New Targets for Antiviral Therapy: Inhibitory Receptors and Immune Checkpoints on Myeloid Cells
Abstract
:1. Introduction
2. Structure of Inhibitory Receptors
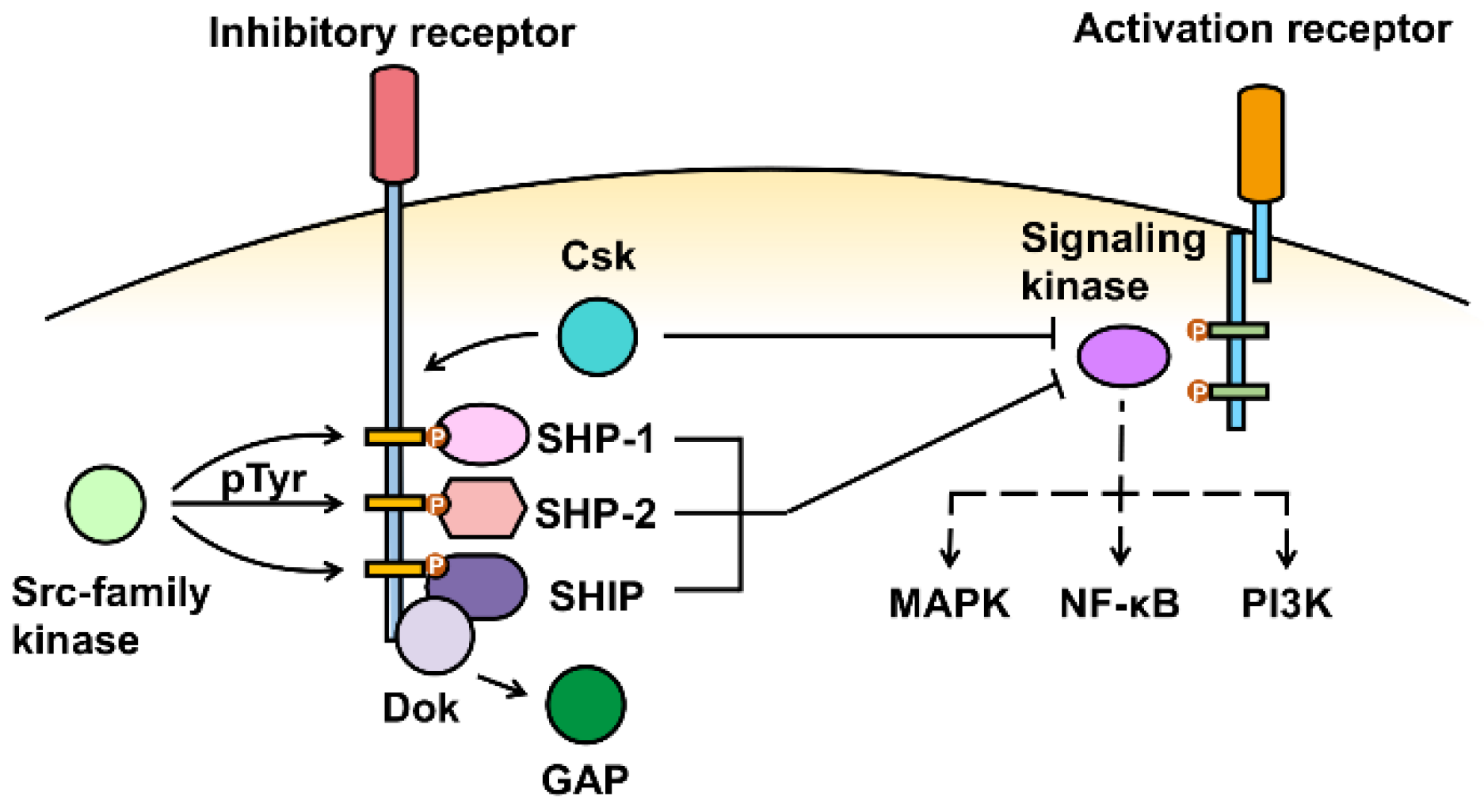
3. Inhibitory Receptor Signaling
4. Viral Interaction with Inhibitory Receptors on Myeloid Cells
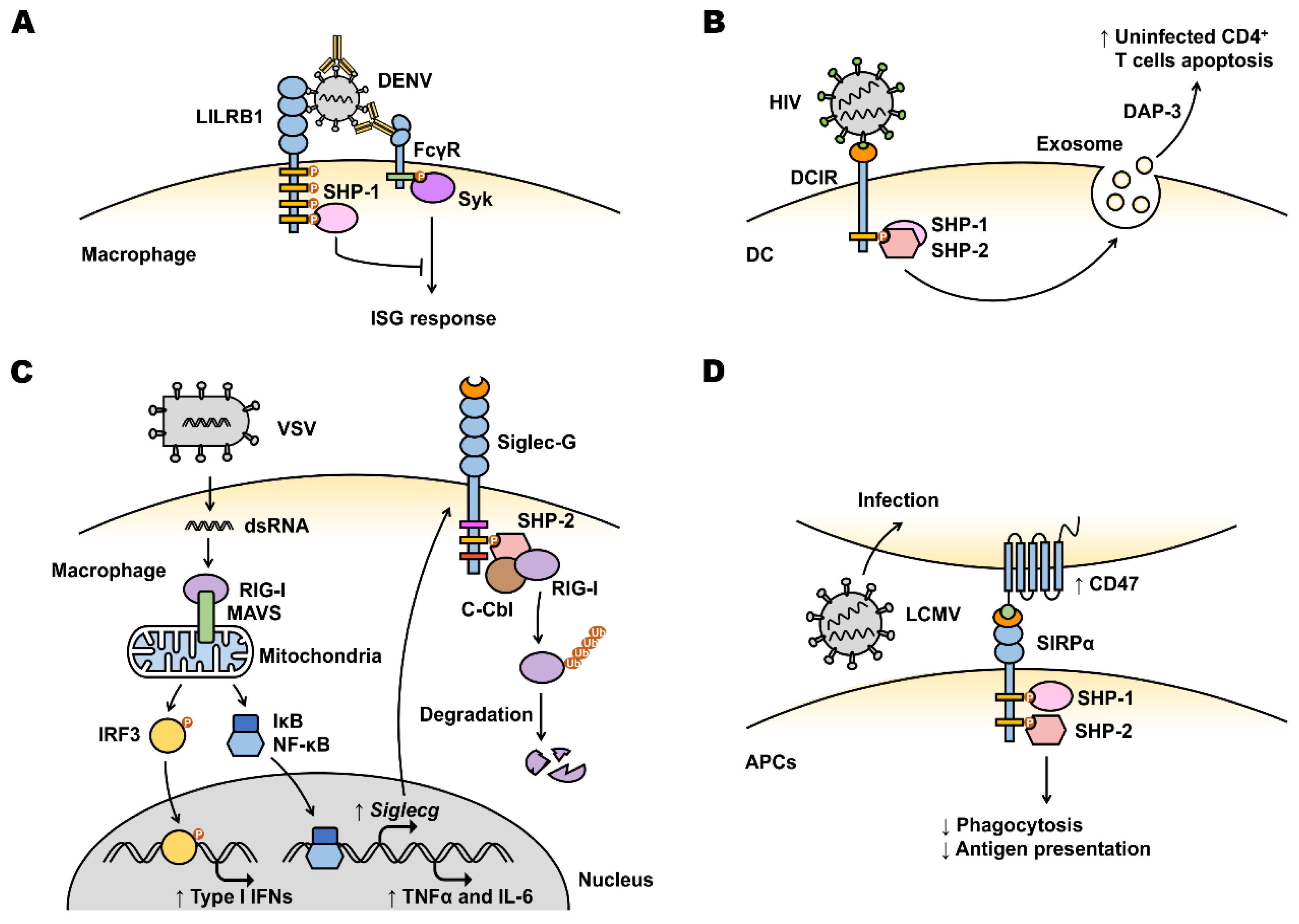
4.1. Viral Manipulation of Inhibitory Receptors on Myeloid Cells to Enhance Infection
4.2. Viral Manipulation of Inhibitory Receptors to Evade Myeloid Cell-Mediated Antiviral Immunity
4.2.1. Myeloid Cell-Mediated Viral Immune Evasion
4.2.2. Myeloid Cell Immune Checkpoints
LILRB
DCIR
Siglecs (Inhibitory)
SIRPα
| Inhibitory Receptors | Myeloid Cell Expression * | Functional Importance | Reference |
|---|---|---|---|
| CD300a | MC, Mac, Mono, DC, N, E, B | Recognizes PtdEth and PtdSer associated with virions and enhance viral entry. | [49] |
| LILRB | Mac, Mono, N, DC, E, B | Antibody-opsonized DENV utilizes LILRB1 to inhibit FcγR signaling in monocytes and inhibit ISG expression. | [100,101] |
| KK10 epitope variant of HIV-1 binds to LILRB2 leading to a tolerogenic phenotype of DCs. | [94] | ||
| HIV elevates LILRB2 expression via IL-10 and inhibits APC capacity of monocytes, thus impairing function of CD4+ and CD8+ T cells. | [70,126] | ||
| DCIR | Mac, Mono, N, DC | HIV-1 utilizes DCIR on DC to infect CD4+ T cells and induces apoptosis in uninfected CD4+ T cells. | [51,52,106] |
| Siglecs | MC, Mac, Mono, N, DC, E, B | HIV-1 membrane gangliosides can be recognized by a variety of inhibitory Siglecs on macrophages including Siglec-3, Siglec-5, Siglec-7 and Siglec-9 and promote infection. | [54] |
| VSV upregulates Siglec-G expression on macrophages, leading to RIG-I degradation and IFN-β response suppression. | [71] | ||
| α2,6-biantennary sialoglycans of HBsAg binding to human Siglec-3 activates Siglec-3 on myeloid cells and induces immunosuppression. | [111] | ||
| SIRPα | Mac, N, DC | CD47-SIRPα blockade by anti-CD47 antibody increased activation of macrophages and DCs, improved CD8+ T cell responses by strengthening APC capacity of DCs, contributing to LCMV control. | [122,123] |
| Blockade of CD47 reduced HIV antigen level and restored CD4+ and CD8+ T cell numbers. | [122] |
4.3. Polymorphism of Inhibitory Receptors on Myeloid Cells Associates with Viral Susceptibility
5. Inhibitory Receptors on Myeloid Cells as Future Therapeutic Target for Antiviral Treatment
6. Concluding Remarks
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Stegelmeier, A.A.; van Vloten, J.P.; Mould, R.C.; Klafuric, E.M.; Minott, J.A.; Wootton, S.K.; Bridle, B.W.; Karimi, K. Myeloid Cells during Viral Infections and Inflammation. Viruses 2019, 11, 168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.P.; Kurt-Jones, E.A.; Shin, O.S.; Manchak, M.D.; Levin, M.J.; Finberg, R.W. Varicella-zoster virus activates inflammatory cytokines in human monocytes and macrophages via Toll-like receptor 2. J. Virol. 2005, 79, 12658–12666. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.P.; Bowen, G.N.; Padden, C.; Cerny, A.; Finberg, R.W.; Newburger, P.E.; Kurt-Jones, E.A. Toll-like receptor-mediated activation of neutrophils by influenza A virus. Blood 2008, 112, 2028–2034. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.; Pamer, E.G. Monocyte recruitment during infection and inflammation. Nat. Rev. Immunol. 2011, 11, 762–774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teijaro, J.R.; Walsh, K.B.; Rice, S.; Rosen, H.; Oldstone, M.B. Mapping the innate signaling cascade essential for cytokine storm during influenza virus infection. Proc. Natl. Acad. Sci. USA 2014, 111, 3799–3804. [Google Scholar] [CrossRef] [Green Version]
- O’Shea, J.J.; Ma, A.; Lipsky, P. Cytokines and autoimmunity. Nat. Rev. Immunol. 2002, 2, 37–45. [Google Scholar] [CrossRef]
- Griseri, T.; McKenzie, B.S.; Schiering, C.; Powrie, F. Dysregulated hematopoietic stem and progenitor cell activity promotes interleukin-23-driven chronic intestinal inflammation. Immunity 2012, 37, 1116–1129. [Google Scholar] [CrossRef] [Green Version]
- Gao, R.; Bhatnagar, J.; Blau, D.M.; Greer, P.; Rollin, D.C.; Denison, A.M.; Deleon-Carnes, M.; Shieh, W.J.; Sambhara, S.; Tumpey, T.M.; et al. Cytokine and chemokine profiles in lung tissues from fatal cases of 2009 pandemic influenza A (H1N1): Role of the host immune response in pathogenesis. Am. J. Pathol. 2013, 183, 1258–1268. [Google Scholar] [CrossRef]
- Rossini, G.; Landini, M.P.; Gelsomino, F.; Sambri, V.; Varani, S. Innate host responses to West Nile virus: Implications for central nervous system immunopathology. World J. Virol. 2013, 2, 49–56. [Google Scholar] [CrossRef]
