Secretory Profile of Adipose-Tissue-Derived Mesenchymal Stem Cells from Cats with Calicivirus-Positive Severe Chronic Gingivostomatitis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Selection Criteria
2.2. Isolation and Culturing of fAd-MSCs
2.3. Production of Secretome from fAd-MSCs
2.4. Characterization of the fAd-MSC-Derived Secretomes by Immunoassay
2.5. Proteomic Profile of the Secretome by Liquid Chromatography High-Resolution Mass Spectrometry and Bioinformatic Analysis
2.6. Statistical Analysis
3. Results
3.1. Selected Cats
3.2. Characterization of the fAd-MSCs
3.3. Characterization of the Secretomes by Immunoassay
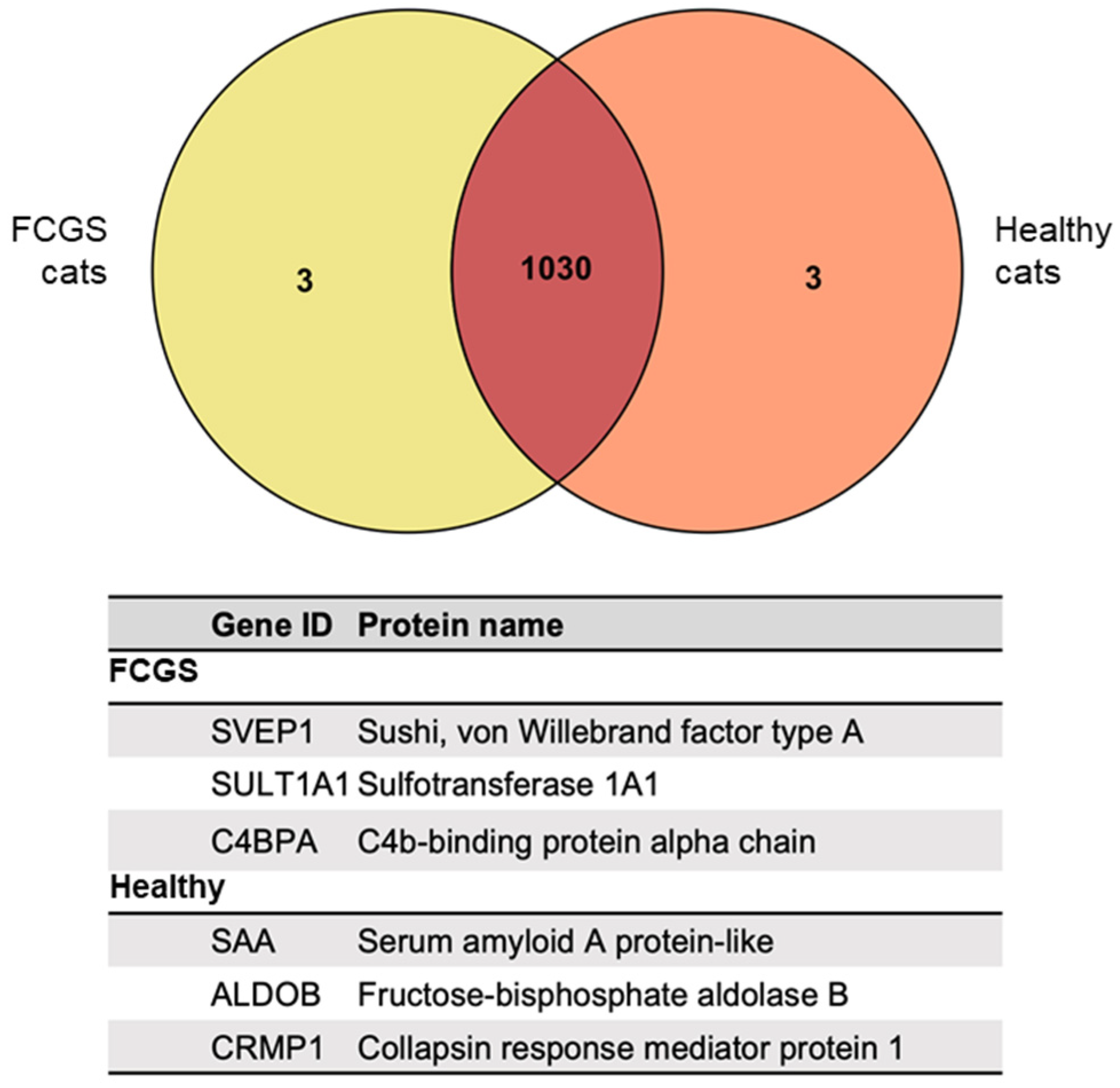
3.4. Proteomic Analysis of the fAd-MSC-Derived Secretomes
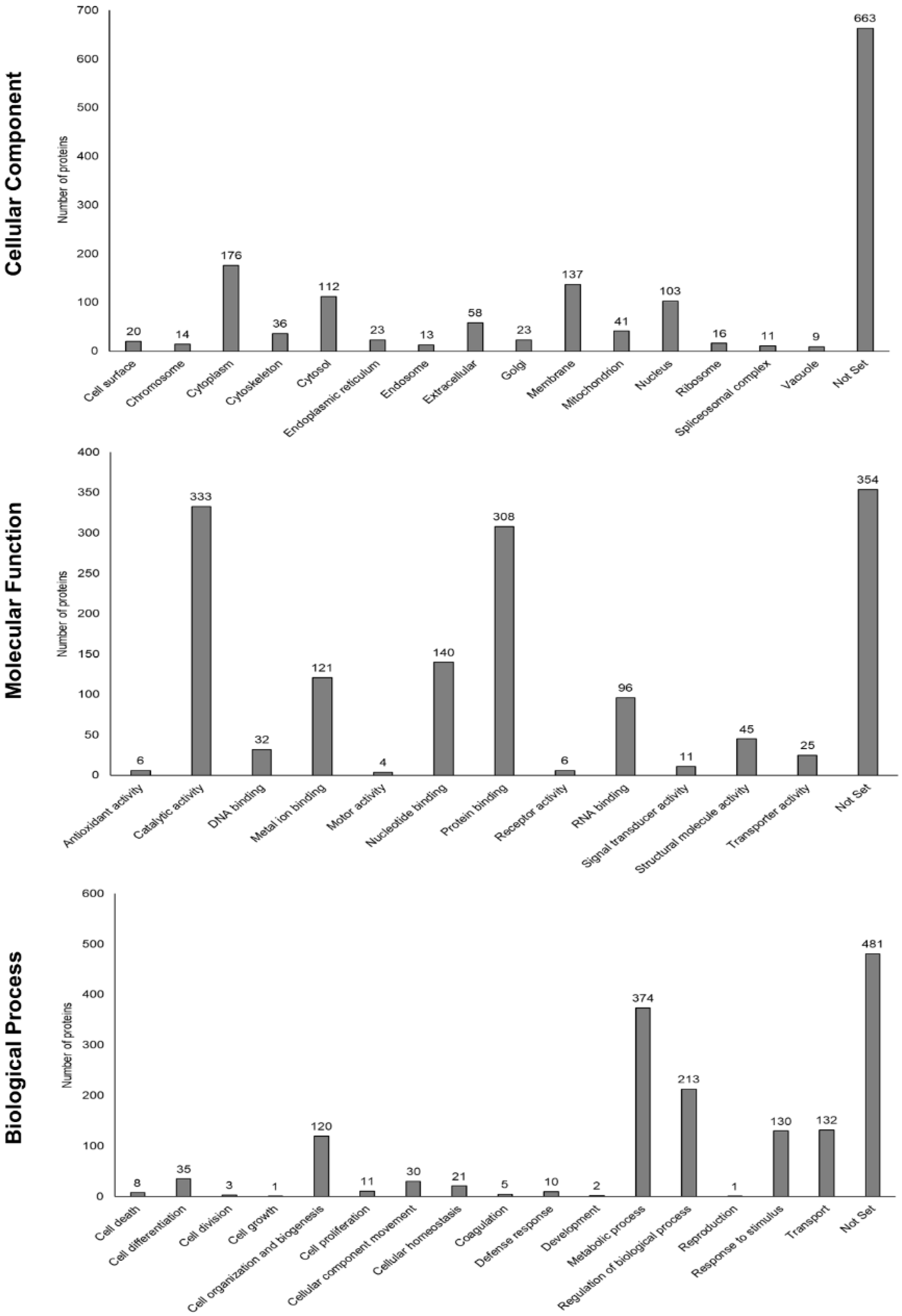
3.5. Functional Enrichment Analysis Based on Gene Ontology and STRING Database
3.6. Label-Free Quantification of the Secretome Proteomes
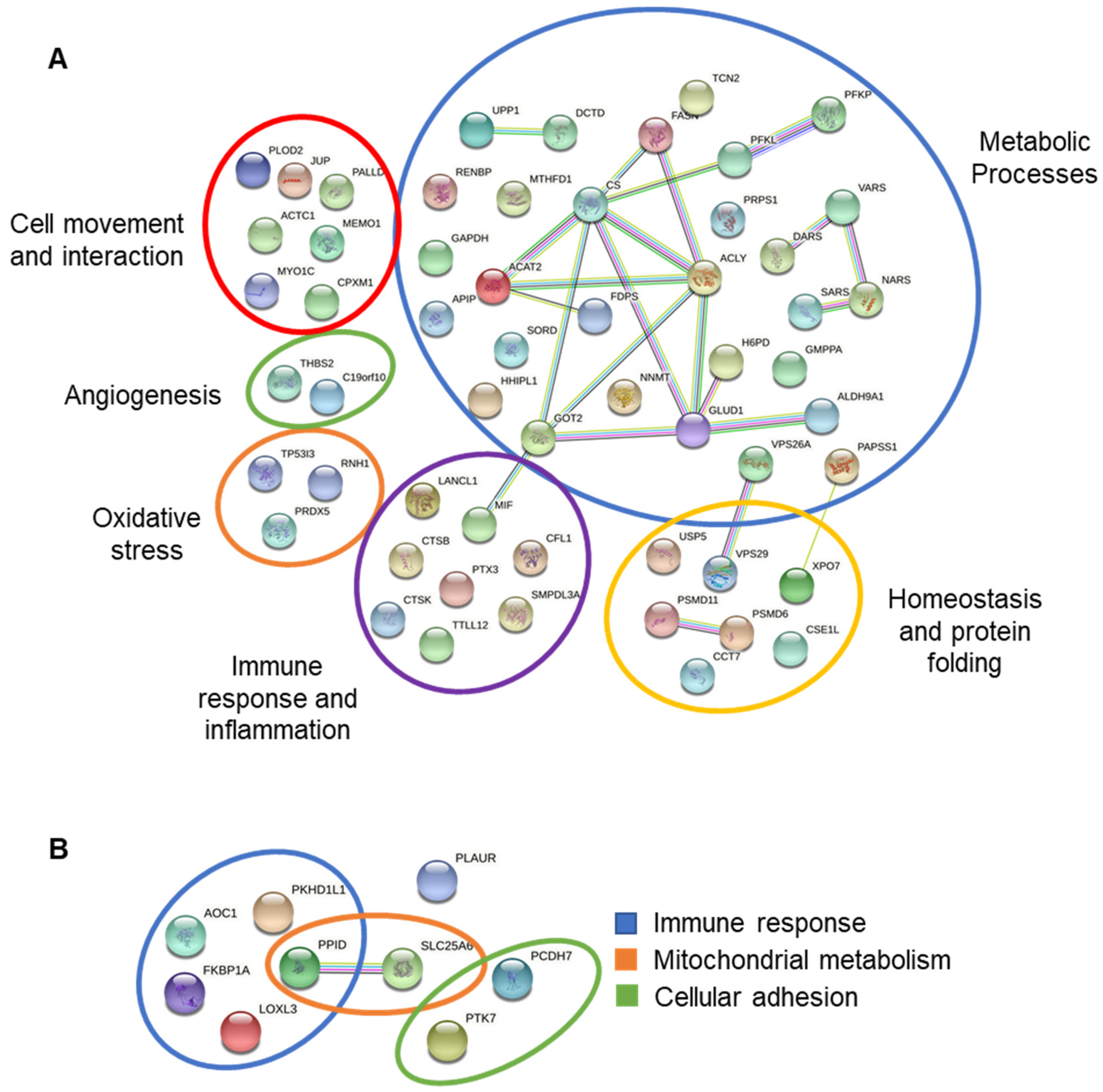
3.7. Interactions between Differentially Expressed Proteins
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Coyne, K.P.; Dawson, S.; Radford, A.D.; Cripps, P.J.; Porter, C.J.; McCracken, C.M.; Gaskell, R.M. Long-term analysis of feline calicivirus prevalence and viral shedding patterns in naturally infected colonies of domestic cats. Vet. Microbiol. 2006, 118, 12–25. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, M.; Manzanilla, E.G.; Lloret, A.; Leon, M.; Thibault, J.C. Prevalence of feline herpesvirus-1, feline calicivirus, Chlamydophila felis and Mycoplasma felis DNA and associated risk factors in cats in Spain with upper respiratory tract disease, conjunctivitis and/or gingivostomatitis. J. Feline Med. Surg. 2017, 19, 461–469. [Google Scholar] [CrossRef] [PubMed]
- Druet, I.; Hennet, P. Relationship between Feline calicivirus Load, Oral Lesions, and Outcome in Feline Chronic Gingivostomatitis (Caudal Stomatitis): Retrospective Study in 104 Cats. Front. Vet. Sci. 2017, 4, 209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, D.B.; Verstraete, F.J.M.; Arzi, B. An Update on Feline Chronic Gingivostomatitis. Vet. Clin. N. Am. Small Anim. Pract. 2020, 50, 973–982. [Google Scholar] [CrossRef]
