, X.; Ning, G.; Wang, W. Comparative genetic analysis of the novel coronavirus (2019-nCoV/SARS-CoV-2) receptor ACE2 in different populations. Cell Discov. 2020, 6, 11. [Google Scholar] [CrossRef] [Green Version]
- Irham, L.M.; Chou, W.H.; Calkins, M.J.; Adikusuma, W.; Hsieh, S.L.; Chang, W.C. Genetic variants that influence SARS-CoV-2 receptor TMPRSS2 expression among population cohorts from multiple continents. Biochem. Biophys. Res. Commun. 2020, 529, 263–269. [Google Scholar] [CrossRef]
- Asselta, R.; Paraboschi, E.M.; Mantovani, A.; Duga, S. ACE2 and TMPRSS2 variants and expression as candidates to sex and country differences in COVID-19 severity in Italy. Aging 2020, 12, 10087–10098. [Google Scholar] [CrossRef]
- Spencer, L.; Olawuni, B.; Singh, P. Gut Virome: Role and Distribution in Health and Gastrointestinal Diseases. Front. Cell Infect. Microbiol. 2022, 12, 836706. [Google Scholar] [CrossRef]
- Yeo, C.; Kaushal, S.; Yeo, D. Enteric involvement of coronaviruses: Is faecal-oral transmission of SARS-CoV-2 possible? Lancet Gastroenterol. Hepatol. 2020, 5, 335–337. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Xu, Y.; Gao, R.; Lu, R.; Han, K.; Wu, G.; Tan, W. Detection of SARS-CoV-2 in Different Types of Clinical Specimens. JAMA 2020, 323, 1843–1844. [Google Scholar] [CrossRef] [Green Version]
- Xiao, F.; Tang, M.; Zheng, X.; Liu, Y.; Li, X.; Shan, H. Evidence for Gastrointestinal Infection of SARS-CoV-2. Gastroenterology 2020, 158, 1831–1833. [Google Scholar] [CrossRef] [PubMed]
- Li, L.Y.; Wu, W.; Chen, S.; Gu, J.W.; Li, X.L.; Song, H.J.; Du, F.; Wang, G.; Zhong, C.Q.; Wang, X.Y.; et al. Digestive system involvement of novel coronavirus infection: Prevention and control infection from a gastroenterology perspective. J. Dig. Dis. 2020, 21, 199–204. [Google Scholar] [CrossRef] [PubMed]
- Jiao, L.; Li, H.; Xu, J.; Yang, M.; Ma, C.; Li, J.; Zhao, S.; Wang, H.; Yang, Y.; Yu, W.; et al. The Gastrointestinal Tract Is an Alternative Route for SARS-CoV-2 Infection in a Nonhuman Primate Model. Gastroenterology 2021, 160, 1647–1661. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Guo, C.; Tang, L.; Hong, Z.; Zhou, J.; Dong, X.; Yin, H.; Xiao, Q.; Tang, Y.; Qu, X.; et al. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol. Hepatol. 2020, 5, 434–435. [Google Scholar] [CrossRef]
- Dieterich, W.; Schink, M.; Zopf, Y. Microbiota in the Gastrointestinal Tract. Med. Sci. 2018, 6, 116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gilbert, J.A.; Lynch, S.V. Community ecology as a framework for human microbiome research. Nat. Med. 2019, 25, 884–889. [Google Scholar] [CrossRef]
- Human Microbiome Project, C. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar] [CrossRef] [Green Version]
- Golonka, R.M.; Saha, P.; Yeoh, B.S.; Chattopadhyay, S.; Gewirtz, A.T.; Joe, B.; Vijay-Kumar, M. Harnessing innate immunity to eliminate SARS-CoV-2 and ameliorate COVID-19 disease. Physiol. Genom. 2020, 52, 217–221. [Google Scholar] [CrossRef] [Green Version]
- Xiao, F.; Sun, J.; Xu, Y.; Li, F.; Huang, X.; Li, H.; Zhao, J.; Huang, J.; Zhao, J. Infectious SARS-CoV-2 in Feces of Patient with Severe COVID-19. Emerg. Infect. Dis. 2020, 26, 1920–1922. [Google Scholar] [CrossRef]