- Srikiatkhachorn, A.; Mathew, A.; Rothman, A.L. Immune-mediated cytokine storm and its role in severe dengue. Semin. Immunopathol. 2017, 39, 563–574. [Google Scholar] [CrossRef]
- Rumpret, M.; Drylewicz, J.; Ackermans, L.J.E.; Borghans, J.A.M.; Medzhitov, R.; Meyaard, L. Functional categories of immune inhibitory receptors. Nat. Rev. Immunol. 2020, 20, 771–780. [Google Scholar] [CrossRef] [PubMed]
- Kuroki, K.; Furukawa, A.; Maenaka, K. Molecular recognition of paired receptors in the immune system. Front. Microbiol. 2012, 3, 429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zarrin, A.A.; Monteiro, R.C. Editorial: The role of inhibitory receptors in inflammation and cancer. Front. Immunol. 2020, 11, 633686. [Google Scholar] [CrossRef] [PubMed]
- Lanier, L.L. NK cell receptors. Annu. Rev. Immunol. 1998, 16, 359–393. [Google Scholar] [CrossRef]
- Sivori, S.; Della Chiesa, M.; Carlomagno, S.; Quatrini, L.; Munari, E.; Vacca, P.; Tumino, N.; Mariotti, F.R.; Mingari, M.C.; Pende, D.; et al. Inhibitory receptors and checkpoints in human NK cells, implications for the immunotherapy of cancer. Front. Immunol. 2020, 11, 2156. [Google Scholar] [CrossRef]
- Schnell, A.; Bod, L.; Madi, A.; Kuchroo, V.K. The yin and yang of co-inhibitory receptors: Toward anti-tumor immunity without autoimmunity. Cell Res. 2020, 30, 285–299. [Google Scholar] [CrossRef] [Green Version]
- Tsubata, T. Ligand recognition determines the role of inhibitory B cell co-receptors in the regulation of B cell homeostasis and autoimmunity. Front. Immunol. 2018, 9, 2276. [Google Scholar] [CrossRef]
- Nakamura, K.; Smyth, M.J. Myeloid immunosuppression and immune checkpoints in the tumor microenvironment. Cell Mol. Immunol. 2020, 17, 1–12. [Google Scholar] [CrossRef]
- Van den Berg, T.K.; Valerius, T. Myeloid immune-checkpoint inhibition enters the clinical stage. Nat. Rev. Clin. Oncol. 2019, 16, 275–276. [Google Scholar] [CrossRef]
- Lin, C.H.; Yeh, Y.C.; Yang, K.D. Functions and therapeutic targets of Siglec-mediated infections, inflammations and cancers. J. Med. Assoc. 2021, 120, 5–24. [Google Scholar] [CrossRef]
- Ong, E.Z.; Chan, K.R.; Ooi, E.E. Viral manipulation of host inhibitory receptor signaling for immune evasion. PLoS Pathog. 2016, 12, e1005776. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ravetch, J.V.; Lanier, L.L. Immune inhibitory receptors. Science 2000, 290, 84–89. [Google Scholar] [CrossRef] [PubMed]
- Shlapatska, L.M.; Mikhalap, S.V.; Berdova, A.G.; Zelensky, O.M.; Yun, T.J.; Nichols, K.E.; Clark, E.A.; Sidorenko, S.P. CD150 association with either the SH2-containing inositol phosphatase or the SH2-containing protein tyrosine phosphatase is regulated by the adaptor protein SH2D1A. J. Immunol. 2001, 166, 5480–5487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Munitz, A. Inhibitory receptors on myeloid cells: New targets for therapy? Pharmacol. Ther. 2010, 125, 128–137. [Google Scholar] [CrossRef]
- Bournazos, S. IgG Fc receptors: Evolutionary considerations. Curr. Top. Microbiol. Immunol. 2019, 423, 1–11. [Google Scholar]
- Borges, L.; Cosman, D. LIRs/ILTs/MIRs, inhibitory and stimulatory Ig-superfamily receptors expressed in myeloid and lymphoid cells. Cytokine Growth Factor Rev. 2000, 11, 209–217. [Google Scholar] [CrossRef]
- Meyaard, L.; Adema, G.J.; Chang, C.; Woollatt, E.; Sutherland, G.R.; Lanier, L.L.; Phillips, J.H. LAIR-1, a novel inhibitory receptor expressed on human mononuclear leukocytes. Immunity 1997, 7, 283–290. [Google Scholar] [CrossRef]
- Barclay, A.N.; Van den Berg, T.K. The interaction between signal regulatory protein alpha (SIRPalpha) and CD47: Structure, function, and therapeutic target. Annu. Rev. Immunol. 2014, 32, 25–50. [Google Scholar] [CrossRef]
- Borrego, F. The CD300 molecules: An emerging family of regulators of the immune system. Blood 2013, 121, 1951–1960. [Google Scholar] [CrossRef] [Green Version]
- Crocker, P.R.; Paulson, J.C.; Varki, A. Siglecs and their roles in the immune system. Nat. Rev. Immunol. 2007, 7, 255–266. [Google Scholar] [CrossRef]
- Robinson, M.J.; Sancho, D.; Slack, E.C.; LeibundGut-Landmann, S.; Reis e Sousa, C. Myeloid C-type lectins in innate immunity. Nat. Immunol. 2006, 7, 1258–1265. [Google Scholar] [CrossRef] [PubMed]
- Kanazawa, N.; Tashiro, K.; Miyachi, Y. Signaling and immune regulatory role of the dendritic cell immunoreceptor (DCIR) family lectins: DCIR, DCAR, dectin-2 and BDCA-2. Immunobiology 2004, 209, 179–190. [Google Scholar] [CrossRef] [PubMed]