- Peralta, S.; Carney, P.C. Feline chronic gingivostomatitis is more prevalent in shared households and its risk correlates with the number of cohabiting cats. J. Feline Med. Surg. 2019, 21, 1165–1171. [Google Scholar] [CrossRef]
- Harbour, D.A.; Howard, P.E.; Gaskell, R.M. Isolation of feline calicivirus and feline herpesvirus from domestic cats 1980 to 1989. Vet. Rec. 1991, 128, 77–80. [Google Scholar] [CrossRef]
- Thomas, S.; Lappin, D.F.; Spears, J.; Bennett, D.; Nile, C.; Riggio, M.P. Prevalence of feline calicivirus in cats with odontoclastic resorptive lesions and chronic gingivostomatitis. Res. Vet. Sci. 2017, 111, 124–126. [Google Scholar] [CrossRef] [Green Version]
- Winer, J.N.; Arzi, B.; Verstraete, F.J. Therapeutic Management of Feline Chronic Gingivostomatitis: A Systematic Review of the Literature. Front. Vet. Sci. 2016, 3, 54. [Google Scholar] [CrossRef]
- Arzi, B.; Mills-Ko, E.; Verstraete, F.J.; Kol, A.; Walker, N.J.; Badgley, M.R.; Fazel, N.; Murphy, W.J.; Vapniarsky, N.; Borjesson, D.L. Therapeutic Efficacy of Fresh, Autologous Mesenchymal Stem Cells for Severe Refractory Gingivostomatitis in Cats. Stem Cells Transl. Med. 2016, 5, 75–86. [Google Scholar] [CrossRef] [Green Version]
- Arzi, B.; Clark, K.C.; Sundaram, A.; Spriet, M.; Verstraete, F.; Walker, N.J.; Loscar, M.R.; Fazel, N.; Murphy, W.J.; Vapniarsky, N.; et al. Therapeutic Efficacy of Fresh, Allogeneic Mesenchymal Stem Cells for Severe Refractory Feline Chronic Gingivostomatitis. Stem Cells Transl. Med. 2017, 6, 1710–1722. [Google Scholar] [CrossRef]
- Villatoro, A.J.; Alcoholado, C.; Martin-Astorga, M.C.; Fernandez, V.; Cifuentes, M.; Becerra, J. Comparative analysis and characterization of soluble factors and exosomes from cultured adipose tissue and bone marrow mesenchymal stem cells in canine species. Vet. Immunol. Immunopathol. 2019, 208, 6–15. [Google Scholar] [CrossRef] [PubMed]
- Makridakis, M.; Roubelakis, M.G.; Vlahou, A. Stem cells: Insights into the secretome. Biochim. Biophys. Acta 2013, 1834, 2380–2384. [Google Scholar] [CrossRef] [PubMed]