- Mazza, S.; Sorce, A.; Peyvandi, F.; Vecchi, M.; Caprioli, F. A fatal case of COVID-19 pneumonia occurring in a patient with severe acute ulcerative colitis. Gut 2020, 69, 1148–1149. [Google Scholar] [CrossRef]
- Farsi, Y.; Tahvildari, A.; Arbabi, M.; Vazife, F.; Sechi, L.A.; Shahidi Bonjar, A.H.; Jamshidi, P.; Nasiri, M.J.; Mirsaeidi, M. Diagnostic, Prognostic, and Therapeutic Roles of Gut Microbiota in COVID-19: A Comprehensive Systematic Review. Front. Cell Infect. Microbiol. 2022, 12, 804644. [Google Scholar] [CrossRef] [PubMed]
- Budden, K.F.; Gellatly, S.L.; Wood, D.L.; Cooper, M.A.; Morrison, M.; Hugenholtz, P.; Hansbro, P.M. Emerging pathogenic links between microbiota and the gut-lung axis. Nat. Rev. Microbiol. 2017, 15, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Reinold, J.; Farahpour, F.; Schoerding, A.K.; Fehring, C.; Dolff, S.; Konik, M.; Korth, J.; van Baal, L.; Buer, J.; Witzke, O.; et al. The Fungal Gut Microbiome Exhibits Reduced Diversity and Increased Relative Abundance of Ascomycota in Severe COVID-19 Illness and Distinct Interconnected Communities in SARS-CoV-2 Positive Patients. Front. Cell Infect. Microbiol. 2022, 12, 848650. [Google Scholar] [CrossRef] [PubMed]
- Hazan, S.; Stollman, N.; Bozkurt, H.S.; Dave, S.; Papoutsis, A.J.; Daniels, J.; Barrows, B.D.; Quigley, E.M.; Borody, T.J. Lost microbes of COVID-19: Bifidobacterium, Faecalibacterium depletion and decreased microbiome diversity associated with SARS-CoV-2 infection severity. BMJ Open Gastroenterol. 2022, 9, e000871. [Google Scholar] [CrossRef]
- Sankova, M.V.; Nikolenko, V.N.; Oganesyan, M.V.; Bakhmet, A.A.; Gavryushova, L.V.; Sankov, S.V.; Sinelnikov, M.Y. Current drug targets for gut microbiota biocorrection during the SARS-CoV-2 pandemic: A systematic review. Curr. Drug Targets 2022. [Google Scholar] [CrossRef]
- Cheung, C.Y.; Poon, L.L.; Ng, I.H.; Luk, W.; Sia, S.F.; Wu, M.H.; Chan, K.H.; Yuen, K.Y.; Gordon, S.; Guan, Y.; et al. Cytokine responses in severe acute respiratory syndrome coronavirus-infected macrophages in vitro: Possible relevance to pathogenesis. J. Virol. 2005, 79, 7819–7826. [Google Scholar] [CrossRef] [Green Version]
- Lau, S.K.P.; Lau, C.C.Y.; Chan, K.H.; Li, C.P.Y.; Chen, H.; Jin, D.Y.; Chan, J.F.W.; Woo, P.C.Y.; Yuen, K.Y. Delayed induction of proinflammatory cytokines and suppression of innate antiviral response by the novel Middle East respiratory syndrome coronavirus: Implications for pathogenesis and treatment. J. Gen. Virol. 2013, 94, 2679–2690. [Google Scholar] [CrossRef]
- Law, H.K.; Cheung, C.Y.; Ng, H.Y.; Sia, S.F.; Chan, Y.O.; Luk, W.; Nicholls, J.M.; Peiris, J.S.; Lau, Y.L. Chemokine up-regulation in SARS-coronavirus-infected, monocyte-derived human dendritic cells. Blood 2005, 106, 2366–2374. [Google Scholar] [CrossRef] [Green Version]
- Mathew, D.; Giles, J.R.; Baxter, A.E.; Oldridge, D.A.; Greenplate, A.R.; Wu, J.E.; Alanio, C.; Kuri-Cervantes, L.; Pampena, M.B.; D’Andrea, K.; et al. Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science 2020, 369, eabc8511. [Google Scholar] [CrossRef]
- Wilk, A.J.; Rustagi, A.; Zhao, N.Q.; Roque, J.; Martinez-Colon, G.J.; McKechnie, J.L.; Ivison, G.T.; Ranganath, T.; Vergara, R.; Hollis, T.; et al. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat. Med. 2020, 26, 1070–1076. [Google Scholar] [CrossRef]