- Abramson, J.; Xu, R.; Pecht, I. An unusual inhibitory receptor—The mast cell function-associated antigen (MAFA). Mol. Immunol. 2002, 38, 1307–1313. [Google Scholar] [CrossRef]
- Fong, D.C.; Malbec, O.; Arock, M.; Cambier, J.C.; Fridman, W.H.; Daeron, M. Selective in vivo recruitment of the phosphatidylinositol phosphatase SHIP by phosphorylated Fc gammaRIIB during negative regulation of IgE-dependent mouse mast cell activation. Immunol. Lett. 1996, 54, 83–91. [Google Scholar] [CrossRef]
- Xu, X.; Masubuchi, T.; Cai, Q.; Zhao, Y.; Hui, E. Molecular features underlying differential SHP1/SHP2 binding of immune checkpoint receptors. eLife 2021, 10, e74276. [Google Scholar] [CrossRef]
- Lu, W.; Gong, D.; Bar-Sagi, D.; Cole, P.A. Site-specific incorporation of a phosphotyrosine mimetic reveals a role for tyrosine phosphorylation of SHP-2 in cell signaling. Mol. Cell 2001, 8, 759–769. [Google Scholar] [CrossRef]
- Zhang, Z.; Shen, K.; Lu, W.; Cole, P.A. The role of C-terminal tyrosine phosphorylation in the regulation of SHP-1 explored via expressed protein ligation. J. Biol. Chem. 2003, 278, 4668–4674. [Google Scholar] [CrossRef] [Green Version]
- Veillette, A.; Thibaudeau, E.; Latour, S. High expression of inhibitory receptor SHPS-1 and its association with protein-tyrosine phosphatase SHP-1 in macrophages. J. Biol. Chem. 1998, 273, 22719–22728. [Google Scholar] [CrossRef] [Green Version]
- Sayos, J.; Martinez-Barriocanal, A.; Kitzig, F.; Bellon, T.; Lopez-Botet, M. Recruitment of C-terminal Src kinase by the leukocyte inhibitory receptor CD85j. Biochem. Biophys. Res. Commun. 2004, 324, 640–647. [Google Scholar] [CrossRef]
- Zhang, S.; Phillips, J.H. Identification of tyrosine residues crucial for CD200R-mediated inhibition of mast cell activation. J. Leukoc. Biol. 2006, 79, 363–368. [Google Scholar] [CrossRef]
- Dietrich, J.; Nakajima, H.; Colonna, M. Human inhibitory and activating Ig-like receptors which modulate the function of myeloid cells. Microbes Infect. 2000, 2, 323–329. [Google Scholar] [CrossRef]
- Brown, E.J. Leukocyte migration: Dismantling inhibition. Trends Cell Biol. 2005, 15, 393–395. [Google Scholar] [CrossRef] [PubMed]
- Cannons, J.L.; Tangye, S.G.; Schwartzberg, P.L. SLAM family receptors and SAP adaptors in immunity. Annu. Rev. Immunol. 2011, 29, 665–705. [Google Scholar] [CrossRef]
- Dimitrov, D.S. Virus entry: Molecular mechanisms and biomedical applications. Nat. Rev. Microbiol. 2004, 2, 109–122. [Google Scholar] [CrossRef] [PubMed]
- Vitalle, J.; Terren, I.; Orrantia, A.; Zenarruzabeitia, O.; Borrego, F. CD300 receptor family in viral infections. Eur. J. Immunol. 2019, 49, 364–374. [Google Scholar] [CrossRef] [Green Version]
- Abdallah, F.; Coindre, S.; Gardet, M.; Meurisse, F.; Naji, A.; Suganuma, N.; Abi-Rached, L.; Lambotte, O.; Favier, B. Leukocyte immunoglobulin-like receptors in regulating the immune response in infectious diseases: A window of opportunity to pathogen persistence and a sound target in therapeutics. Front. Immunol. 2021, 12, 717998. [Google Scholar] [CrossRef]
- Bermejo-Jambrina, M.; Eder, J.; Helgers, L.C.; Hertoghs, N.; Nijmeijer, B.M.; Stunnenberg, M.; Geijtenbeek, T.B.H. C-type lectin receptors in antiviral immunity and viral escape. Front. Immunol. 2018, 9, 590. [Google Scholar] [CrossRef]
- Chang, Y.C.; Nizet, V. Siglecs at the host-pathogen interface. Adv. Exp. Med. Biol. 2020, 1204, 197–214. [Google Scholar]
- Carnec, X.; Meertens, L.; Dejarnac, O.; Perera-Lecoin, M.; Hafirassou, M.L.; Kitaura, J.; Ramdasi, R.; Schwartz, O.; Amara, A. The phosphatidylserine and phosphatidylethanolamine receptor CD300a binds dengue virus and enhances infection. J. Virol. 2016, 90, 92–102. [Google Scholar] [CrossRef] [Green Version]
- Troegeler, A.; Mercier, I.; Cougoule, C.; Pietretti, D.; Colom, A.; Duval, C.; Vu Manh, T.P.; Capilla, F.; Poincloux, R.; Pingris, K.; et al. C-type lectin receptor DCIR modulates immunity to tuberculosis by sustaining type I interferon signaling in dendritic cells. Proc. Natl. Acad. Sci. USA 2017, 114, E540–E549. [Google Scholar] [CrossRef] [Green Version]
- Gummuluru, S.; Rogel, M.; Stamatatos, L.; Emerman, M. Binding of human immunodeficiency virus type 1 to immature dendritic cells can occur independently of DC-SIGN and mannose binding C-type lectin receptors via a cholesterol-dependent pathway. J. Virol. 2003, 77, 12865–12874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lambert, A.A.; Gilbert, C.; Richard, M.; Beaulieu, A.D.; Tremblay, M.J. The C-type lectin surface receptor DCIR acts as a new attachment factor for HIV-1 in dendritic cells and contributes to trans- and cis-infection pathways. Blood 2008, 112, 1299–1307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bull, C.; Nason, R.; Sun, L.; Van Coillie, J.; Madriz Sorensen, D.; Moons, S.J.; Yang, Z.; Arbitman, S.; Fernandes, S.M.; Furukawa, S.; et al. Probing the binding specificities of human Siglecs by cell-based glycan arrays. Proc. Natl. Acad. Sci. USA 2021, 118, e2026102118. [Google Scholar] [CrossRef]