- Villatoro, A.J.; Martín-Astorga, M.C.; Alcoholado, C.; Sánchez-Martín, M.; Becerra, J. Proteomic Analysis of the Secretome and Exosomes of Feline Adipose-Derived Mesenchymal Stem Cells. Animals 2021, 11, 295. [Google Scholar] [CrossRef]
- Kornicka, K.; Houston, J.; Marycz, K. Dysfunction of Mesenchymal Stem Cells Isolated from Metabolic Syndrome and Type 2 Diabetic Patients as Result of Oxidative Stress and Autophagy may Limit Their Potential Therapeutic Use. Stem Cell Rev. Rep. 2018, 14, 337–345. [Google Scholar] [CrossRef] [Green Version]
- Cheng, R.J.; Xiong, A.J.; Li, Y.H.; Pan, S.Y.; Zhang, Q.P.; Zhao, Y.; Liu, Y.; Marion, T.N. Mesenchymal Stem Cells: Allogeneic MSC May Be Immunosuppressive but Autologous MSC Are Dysfunctional in Lupus Patients. Front. Cell Dev. Biol. 2019, 7, 285. [Google Scholar] [CrossRef]
- Pérez, L.M.; de Lucas, B.; Gálvez, B.G. Unhealthy Stem Cells: When Health Conditions Upset Stem Cell Properties. Cell Physiol. Biochem. 2018, 46, 1999–2016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papadaki, H.A.; Kritikos, H.D.; Gemetzi, C.; Koutala, H.; Marsh, J.C.; Boumpas, D.T.; Eliopoulos, G.D. Bone marrow progenitor cell reserve and function and stromal cell function are defective in rheumatoid arthritis: Evidence for a tumor necrosis factor alpha-mediated effect. Blood 2002, 99, 1610–1619. [Google Scholar] [CrossRef] [Green Version]
- Marycz, K.; Weiss, C.; Śmieszek, A.; Kornicka, K. Evaluation of Oxidative Stress and Mitophagy during Adipogenic Differentiation of Adipose-Derived Stem Cells Isolated from Equine Metabolic Syndrome (EMS) Horses. Stem Cells Int. 2018, 2018, 5340756. [Google Scholar] [CrossRef]
- Marycz, K.; Kornicka, K.; Basinska, K.; Czyrek, A. Equine Metabolic Syndrome Affects Viability, Senescence, and Stress Factors of Equine Adipose-Derived Mesenchymal Stromal Stem Cells: New Insight into EqASCs Isolated from EMS Horses in the Context of Their Aging. Oxid. Med. Cell. Longev. 2016, 2016, 4710326. [Google Scholar] [CrossRef] [Green Version]
- Villatoro, A.J.; Martín-Astorga, M.C.; Alcoholado, C.; Cárdenas, C.; Fariñas, F.; Becerra, J.; Visser, R. Altered Proteomic Profile of Adipose Tissue-Derived Mesenchymal Stem Cell Exosomes from Cats with Severe Chronic Gingivostomatitis. Animals 2021, 11, 2466. [Google Scholar] [CrossRef]