- He, L.; Ding, Y.; Zhang, Q.; Che, X.; He, Y.; Shen, H.; Wang, H.; Li, Z.; Zhao, L.; Geng, J.; et al. Expression of elevated levels of pro-inflammatory cytokines in SARS-CoV-infected ACE2+ cells in SARS patients: Relation to the acute lung injury and pathogenesis of SARS. J. Pathol. 2006, 210, 288–297. [Google Scholar] [CrossRef] [PubMed]
- Moore, J.B.; June, C.H. Cytokine release syndrome in severe COVID-19. Science 2020, 368, 473–474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Group, R.C.; Horby, P.; Lim, W.S.; Emberson, J.R.; Mafham, M.; Bell, J.L.; Linsell, L.; Staplin, N.; Brightling, C.; Ustianowski, A.; et al. Dexamethasone in Hospitalized Patients with Covid-19. N. Engl. J. Med. 2021, 384, 693–704. [Google Scholar]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Z.; Cai, T.; Fan, L.; Lou, K.; Hua, X.; Huang, Z.; Gao, G. Clinical value of immune-inflammatory parameters to assess the severity of coronavirus disease 2019. Int. J. Infect. Dis. 2020, 95, 332–339. [Google Scholar] [CrossRef]
- Del Valle, D.M.; Kim-Schulze, S.; Huang, H.H.; Beckmann, N.D.; Nirenberg, S.; Wang, B.; Lavin, Y.; Swartz, T.H.; Madduri, D.; Stock, A.; et al. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat. Med. 2020, 26, 1636–1643. [Google Scholar] [CrossRef]
- Wan, Y.; Shang, J.; Graham, R.; Baric, R.S.; Li, F. Receptor Recognition by the Novel Coronavirus from Wuhan: An Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. J. Virol. 2020, 94, e00127-20. [Google Scholar] [CrossRef] [Green Version]
- Jeyanathan, M.; Afkhami, S.; Smaill, F.; Miller, M.S.; Lichty, B.D.; Xing, Z. Immunological considerations for COVID-19 vaccine strategies. Nat. Rev. Immunol. 2020, 20, 615–632. [Google Scholar] [CrossRef]
- Sterlin, D.; Mathian, A.; Miyara, M.; Mohr, A.; Anna, F.; Claer, L.; Quentric, P.; Fadlallah, J.; Devilliers, H.; Ghillani, P.; et al. IgA dominates the early neutralizing antibody response to SARS-CoV-2. Sci. Transl. Med. 2021, 13, eabd2223. [Google Scholar] [CrossRef]
- Wang, J.; Li, F.; Wei, H.; Lian, Z.X.; Sun, R.; Tian, Z. Respiratory influenza virus infection induces intestinal immune injury via microbiota-mediated Th17 cell-dependent inflammation. J. Exp. Med. 2014, 211, 2397–2410. [Google Scholar] [CrossRef] [Green Version]
- Papadakis, K.A.; Prehn, J.; Nelson, V.; Cheng, L.; Binder, S.W.; Ponath, P.D.; Andrew, D.P.; Targan, S.R. The role of thymus-expressed chemokine and its receptor CCR9 on lymphocytes in the regional specialization of the mucosal immune system. J. Immunol. 2000, 165, 5069–5076. [Google Scholar] [CrossRef] [PubMed]
- Stenstad, H.; Ericsson, A.; Johansson-Lindbom, B.; Svensson, M.; Marsal, J.; Mack, M.; Picarella, D.; Soler, D.; Marquez, G.; Briskin, M.; et al. Gut-associated lymphoid tissue-primed CD4+ T cells display CCR9-dependent and -independent homing to the small intestine. Blood 2006, 107, 3447–3454. [Google Scholar] [CrossRef] [PubMed]
- Varga, Z.; Flammer, A.J.; Steiger, P.; Haberecker, M.; Andermatt, R.; Zinkernagel, A.S.; Mehra, M.R.; Schuepbach, R.A.; Ruschitzka, F.; Moch, H. Endothelial cell infection and endotheliitis in COVID-19. Lancet 2020, 395, 1417–1418. [Google Scholar] [CrossRef]