- Zou, Z.; Chastain, A.; Moir, S.; Ford, J.; Trandem, K.; Martinelli, E.; Cicala, C.; Crocker, P.; Arthos, J.; Sun, P.D. Siglecs facilitate HIV-1 infection of macrophages through adhesion with viral sialic acids. PLoS ONE 2011, 6, e24559. [Google Scholar] [CrossRef] [Green Version]
- Ferreira, A.R.; Ramos, B.; Nunes, A.; Ribeiro, D. Hepatitis C virus: Evading the intracellular innate immunity. J. Clin. Med. 2020, 9, 790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuipery, A.; Gehring, A.J.; Isogawa, M. Mechanisms of HBV immune evasion. Antivir. Res 2020, 179, 104816. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Langer, S.; Zhang, Z.; Herbert, K.M.; Yoh, S.; Konig, R.; Chanda, S.K. Sensor sensibility-HIV-1 and the innate immune response. Cells 2020, 9, 254. [Google Scholar] [CrossRef] [Green Version]
- Berry, R.; Watson, G.M.; Jonjic, S.; Degli-Esposti, M.A.; Rossjohn, J. Modulation of innate and adaptive immunity by cytomegaloviruses. Nat. Rev. Immunol. 2020, 20, 113–127. [Google Scholar] [CrossRef]
- Veglia, F.; Sanseviero, E.; Gabrilovich, D.I. Myeloid-derived suppressor cells in the era of increasing myeloid cell diversity. Nat. Rev. Immunol. 2021, 21, 485–498. [Google Scholar] [CrossRef]
- Gabrilovich, D.I.; Nagaraj, S. Myeloid-derived suppressor cells as regulators of the immune system. Nat. Rev. Immunol. 2009, 9, 162–174. [Google Scholar] [CrossRef]
- Tacke, R.S.; Lee, H.C.; Goh, C.; Courtney, J.; Polyak, S.J.; Rosen, H.R.; Hahn, Y.S. Myeloid suppressor cells induced by hepatitis C virus suppress T-cell responses through the production of reactive oxygen species. Hepatology 2012, 55, 343–353. [Google Scholar] [CrossRef] [PubMed]
- Pallett, L.J.; Gill, U.S.; Quaglia, A.; Sinclair, L.V.; Jover-Cobos, M.; Schurich, A.; Singh, K.P.; Thomas, N.; Das, A.; Chen, A.; et al. Metabolic regulation of hepatitis B immunopathology by myeloid-derived suppressor cells. Nat. Med. 2015, 21, 591–600. [Google Scholar] [CrossRef] [Green Version]
- Vollbrecht, T.; Stirner, R.; Tufman, A.; Roider, J.; Huber, R.M.; Bogner, J.R.; Lechner, A.; Bourquin, C.; Draenert, R. Chronic progressive HIV-1 infection is associated with elevated levels of myeloid-derived suppressor cells. AIDS 2012, 26, F31–F37. [Google Scholar] [CrossRef] [Green Version]
- Nelemans, T.; Kikkert, M. Viral innate immune evasion and the pathogenesis of emerging RNA virus infections. Viruses 2019, 11, 961. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Telcian, A.G.; Laza-Stanca, V.; Edwards, M.R.; Harker, J.A.; Wang, H.; Bartlett, N.W.; Mallia, P.; Zdrenghea, M.T.; Kebadze, T.; Coyle, A.J.; et al. RSV-induced bronchial epithelial cell PD-L1 expression inhibits CD8+ T cell nonspecific antiviral activity. J. Infect. Dis. 2011, 203, 85–94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lafon, M.; Megret, F.; Meuth, S.G.; Simon, O.; Velandia Romero, M.L.; Lafage, M.; Chen, L.; Alexopoulou, L.; Flavell, R.A.; Prehaud, C.; et al. Detrimental contribution of the immuno-inhibitor B7-H1 to rabies virus encephalitis. J. Immunol. 2008, 180, 7506–7515. [Google Scholar] [CrossRef]
- Sander, L.E.; Sackett, S.D.; Dierssen, U.; Beraza, N.; Linke, R.P.; Muller, M.; Blander, J.M.; Tacke, F.; Trautwein, C. Hepatic acute-phase proteins control innate immune responses during infection by promoting myeloid-derived suppressor cell function. J. Exp. Med. 2010, 207, 1453–1464. [Google Scholar] [CrossRef] [Green Version]
- Agrati, C.; Sacchi, A.; Bordoni, V.; Cimini, E.; Notari, S.; Grassi, G.; Casetti, R.; Tartaglia, E.; Lalle, E.; D’Abramo, A.; et al. Expansion of myeloid-derived suppressor cells in patients with severe coronavirus disease (COVID-19). Cell Death Differ. 2020, 27, 3196–3207. [Google Scholar] [CrossRef]
- Thompson, E.A.; Cascino, K.; Ordonez, A.A.; Zhou, W.; Vaghasia, A.; Hamacher-Brady, A.; Brady, N.R.; Sun, I.H.; Wang, R.; Rosenberg, A.Z.; et al. Metabolic programs define dysfunctional immune responses in severe COVID-19 patients. Cell Rep. 2021, 34, 108863. [Google Scholar] [CrossRef]
- Vlad, G.; Piazza, F.; Colovai, A.; Cortesini, R.; Della Pietra, F.; Suciu-Foca, N.; Manavalan, J.S. Interleukin-10 induces the upregulation of the inhibitory receptor ILT4 in monocytes from HIV positive individuals. Hum. Immunol. 2003, 64, 483–489. [Google Scholar] [CrossRef]