- Hennet, P.R.; Camy, G.A.; McGahie, D.M.; Albouy, M.V. Comparative efficacy of a recombinant feline interferon omega in refractory cases of calicivirus-positive cats with caudal stomatitis: A randomised, multi-centre, controlled, double-blind study in 39 cats. J. Feline Med. Surg. 2011, 13, 577–587. [Google Scholar] [CrossRef] [PubMed]
- Lommer, M.J. Efficacy of cyclosporine for chronic, refractory stomatitis in cats: A randomized, placebo-controlled, double-blinded clinical study. J. Vet. Dent. 2013, 30, 8–17. [Google Scholar] [CrossRef] [PubMed]
- Villatoro, A.J.; Claros, S.; Fernández, V.; Alcoholado, C.; Fariñas, F.; Moreno, A.; Becerra, J.; Andrades, J.A. Safety and efficacy of the mesenchymal stem cell in feline eosinophilic keratitis treatment. BMC Vet. Res. 2018, 14, 116. [Google Scholar] [CrossRef] [Green Version]
- Dominici, M.; Le Blanc, K.; Mueller, I.; Slaper-Cortenbach, I.; Marini, F.; Krause, D.; Deans, R.; Keating, A.; Prockop, D.J.; Horwitz, E. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy 2006, 8, 315–317. [Google Scholar] [CrossRef] [PubMed]
- Thanunchai, M.; Hongeng, S.; Thitithanyanont, A. Mesenchymal Stromal Cells, and Viral Infection. Stem Cells. Int. 2015, 2015, 860950. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martiny, P.; Goggs, R. Biomarker Guided Diagnosis of Septic Peritonitis in Dogs. Front. Vet. Sci. 2019, 6, 208. [Google Scholar] [CrossRef]
- Troia, R.; Mascalzoni, G.; Agnoli, C.; Lalonde-Paul, D.; Giunti, M.; Goggs, R. Cytokine and Chemokine Profiling in Cats with Sepsis and Septic Shock. Front. Vet. Sci. 2020, 7, 305. [Google Scholar] [CrossRef]
- Russo, R.C.; Garcia, C.C.; Teixeira, M.M.; Amaral, F.A. The CXCL8/IL-8 chemokine family and its receptors in inflammatory diseases. Expert. Rev. Clin. Immunol. 2014, 10, 593–619. [Google Scholar] [CrossRef] [Green Version]
- Gruen, M.E.; Messenger, K.M.; Thomson, A.E.; Griffith, E.H.; Aldrich, L.A.; Vaden, S.; Duncan, B.; Lascelles, X. Evaluation of serum cytokines in cats with and without degenerative joint disease and associated pain. Vet. Immunol. Immunopathol. 2017, 183, 49–59. [Google Scholar] [CrossRef] [Green Version]