- Zhang, D.; Li, S.; Wang, N.; Tan, H.Y.; Zhang, Z.; Feng, Y. The Cross-Talk Between Gut Microbiota and Lungs in Common Lung Diseases. Front. Microbiol. 2020, 11, 301. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.F.; Kok, K.H.; Zhu, Z.; Chu, H.; To, K.K.; Yuan, S.; Yuen, K.Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes. Infect. 2020, 9, 221–236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Megyeri, K.; Dernovics, A.; Al-Luhaibi, Z.I.I.; Rosztoczy, A. COVID-19-associated diarrhea. World J. Gastroenterol. 2021, 27, 3208–3222. [Google Scholar] [CrossRef]
- Delorme-Axford, E.; Klionsky, D.J. Highlights in the fight against COVID-19: Does autophagy play a role in SARS-CoV-2 infection? Autophagy 2020, 16, 2123–2127. [Google Scholar] [CrossRef]
- Xiao, L.; Sakagami, H.; Miwa, N. ACE2: The key Molecule for Understanding the Pathophysiology of Severe and Critical Conditions of COVID-19: Demon or Angel? Viruses 2020, 12, 491. [Google Scholar] [CrossRef]
- Zhang, T.; Liu, D.; Tian, D.; Xia, L. The roles of nausea and vomiting in COVID-19: Did we miss something? J. Microbiol. Immunol. Infect 2021, 54, 541–546. [Google Scholar] [CrossRef]
- Perisetti, A.; Goyal, H.; Gajendran, M.; Boregowda, U.; Mann, R.; Sharma, N. Prevalence, Mechanisms, and Implications of Gastrointestinal Symptoms in COVID-19. Front. Med. 2020, 7, 588711. [Google Scholar] [CrossRef]
- Brann, D.H.; Tsukahara, T.; Weinreb, C.; Lipovsek, M.; Van den Berge, K.; Gong, B.; Chance, R.; Macaulay, I.C.; Chou, H.J.; Fletcher, R.B.; et al. Non-neuronal expression of SARS-CoV-2 entry genes in the olfactory system suggests mechanisms underlying COVID-19-associated anosmia. Sci. Adv. 2020, 6, eabc5801. [Google Scholar] [CrossRef] [PubMed]
- Aziz, M.; Perisetti, A.; Lee-Smith, W.M.; Gajendran, M.; Bansal, P.; Goyal, H. Taste Changes (Dysgeusia) in COVID-19: A Systematic Review and Meta-analysis. Gastroenterology 2020, 159, 1132–1133. [Google Scholar] [CrossRef] [PubMed]
- Cox, R.J.; Brokstad, K.A. Not just antibodies: B cells and T cells mediate immunity to COVID-19. Nat. Rev. Immunol. 2020, 20, 581–582. [Google Scholar] [CrossRef] [PubMed]
- Grifoni, A.; Weiskopf, D.; Ramirez, S.I.; Mateus, J.; Dan, J.M.; Moderbacher, C.R.; Rawlings, S.A.; Sutherland, A.; Premkumar, L.; Jadi, R.S.; et al. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell 2020, 181, 1489–1501. [Google Scholar] [CrossRef]
- Sekine, T.; Perez-Potti, A.; Rivera-Ballesteros, O.; Stralin, K.; Gorin, J.B.; Olsson, A.; Llewellyn-Lacey, S.; Kamal, H.; Bogdanovic, G.; Muschiol, S.; et al. Robust T Cell Immunity in Convalescent Individuals with Asymptomatic or Mild COVID-19. Cell 2020, 183, 158–168. [Google Scholar] [CrossRef]
- Ashraf, M.U.; Kim, Y.; Kumar, S.; Seo, D.; Ashraf, M.; Bae, Y.S. COVID-19 Vaccines (Revisited) and Oral-Mucosal Vector System as a Potential Vaccine Platform. Vaccines 2021, 9, 171. [Google Scholar] [CrossRef]
- Zhang, H.; Kang, Z.; Gong, H.; Xu, D.; Wang, J.; Li, Z.; Li, Z.; Cui, X.; Xiao, J.; Zhan, J.; et al. Digestive system is a potential route of COVID-19: An analysis of single-cell coexpression pattern of key proteins in viral entry process. Gut 2020, 69, 1010–1018. [Google Scholar] [CrossRef]