- Chen, W.; Han, C.; Xie, B.; Hu, X.; Yu, Q.; Shi, L.; Wang, Q.; Li, D.; Wang, J.; Zheng, P.; et al. Induction of Siglec-G by RNA viruses inhibits the innate immune response by promoting RIG-I degradation. Cell 2013, 152, 467–478. [Google Scholar] [CrossRef] [Green Version]
- Saheb Sharif-Askari, N.; Saheb Sharif-Askari, F.; Mdkhana, B.; Al Heialy, S.; Alsafar, H.S.; Hamoudi, R.; Hamid, Q.; Halwani, R. Enhanced expression of immune checkpoint receptors during SARS-CoV-2 viral infection. Mol. Methods Clin. Dev. 2021, 20, 109–121. [Google Scholar] [CrossRef] [PubMed]
- Waldman, A.D.; Fritz, J.M.; Lenardo, M.J. A guide to cancer immunotherapy: From T cell basic science to clinical practice. Nat. Rev. Immunol. 2020, 20, 651–668. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Wang, X.; Jin, T.; Tian, Y.; Dai, C.; Widarma, C.; Song, R.; Xu, F. Immune checkpoint molecules in natural killer cells as potential targets for cancer immunotherapy. Signal. Transduct. Target. 2020, 5, 250. [Google Scholar] [CrossRef] [PubMed]
- Samaridis, J.; Colonna, M. Cloning of novel immunoglobulin superfamily receptors expressed on human myeloid and lymphoid cells: Structural evidence for new stimulatory and inhibitory pathways. Eur. J. Immunol. 1997, 27, 660–665. [Google Scholar] [CrossRef]
- Katz, H.R. Inhibition of inflammatory responses by leukocyte Ig-like receptors. Adv. Immunol. 2006, 91, 251–272. [Google Scholar]
- Mori, Y.; Tsuji, S.; Inui, M.; Sakamoto, Y.; Endo, S.; Ito, Y.; Fujimura, S.; Koga, T.; Nakamura, A.; Takayanagi, H.; et al. Inhibitory immunoglobulin-like receptors LILRB and PIR-B negatively regulate osteoclast development. J. Immunol. 2008, 181, 4742–4751. [Google Scholar] [CrossRef] [Green Version]
- Kang, X.; Kim, J.; Deng, M.; John, S.; Chen, H.; Wu, G.; Phan, H.; Zhang, C.C. Inhibitory leukocyte immunoglobulin-like receptors: Immune checkpoint proteins and tumor sustaining factors. Cell Cycle 2016, 15, 25–40. [Google Scholar] [CrossRef] [Green Version]
- McIntire, R.H.; Sifers, T.; Platt, J.S.; Ganacias, K.G.; Langat, D.K.; Hunt, J.S. Novel HLA-G-binding leukocyte immunoglobulin-like receptor (LILR) expression patterns in human placentas and umbilical cords. Placenta 2008, 29, 631–638. [Google Scholar] [CrossRef] [Green Version]
- Kim-Schulze, S.; Seki, T.; Vlad, G.; Scotto, L.; Fan, J.; Colombo, P.C.; Liu, J.; Cortesini, R.; Suciu-Foca, N. Regulation of ILT3 gene expression by processing of precursor transcripts in human endothelial cells. Am. J. Transpl. 2006, 6, 76–82. [Google Scholar] [CrossRef]
- Van der Touw, W.; Chen, H.M.; Pan, P.Y.; Chen, S.H. LILRB receptor-mediated regulation of myeloid cell maturation and function. Cancer Immunol. Immunother. 2017, 66, 1079–1087. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beyer, M.; Mallmann, M.R.; Xue, J.; Staratschek-Jox, A.; Vorholt, D.; Krebs, W.; Sommer, D.; Sander, J.; Mertens, C.; Nino-Castro, A.; et al. High-resolution transcriptome of human macrophages. PLoS ONE 2012, 7, e45466. [Google Scholar] [CrossRef] [PubMed]
- Beinhauer, B.G.; McBride, J.M.; Graf, P.; Pursch, E.; Bongers, M.; Rogy, M.; Korthauer, U.; de Vries, J.E.; Aversa, G.; Jung, T. Interleukin 10 regulates cell surface and soluble LIR-2 (CD85d) expression on dendritic cells resulting in T cell hyporesponsiveness in vitro. Eur. J. Immunol. 2004, 34, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Manavalan, J.S.; Rossi, P.C.; Vlad, G.; Piazza, F.; Yarilina, A.; Cortesini, R.; Mancini, D.; Suciu-Foca, N. High expression of ILT3 and ILT4 is a general feature of tolerogenic dendritic cells. Transpl. Immunol. 2003, 11, 245–258. [Google Scholar] [CrossRef]
- Banchereau, J.; Zurawski, S.; Thompson-Snipes, L.; Blanck, J.P.; Clayton, S.; Munk, A.; Cao, Y.; Wang, Z.; Khandelwal, S.; Hu, J.; et al. Immunoglobulin-like transcript receptors on human dermal CD14+ dendritic cells act as a CD8-antagonist to control cytotoxic T cell priming. Proc. Natl. Acad. Sci. USA 2012, 109, 18885–18890. [Google Scholar] [CrossRef] [Green Version]
- Barkal, A.A.; Weiskopf, K.; Kao, K.S.; Gordon, S.R.; Rosental, B.; Yiu, Y.Y.; George, B.M.; Markovic, M.; Ring, N.G.; Tsai, J.M.; et al. Engagement of MHC class I by the inhibitory receptor LILRB1 suppresses macrophages and is a target of cancer immunotherapy. Nat. Immunol. 2018, 19, 76–84. [Google Scholar] [CrossRef] [PubMed]