- Parys, M.; Yuzbasiyan-Gurkan, V.; Kruger, J. Serum Cytokine Profiling in Cats with Acute Idiopathic Cystitis. J. Vet. Intern. Med. 2018, 32, 274–279. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.; Lee, S.; Kim, S.; Kim, D.; Ahn, J.H.; Ahn, K. Human cytomegalovirus clinical strain-specific microRNA miR-UL148D targets the human chemokine RANTES during infection. PLoS Pathog. 2012, 8, e1002577. [Google Scholar] [CrossRef] [PubMed]
- Frink, M.; Hsieh, Y.C.; Hsieh, C.H.; Pape, H.C.; Choudhry, M.A.; Schwacha, M.G.; Chaudry, I.H. Keratinocyte-derived chemokine plays a critical role in the induction of systemic inflammation and tissue damage after trauma-hemorrhage. Shock 2007, 28, 576–581. [Google Scholar] [CrossRef] [PubMed]
- Harrell, C.R.; Popovska Jovicic, B.; Djonov, V.; Volarevic, V. Molecular Mechanisms Responsible for Mesenchymal Stem Cell-Based Treatment of Viral Diseases. Pathogens 2021, 10, 409. [Google Scholar] [CrossRef] [PubMed]
- Villatoro, A.J.; Alcoholado, C.; Martín-Astorga, M.; Rubio, N.; Blanco, J.; Garrido, C.P.; Becerra, J. Suicide gene therapy by canine mesenchymal stem cell transduced with thymidine kinase in a u-87 glioblastoma murine model: Secretory profile and antitumor activity. PLoS ONE 2022, 17, e0264001. [Google Scholar] [CrossRef] [PubMed]
- Arakaki, T.L.; Pezza, J.A.; Cronin, M.A.; Hopkins, C.E.; Zimmer, D.B.; Tolan, D.R.; Allen, K.N. Structure of human brain fructose 1,6-(bis)phosphate aldolase: Linking isozyme structure with function. Protein Sci. 2004, 13, 3077–3084. [Google Scholar] [CrossRef]
- Monferrer, E.; Sanegre, S.; Vieco-Martí, I.; López-Carrasco, A.; Fariñas, F.; Villatoro, A.; Abanades, S.; Mañes, S.; de la Cruz-Merino, L.; Noguera, R.; et al. Immunometabolism Modulation in Therapy. Biomedicines 2021, 9, 798. [Google Scholar] [CrossRef]
- Li, Q.; Li, Y.; Xu, J.; Wang, S.; Xu, Y.; Li, X.; Cai, S. Aldolase B Overexpression is Associated with Poor Prognosis and Promotes Tumor Progression by Epithelial-Mesenchymal Transition in Colorectal Adenocarcinoma. Cell. Physiol. Biochem. 2017, 42, 397–406. [Google Scholar] [CrossRef]