- Hou, Y.J.; Okuda, K.; Edwards, C.E.; Martinez, D.R.; Asakura, T.; Dinnon, K.H., 3rd; Kato, T.; Lee, R.E.; Yount, B.L.; Mascenik, T.M.; et al. SARS-CoV-2 Reverse Genetics Reveals a Variable Infection Gradient in the Respiratory Tract. Cell 2020, 182, 429–446. [Google Scholar] [CrossRef]
- Sims, A.C.; Baric, R.S.; Yount, B.; Burkett, S.E.; Collins, P.L.; Pickles, R.J. Severe acute respiratory syndrome coronavirus infection of human ciliated airway epithelia: Role of ciliated cells in viral spread in the conducting airways of the lungs. J. Virol 2005, 79, 15511–15524. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.; Zhong, Y.; Chen, J.; Liu, Y.; Tang, C.; Wang, X.; Zhang, Y.; Wang, P.; Logan, S.M.; Chen, W.; et al. Oral immunization of mice with a multivalent therapeutic subunit vaccine protects against Helicobacter pylori infection. Vaccine 2020, 38, 3031–3041. [Google Scholar] [CrossRef]
- Wang, S.; Geng, N.; Zhou, D.; Qu, Y.; Shi, M.; Xu, Y.; Liu, K.; Liu, Y.; Liu, J. Oral Immunization of Chickens With Recombinant Lactobacillus plantarum Vaccine Against Early ALV-J Infection. Front. Immunol. 2019, 10, 2299. [Google Scholar] [CrossRef] [PubMed]
- Mustafa, A.D.; Kalyanasundram, J.; Sabidi, S.; Song, A.A.; Abdullah, M.; Abdul Rahim, R.; Yusoff, K. Proof of concept in utilizing in-trans surface display system of Lactobacillus plantarum as mucosal tuberculosis vaccine via oral administration in mice. BMC Biotechnol. 2018, 18, 63. [Google Scholar] [CrossRef]
- Baker, P.J. Advantages of an Oral Vaccine to Control the COVID-19 Pandemic. Am. J. Med. 2022, 135, 133–134. [Google Scholar] [CrossRef] [PubMed]
- Shakoor, S.; Rao, A.Q.; Shahid, N.; Yaqoob, A.; Samiullah, T.R.; Shakoor, S.; Latif, A.; Tabassum, B.; Khan, M.A.U.; Shahid, A.A.; et al. Role of oral vaccines as an edible tool to prevent infectious diseases. Acta Virol. 2019, 63, 245–252. [Google Scholar] [CrossRef]
- Savoie, K. Edible vaccine success. Nat. Biotechnol. 2000, 18, 367. [Google Scholar] [CrossRef] [PubMed]
- del Rio, B.; Dattwyler, R.J.; Aroso, M.; Neves, V.; Meirelles, L.; Seegers, J.F.; Gomes-Solecki, M. Oral immunization with recombinant lactobacillus plantarum induces a protective immune response in mice with Lyme disease. Clin. Vaccine Immunol. 2008, 15, 1429–1435. [Google Scholar] [CrossRef] [Green Version]
- Pitcovski, J.; Gruzdev, N.; Abzach, A.; Katz, C.; Ben-Adiva, R.; Brand-Shwartz, M.; Yadid, I.; Ratzon-Ashkenazi, E.; Emquies, K.; Israeli, H.; et al. Oral subunit SARS-CoV-2 vaccine induces systemic neutralizing IgG, IgA and cellular immune responses and can boost neutralizing antibody responses primed by an injected vaccine. Vaccine 2022, 40, 1098–1107. [Google Scholar] [CrossRef]
- Gao, T.; Ren, Y.; Li, S.; Lu, X.; Lei, H. Immune response induced by oral administration with a Saccharomyces cerevisiae-based SARS-CoV-2 vaccine in mice. Microb. Cell Fact. 2021, 20, 95. [Google Scholar] [CrossRef]
- Bellier, B.; Saura, A.; Lujan, L.A.; Molina, C.R.; Lujan, H.D.; Klatzmann, D. A Thermostable Oral SARS-CoV-2 Vaccine Induces Mucosal and Protective Immunity. Front. Immunol. 2022, 13, 837443. [Google Scholar] [CrossRef]
- Ballini, A.; Santacroce, L.; Cantore, S.; Bottalico, L.; Dipalma, G.; Topi, S.; Saini, R.; De Vito, D.; Inchingolo, F. Probiotics Efficacy on Oxidative Stress Values in Inflammatory Bowel Disease: A Randomized Double-Blinded Placebo-Controlled Pilot Study. Endocr. Metab. Immune Disord. Drug Targets 2019, 19, 373–381. [Google Scholar] [CrossRef]