- Colonna, M.; Samaridis, J.; Cella, M.; Angman, L.; Allen, R.L.; O’Callaghan, C.A.; Dunbar, R.; Ogg, G.S.; Cerundolo, V.; Rolink, A. Human myelomonocytic cells express an inhibitory receptor for classical and nonclassical MHC class I molecules. J. Immunol. 1998, 160, 3096–10300. [Google Scholar]
- Tenca, C.; Merlo, A.; Merck, E.; Bates, E.E.; Saverino, D.; Simone, R.; Zarcone, D.; Trinchieri, G.; Grossi, C.E.; Ciccone, E. CD85j (leukocyte Ig-like receptor-1/Ig-like transcript 2) inhibits human osteoclast-associated receptor-mediated activation of human dendritic cells. J. Immunol. 2005, 174, 6757–6763. [Google Scholar] [CrossRef] [Green Version]
- Young, N.T.; Waller, E.C.; Patel, R.; Roghanian, A.; Austyn, J.M.; Trowsdale, J. The inhibitory receptor LILRB1 modulates the differentiation and regulatory potential of human dendritic cells. Blood 2008, 111, 3090–3096. [Google Scholar] [CrossRef]
- Kubagawa, H.; Burrows, P.D.; Cooper, M.D. A novel pair of immunoglobulin-like receptors expressed by B cells and myeloid cells. Proc. Natl. Acad. Sci. USA 1997, 94, 5261–5266. [Google Scholar] [CrossRef] [Green Version]
- Endo, S.; Sakamoto, Y.; Kobayashi, E.; Nakamura, A.; Takai, T. Regulation of cytotoxic T lymphocyte triggering by PIR-B on dendritic cells. Proc. Natl. Acad. Sci. USA 2008, 105, 14515–14520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, G.; Pan, P.Y.; Eisenstein, S.; Divino, C.M.; Lowell, C.A.; Takai, T.; Chen, S.H. Paired immunoglobin-like receptor-B regulates the suppressive function and fate of myeloid-derived suppressor cells. Immunity 2011, 34, 385–395. [Google Scholar] [CrossRef] [Green Version]
- Zhang, F.; Zheng, J.; Kang, X.; Deng, M.; Lu, Z.; Kim, J.; Zhang, C. Inhibitory leukocyte immunoglobulin-like receptors in cancer development. Sci. China Life Sci. 2015, 58, 1216–1225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lichterfeld, M.; Kavanagh, D.G.; Williams, K.L.; Moza, B.; Mui, S.K.; Miura, T.; Sivamurthy, R.; Allgaier, R.; Pereyra, F.; Trocha, A.; et al. A viral CTL escape mutation leading to immunoglobulin-like transcript 4-mediated functional inhibition of myelomonocytic cells. J. Exp. Med. 2007, 204, 2813–2824. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lichterfeld, M.; Yu, X.G. The emerging role of leukocyte immunoglobulin-like receptors (LILRs) in HIV-1 infection. J. Leukoc. Biol. 2012, 91, 27–33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, J.; Goedert, J.J.; Sundberg, E.J.; Cung, T.D.; Burke, P.S.; Martin, M.P.; Preiss, L.; Lifson, J.; Lichterfeld, M.; Carrington, M.; et al. HLA-B*35-Px-mediated acceleration of HIV-1 infection by increased inhibitory immunoregulatory impulses. J. Exp. Med. 2009, 206, 2959–2966. [Google Scholar] [CrossRef] [Green Version]
- Huang, J.; Al-Mozaini, M.; Rogich, J.; Carrington, M.F.; Seiss, K.; Pereyra, F.; Lichterfeld, M.; Yu, X.G. Systemic inhibition of myeloid dendritic cells by circulating HLA class I molecules in HIV-1 infection. Retrovirology 2012, 9, 11. [Google Scholar] [CrossRef] [Green Version]
- Huang, J.; Burke, P.; Yang, Y.; Seiss, K.; Beamon, J.; Cung, T.; Toth, I.; Pereyra, F.; Lichterfeld, M.; Yu, X.G. Soluble HLA-G inhibits myeloid dendritic cell function in HIV-1 infection by interacting with leukocyte immunoglobulin-like receptor B2. J. Virol. 2010, 84, 10784–10791. [Google Scholar] [CrossRef] [Green Version]
- Bashirova, A.A.; Martin-Gayo, E.; Jones, D.C.; Qi, Y.; Apps, R.; Gao, X.; Burke, P.S.; Taylor, C.J.; Rogich, J.; Wolinsky, S.; et al. LILRB2 interaction with HLA class I correlates with control of HIV-1 infection. PLoS Genet. 2014, 10, e1004196. [Google Scholar] [CrossRef] [Green Version]
- Chan, K.R.; Ong, E.Z.; Tan, H.C.; Zhang, S.L.; Zhang, Q.; Tang, K.F.; Kaliaperumal, N.; Lim, A.P.; Hibberd, M.L.; Chan, S.H.; et al. Leukocyte immunoglobulin-like receptor B1 is critical for antibody-dependent dengue. Proc. Natl. Acad. Sci. USA 2014, 111, 2722–2727. [Google Scholar] [CrossRef] [Green Version]
- Nimmerjahn, F.; Lux, A. LILR-B1 blocks activating FcgammaR signaling to allow antibody dependent enhancement of dengue virus infection. Proc. Natl. Acad. Sci. USA 2014, 111, 2404–2405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bates, E.E.; Fournier, N.; Garcia, E.; Valladeau, J.; Durand, I.; Pin, J.J.; Zurawski, S.M.; Patel, S.; Abrams, J.S.; Lebecque, S.; et al. APCs express DCIR, a novel C-type lectin surface receptor containing an immunoreceptor tyrosine-based inhibitory motif. J. Immunol. 1999, 163, 1973–1983. [Google Scholar] [PubMed]