- Peng, S.Y.; Lai, P.L.; Pan, H.W.; Hsiao, L.P.; Hsu, H.C. Aberrant expression of the glycolytic enzymes aldolase B and type II hexokinase in hepatocellular carcinoma are predictive markers for advanced stage, early recurrence and poor prognosis. Oncol. Rep. 2008, 19, 1045–1053. [Google Scholar] [CrossRef]
- Cnops, L.; Van de Plas, B.; Arckens, L. Age-dependent expression of collapsin response mediator proteins (CRMPs) in cat visual cortex. Eur. J. Neurosci. 2004, 19, 2345–2351. [Google Scholar] [CrossRef]
- Fumio, N.; Toshio, O.; Yoshio, G. Collapsin Response Mediator Proteins: Their Biological Functions and Pathophysiology in Neuronal Development and Regeneration. Front. Cell. Neurosci. 2020, 14. [Google Scholar] [CrossRef]
- Yuki, M.; Aoyama, R.; Nakagawa, M.; Hirano, T.; Naitoh, E.; Kainuma, D. A Clinical Investigation on Serum Amyloid A Concentration in Client-Owned Healthy and Diseased Cats in a Primary Care Animal Hospital. Vet. Sci. 2020, 7, 45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Samuelov, L.; Li, Q.; Bochner, R.; Najor, N.A.; Albrecht, L.; Malchin, N.; Goldsmith, T.; Grafi-Cohen, M.; Vodo, D.; Fainberg, G.; et al. SVEP1 plays a crucial role in epidermal differentiation. Exp. Dermatol. 2017, 26, 423–430. [Google Scholar] [CrossRef] [Green Version]
- Morooka, N.; Futaki, S.; Sato-Nishiuchi, R.; Nishino, M.; Totani, Y.; Shimono, C.; Nakano, I.; Nakajima, H.; Mochizuki, N.; Sekiguchi, K. Polydom Is an Extracellular Matrix Protein Involved in Lymphatic Vessel Remodeling. Cir. Res. 2017, 120, 1276–1288. [Google Scholar] [CrossRef] [PubMed]
- Yalcin, E.B.; More, V.; Neira, K.L.; Lu, Z.J.; Cherrington, N.J.; Slitt, A.L.; King, R.S. Downregulation of Sulfotransferase Expression and Activity in Diseased Human Livers. Drug Metab. Dispos. 2013, 41, 1642–1650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iqbal, A.; Ziyi, P.; Yu, H.; Jialing, L.; Haochen, W.; Jing, F.; Ping, J.; Zhihui, Z. C4BPA: A Novel Co-Regulator of Immunity and Fat Metabolism in the Bovine Mammary Epithelial Cells. Front. Genet. 2022, 12, 830566. [Google Scholar] [CrossRef] [PubMed]
- Bailey, C.C.; Zhong, G.; Huang, I.C.; Farzan, M. IFITM-Family Proteins: The Cell’s First Line of Antiviral Defense. Annu. Rev. Virol. 2014, 1, 261–283. [Google Scholar] [CrossRef] [Green Version]