- Santacroce, L.; Charitos, I.A.; Bottalico, L. A successful history: Probiotics and their potential as antimicrobials. Expert Rev. Anti Infect. Ther 2019, 17, 635–645. [Google Scholar] [CrossRef] [PubMed]
- Santacroce, L.; Inchingolo, F.; Topi, S.; Del Prete, R.; Di Cosola, M.; Charitos, I.A.; Montagnani, M. Potential beneficial role of probiotics on the outcome of COVID-19 patients: An evolving perspective. Diabetes Metab. Syndr. 2021, 15, 295–301. [Google Scholar] [CrossRef] [PubMed]
- Mu, Q.; Tavella, V.J.; Luo, X.M. Role of Lactobacillus reuteri in Human Health and Diseases. Front. Microbiol. 2018, 9, 757. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Song, Z.G.; Liu, C.; Tan, S.; Lin, S.; Zhu, J.; Dai, F.H.; Gao, J.; She, J.L.; Mei, Z.; et al. Gut microbiome alterations and gut barrier dysfunction are associated with host immune homeostasis in COVID-19 patients. BMC Med. 2022, 20, 24. [Google Scholar] [CrossRef] [PubMed]
- Zuo, T.; Zhang, F.; Lui, G.C.Y.; Yeoh, Y.K.; Li, A.Y.L.; Zhan, H.; Wan, Y.; Chung, A.C.K.; Cheung, C.P.; Chen, N.; et al. Alterations in Gut Microbiota of Patients With COVID-19 During Time of Hospitalization. Gastroenterology 2020, 159, 944–955. [Google Scholar] [CrossRef]
- Wu, Y.; Cheng, X.; Jiang, G.; Tang, H.; Ming, S.; Tang, L.; Lu, J.; Guo, C.; Shan, H.; Huang, X. Altered oral and gut microbiota and its association with SARS-CoV-2 viral load in COVID-19 patients during hospitalization. NPJ Biofilms Microbiomes 2021, 7, 61. [Google Scholar] [CrossRef]
- Rajput, S.; Paliwal, D.; Naithani, M.; Kothari, A.; Meena, K.; Rana, S. COVID-19 and Gut Microbiota: A Potential Connection. Indian J. Clin. Biochem. 2021, 36, 266–277. [Google Scholar] [CrossRef]
- d’Ettorre, G.; Ceccarelli, G.; Marazzato, M.; Campagna, G.; Pinacchio, C.; Alessandri, F.; Ruberto, F.; Rossi, G.; Celani, L.; Scagnolari, C.; et al. Challenges in the Management of SARS-CoV2 Infection: The Role of Oral Bacteriotherapy as Complementary Therapeutic Strategy to Avoid the Progression of COVID-19. Front. Med. 2020, 7, 389. [Google Scholar] [CrossRef]
- de Ponte, M.C.; Cardoso, V.G.; Goncalves, G.L.; Costa-Pessoa, J.M.; Oliveira-Souza, M. Early type 1 diabetes aggravates renal ischemia/reperfusion-induced acute kidney injury. Sci. Rep. 2021, 11, 19028. [Google Scholar] [CrossRef]
- Villapol, S. Gastrointestinal symptoms associated with COVID-19: Impact on the gut microbiome. Transl. Res. 2020, 226, 57–69. [Google Scholar] [CrossRef]
- Chen, J.; Vitetta, L. Modulation of Gut Microbiota for the Prevention and Treatment of COVID-19. J. Clin. Med. 2021, 10, 2903. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, T.-H.; Hsu, M.-T.; Lee, M.-Y.; Chou, C.-K. Gastrointestinal Involvement in SARS-CoV-2 Infection. Viruses 2022, 14, 1188. https://doi.org/10.3390/v14061188
Chen T-H, Hsu M-T, Lee M-Y, Chou C-K. Gastrointestinal Involvement in SARS-CoV-2 Infection. Viruses. 2022; 14(6):1188. https://doi.org/10.3390/v14061188
Chicago/Turabian StyleChen, Tsung-Hsien, Ming-Tse Hsu, Ming-Yang Lee, and Chu-Kuang Chou. 2022. "Gastrointestinal Involvement in SARS-CoV-2 Infection" Viruses 14, no. 6: 1188. https://doi.org/10.3390/v14061188