- Meyer-Wentrup, F.; Cambi, A.; Joosten, B.; Looman, M.W.; de Vries, I.J.; Figdor, C.G.; Adema, G.J. DCIR is endocytosed into human dendritic cells and inhibits TLR8-mediated cytokine production. J. Leukoc. Biol. 2009, 85, 518–525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, X.; Shen, Y.; Hu, W.; Chen, J.; Wu, T.; Sun, X.; Yu, J.; Wu, T.; Chen, W. DCIR negatively regulates CpG-ODN-induced IL-1beta and IL-6 production. Mol. Immunol. 2015, 68, 641–647. [Google Scholar] [CrossRef]
- Lambert, A.A.; Barabe, F.; Gilbert, C.; Tremblay, M.J. DCIR-mediated enhancement of HIV-1 infection requires the ITIM-associated signal transduction pathway. Blood 2011, 117, 6589–6599. [Google Scholar] [CrossRef] [PubMed]
- Mfunyi, C.M.; Vaillancourt, M.; Vitry, J.; Nsimba Batomene, T.R.; Posvandzic, A.; Lambert, A.A.; Gilbert, C. Exosome release following activation of the dendritic cell immunoreceptor: A potential role in HIV-1 pathogenesis. Virology 2015, 484, 103–112. [Google Scholar] [CrossRef] [Green Version]
- Meyer-Wentrup, F.; Benitez-Ribas, D.; Tacken, P.J.; Punt, C.J.; Figdor, C.G.; de Vries, I.J.; Adema, G.J. Targeting DCIR on human plasmacytoid dendritic cells results in antigen presentation and inhibits IFN-alpha production. Blood 2008, 111, 4245–4253. [Google Scholar] [CrossRef] [Green Version]
- Lenza, M.P.; Atxabal, U.; Oyenarte, I.; Jimenez-Barbero, J.; Ereno-Orbea, J. Current status on therapeutic molecules targeting siglec receptors. Cells 2020, 9, 2691. [Google Scholar] [CrossRef]
- Macauley, M.S.; Crocker, P.R.; Paulson, J.C. Siglec-mediated regulation of immune cell function in disease. Nat. Rev. Immunol. 2014, 14, 653–666. [Google Scholar] [CrossRef] [Green Version]
- Hoffmann, A.; Kerr, S.; Jellusova, J.; Zhang, J.; Weisel, F.; Wellmann, U.; Winkler, T.H.; Kneitz, B.; Crocker, P.R.; Nitschke, L. Siglec-G is a B1 cell-inhibitory receptor that controls expansion and calcium signaling of the B1 cell population. Nat. Immunol. 2007, 8, 695–704. [Google Scholar] [CrossRef]
- Tsai, T.Y.; Huang, M.T.; Sung, P.S.; Peng, C.Y.; Tao, M.H.; Yang, H.I.; Chang, W.C.; Yang, A.S.; Yu, C.M.; Lin, Y.P.; et al. SIGLEC-3 (CD33) serves as an immune checkpoint receptor for HBV infection. J. Clin. Investig. 2021, 131, e141965. [Google Scholar] [CrossRef] [PubMed]
- Tsai, R.K.; Discher, D.E. Inhibition of “self” engulfment through deactivation of myosin-II at the phagocytic synapse between human cells. J. Cell Biol. 2008, 180, 989–1003. [Google Scholar] [CrossRef] [Green Version]
- Matozaki, T.; Murata, Y.; Okazawa, H.; Ohnishi, H. Functions and molecular mechanisms of the CD47-SIRPalpha signalling pathway. Trends Cell Biol. 2009, 19, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Wen, W.; Tang, L.; Qin, C.J.; Lin, Y.; Zhang, H.L.; Wu, H.; Ashton, C.; Wu, H.P.; Ding, J.; et al. Inhibition of SIRPalpha in dendritic cells potentiates potent antitumor immunity. Oncoimmunology 2016, 5, e1183850. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Folkes, A.S.; Feng, M.; Zain, J.M.; Abdulla, F.; Rosen, S.T.; Querfeld, C. Targeting CD47 as a cancer therapeutic strategy: The cutaneous T-cell lymphoma experience. Curr. Opin. Oncol. 2018, 30, 332–667. [Google Scholar] [CrossRef] [PubMed]
- Veillette, A.; Chen, J. SIRPalpha-CD47 immune checkpoint blockade in anticancer therapy. Trends Immunol. 2018, 39, 173–184. [Google Scholar] [CrossRef]
- Pengam, S.; Durand, J.; Usal, C.; Gauttier, V.; Dilek, N.; Martinet, B.; Daguin, V.; Mary, C.; Thepenier, V.; Teppaz, G.; et al. SIRPalpha/CD47 axis controls the maintenance of transplant tolerance sustained by myeloid-derived suppressor cells. Am. J. Transpl. 2019, 19, 3263–3275. [Google Scholar] [CrossRef]
- Willingham, S.B.; Volkmer, J.P.; Gentles, A.J.; Sahoo, D.; Dalerba, P.; Mitra, S.S.; Wang, J.; Contreras-Trujillo, H.; Martin, R.; Cohen, J.D.; et al. The CD47-signal regulatory protein alpha (SIRPa) interaction is a therapeutic target for human solid tumors. Proc. Natl. Acad. Sci. USA 2012, 109, 6662–6667. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.; Shao, R.; Huang, H.; Wang, X.; Rong, Z.; Lin, Y. Engineering macrophages to phagocytose cancer cells by blocking the CD47/SIRPa axis. Cancer Med. 2019, 8, 4245–4253. [Google Scholar] [CrossRef] [Green Version]
- Sikic, B.I.; Lakhani, N.; Patnaik, A.; Shah, S.A.; Chandana, S.R.; Rasco, D.; Colevas, A.D.; O’Rourke, T.; Narayanan, S.; Papadopoulos, K.; et al. First-in-human, first-in-class phase I trial of the anti-CD47 antibody Hu5F9-G4 in patients with advanced cancers. J. Clin. Oncol. 2019, 37, 946–953. [Google Scholar] [CrossRef]
- Cham, L.B.; Adomati, T.; Li, F.; Ali, M.; Lang, K.S. CD47 as a potential target to therapy for infectious diseases. Antibodies 2020, 9, 44. [Google Scholar] [CrossRef] [PubMed]