- Schäfer, R.; Spohn, G.; Bechtel, M.; Bojkova, D.; Baer, P.C.; Kuçi, S.; Seifried, E.; Ciesek, S.; Cinatl, J. Human Mesenchymal Stromal Cells Are Resistant to SARS-CoV-2 Infection under Steady-State, Inflammatory Conditions and in the Presence of SARS-CoV-2-Infected Cells. Stem Cell Rep. 2021, 16, 419–427. [Google Scholar] [CrossRef]
- Schoggins, J.W. Interferon-Stimulated Genes: What Do They All Do? Annu. Rev. Virol. 2019, 6, 567–584. [Google Scholar] [CrossRef]
- Wu, X.; Dao Thi, V.L.; Huang, Y.; Billerbeck, E.; Saha, D.; Hoffmann, H.H.; Wang, Y.; Silva, L.; Sarbanes, S.; Sun, T.; et al. Intrinsic Immunity Shapes Viral Resistance of Stem Cells. Cell 2018, 172, 423–438.e25. [Google Scholar] [CrossRef]
- Peñaflor-Téllez, Y.; Trujillo-Uscanga, A.; Escobar-Almazán, J.A.; Gutiérrez-Escolano, A.L. Immune Response Modulation by Caliciviruses. Front. Immunol. 2019, 10, 2334. [Google Scholar] [CrossRef]
- Trzil, J.E.; Masseau, I.; Webb, T.L.; Chang, C.H.; Dodam, J.R.; Cohn, L.A.; Liu, H.; Quimby, J.M.; Dow, S.W.; Reinero, C.R. Long-term evaluation of mesenchymal stem cell therapy in a feline model of chronic allergic asthma. Clin. Exp. Allergy 2014, 44, 1546–1557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dias, I.E.; Pinto, P.O.; Barros, L.C.; Viegas, C.A.; Dias, I.R.; Carvalho, P.P. Mesenchymal stem cells therapy in companion animals: Useful for immune-mediated diseases? BMC Vet. Res. 2019, 15, 358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Webb, T.L.; Webb, C.B. Stem cell therapy in cats with chronic enteropathy: A proof-of-concept study. J. Feline Med. Surg. 2015, 17, 901–908. [Google Scholar] [CrossRef] [PubMed]
- Arzi, B.; Peralta, S.; Fiani, N.; Vapniarsky, N.; Taechangam, N.; Delatorre, U.; Clark, K.C.; Walker, N.J.; Loscar, M.R.; Lommer, M.J.; et al. A multicenter experience using adipose-derived mesenchymal stem cell therapy for cats with chronic, non-responsive gingivostomatitis. Stem Cell Res. Ther. 2020, 11, 115. [Google Scholar] [CrossRef]
- Li, C.; Zhao, H.; Cheng, L.; Wang, B. Allogeneic vs. autologous mesenchymal stem/stromal cells in their medication practice. Cell Biosci. 2021, 11, 187. [Google Scholar] [CrossRef] [PubMed]
- Kol, A.; Arzi, B.; Athanasiou, K.A.; Farmer, D.L.; Nolta, J.A.; Rebhun, R.B.; Chen, X.; Griffiths, L.G.; Verstraete, F.J.; Murphy, C.J.; et al. Companion animals: Translational scientist’s new best friends. Sci. Transl. Med. 2015, 7, 308ps21. [Google Scholar] [CrossRef] [Green Version]


| Group | Sex | Weight (kg) | Age (Years) | SDAI Score |
|---|---|---|---|---|
| FCGS patients | ♂ | 2.9 | 2.5 | 23 |
| ♂ | 3.3 | 11.5 | 23 | |
| ♂ | 3.4 | 9 | 22 | |
| ♀ | 3.1 | 4 | 23 | |
| ♀ | 4.5 | 5 | 21 | |
| Average | 3.4 ± 0.6 | 6.4 ± 3.7 | 22.4 ± 0.9 | |
| Healthy cats | ♂ | 4.1 | 3 | |
| ♂ | 4 | 4 | ||
| ♂ | 3.8 | 6 | ||
| ♀ | 3.7 | 3 | ||
| ♀ | 3.5 | 5 | ||
| Average | 3.8 ± 0.24 | 4.2 ± 1.3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Villatoro, A.J.; Martín-Astorga, M.d.C.; Alcoholado, C.; Kazantseva, L.; Cárdenas, C.; Fariñas, F.; Becerra, J.; Visser, R. Secretory Profile of Adipose-Tissue-Derived Mesenchymal Stem Cells from Cats with Calicivirus-Positive Severe Chronic Gingivostomatitis. Viruses 2022, 14, 1146. https://doi.org/10.3390/v14061146
Villatoro AJ, Martín-Astorga MdC, Alcoholado C, Kazantseva L, Cárdenas C, Fariñas F, Becerra J, Visser R. Secretory Profile of Adipose-Tissue-Derived Mesenchymal Stem Cells from Cats with Calicivirus-Positive Severe Chronic Gingivostomatitis. Viruses. 2022; 14(6):1146. https://doi.org/10.3390/v14061146
Chicago/Turabian StyleVillatoro, Antonio J., María del Carmen Martín-Astorga, Cristina Alcoholado, Liliya Kazantseva, Casimiro Cárdenas, Fernando Fariñas, José Becerra, and Rick Visser. 2022. "Secretory Profile of Adipose-Tissue-Derived Mesenchymal Stem Cells from Cats with Calicivirus-Positive Severe Chronic Gingivostomatitis" Viruses 14, no. 6: 1146. https://doi.org/10.3390/v14061146
APA StyleVillatoro, A. J., Martín-Astorga, M. d. C., Alcoholado, C., Kazantseva, L., Cárdenas, C., Fariñas, F., Becerra, J., & Visser, R. (2022). Secretory Profile of Adipose-Tissue-Derived Mesenchymal Stem Cells from Cats with Calicivirus-Positive Severe Chronic Gingivostomatitis. Viruses, 14(6), 1146. https://doi.org/10.3390/v14061146