- Cham, L.B.; Torrez Dulgeroff, L.B.; Tal, M.C.; Adomati, T.; Li, F.; Bhat, H.; Huang, A.; Lang, P.A.; Moreno, M.E.; Rivera, J.M.; et al. Immunotherapeutic blockade of CD47 inhibitory signaling enhances innate and adaptive immune responses to viral infection. Cell Rep. 2020, 31, 107494. [Google Scholar] [CrossRef] [PubMed]
- Tal, M.C.; Torrez Dulgeroff, L.B.; Myers, L.; Cham, L.B.; Mayer-Barber, K.D.; Bohrer, A.C.; Castro, E.; Yiu, Y.Y.; Lopez Angel, C.; Pham, E.; et al. Upregulation of CD47 is a host checkpoint tesponse to pathogen recognition. mBio 2020, 11, e01293-20. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.T.; Ko, E.J.; Lee, Y.; Lee, Y.N.; Bian, Z.; Liu, Y.; Kang, S.M. CD47 plays a role as a negative regulator in inducing protective immune responses to vaccination against influenza virus. J. Virol. 2016, 90, 6746–6758. [Google Scholar] [CrossRef] [Green Version]
- McLaughlin, K.M.; Bojkova, D.; Kandler, J.D.; Bechtel, M.; Reus, P.; Le, T.; Rothweiler, F.; Wagner, J.U.G.; Weigert, A.; Ciesek, S.; et al. A potential role of the CD47/SIRPalpha axis in COVID-19 pathogenesis. Curr. Issues Mol. Biol. 2021, 43, 1212–1225. [Google Scholar] [CrossRef]
- Chang, C.C.; Ciubotariu, R.; Manavalan, J.S.; Yuan, J.; Colovai, A.I.; Piazza, F.; Lederman, S.; Colonna, M.; Cortesini, R.; Dalla-Favera, R.; et al. Tolerization of dendritic cells by T(S) cells: The crucial role of inhibitory receptors ILT3 and ILT4. Nat. Immunol. 2002, 3, 237–243. [Google Scholar] [CrossRef]
- Yu, K.; Davidson, C.L.; Wojtowicz, A.; Lisboa, L.; Wang, T.; Airo, A.M.; Villard, J.; Buratto, J.; Sandalova, T.; Achour, A.; et al. LILRB1 polymorphisms influence posttransplant HCMV susceptibility and ligand interactions. J. Clin. Investig. 2018, 128, 1523–1537. [Google Scholar] [CrossRef] [Green Version]
- Angata, T. Associations of genetic polymorphisms of Siglecs with human diseases. Glycobiology 2014, 24, 785–793. [Google Scholar] [CrossRef] [Green Version]
- Sharma, P.; Allison, J.P. The future of immune checkpoint therapy. Science 2015, 348, 56–61. [Google Scholar] [CrossRef]
- Sharma, P.; Allison, J.P. Immune checkpoint targeting in cancer therapy: Toward combination strategies with curative potential. Cell 2015, 161, 205–214. [Google Scholar] [CrossRef] [Green Version]
- Cox, M.A.; Nechanitzky, R.; Mak, T.W. Check point inhibitors as therapies for infectious diseases. Curr. Opin. Immunol. 2017, 48, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Dyck, L.; Mills, K.H.G. Immune checkpoints and their inhibition in cancer and infectious diseases. Eur. J. Immunol. 2017, 47, 765–779. [Google Scholar] [CrossRef] [PubMed]
- Chao, M.P.; Alizadeh, A.A.; Tang, C.; Myklebust, J.H.; Varghese, B.; Gill, S.; Jan, M.; Cha, A.C.; Chan, C.K.; Tan, B.T.; et al. Anti-CD47 antibody synergizes with rituximab to promote phagocytosis and eradicate non-Hodgkin lymphoma. Cell 2010, 142, 699–713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeng, D.; Sun, Q.; Chen, A.; Fan, J.; Yang, X.; Xu, L.; Du, P.; Qiu, W.; Zhang, W.; Wang, S.; et al. A fully human anti-CD47 blocking antibody with therapeutic potential for cancer. Oncotarget 2016, 7, 83040–83050. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- LeMessurier, K.S.; Rooney, R.; Ghoneim, H.E.; Liu, B.; Li, K.; Smallwood, H.S.; Samarasinghe, A.E. Influenza A virus directly modulates mouse eosinophil responses. J. Leukoc. Biol. 2020, 108, 151–168. [Google Scholar] [CrossRef] [PubMed]
- Alaoui, L.; Palomino, G.; Zurawski, S.; Zurawski, G.; Coindre, S.; Dereuddre-Bosquet, N.; Lecuroux, C.; Goujard, C.; Vaslin, B.; Bourgeois, C.; et al. Early SIV and HIV infection promotes the LILRB2/MHC-I inhibitory axis in cDCs. Cell Mol. Life Sci. 2018, 75, 1871–1887. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Nicklin, P.; He, Y. New Targets for Antiviral Therapy: Inhibitory Receptors and Immune Checkpoints on Myeloid Cells. Viruses 2022, 14, 1144. https://doi.org/10.3390/v14061144
Liu Y, Nicklin P, He Y. New Targets for Antiviral Therapy: Inhibitory Receptors and Immune Checkpoints on Myeloid Cells. Viruses. 2022; 14(6):1144. https://doi.org/10.3390/v14061144
Chicago/Turabian StyleLiu, Yanni, Paul Nicklin, and Yuan He. 2022. "New Targets for Antiviral Therapy: Inhibitory Receptors and Immune Checkpoints on Myeloid Cells" Viruses 14, no. 6: 1144. https://doi.org/10.3390/v14061144
APA StyleLiu, Y., Nicklin, P., & He, Y. (2022). New Targets for Antiviral Therapy: Inhibitory Receptors and Immune Checkpoints on Myeloid Cells. Viruses, 14(6), 1144. https://doi.org/10.3390/v14061144





