A Review of Omics Studies on Arboviruses: Alphavirus, Orthobunyavirus and Phlebovirus
Abstract
1. Introduction
2. An Overview of the Genera
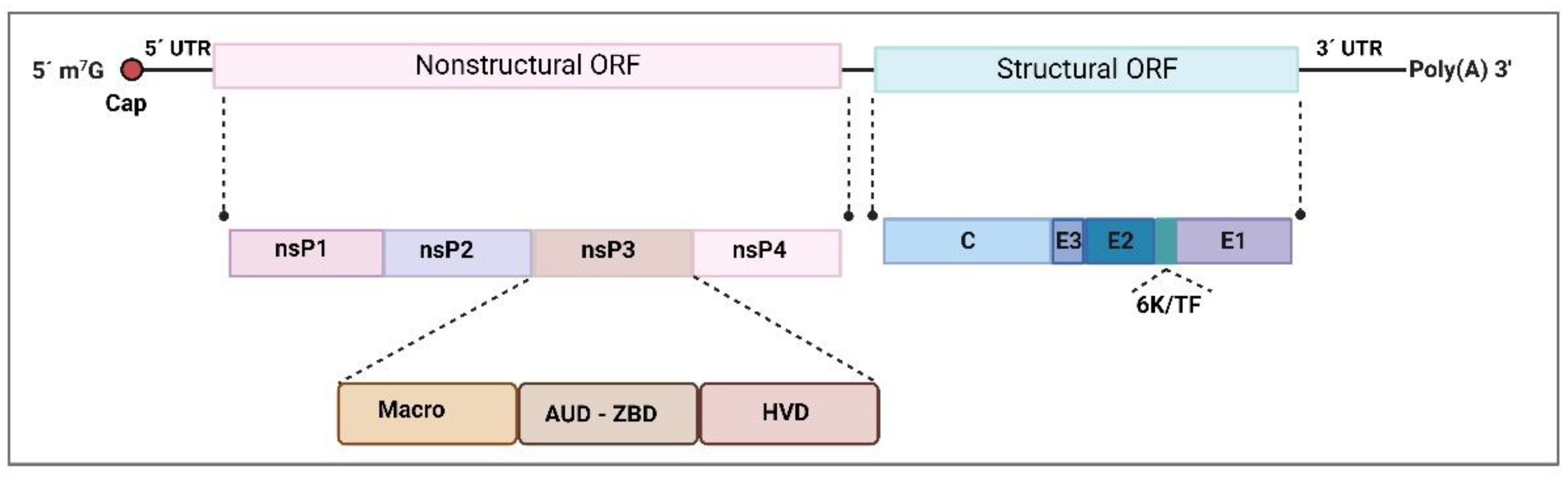
2.1. Togaviridae—Alphavirus
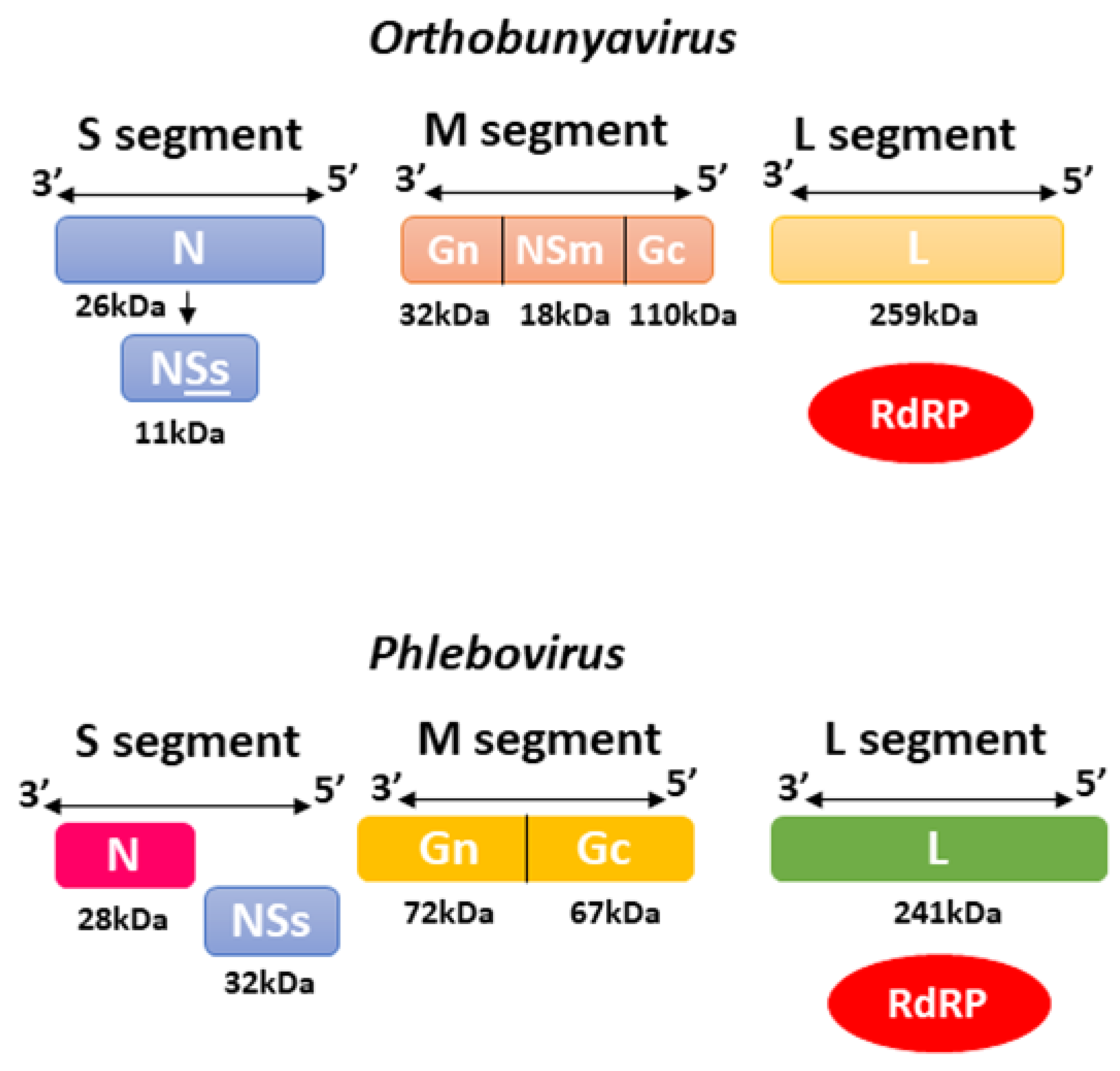
2.2. Bunyavirales—Orthobunyavirus and Phlebovirus
3. Omic’s Study across Members of the Togaviridae Family
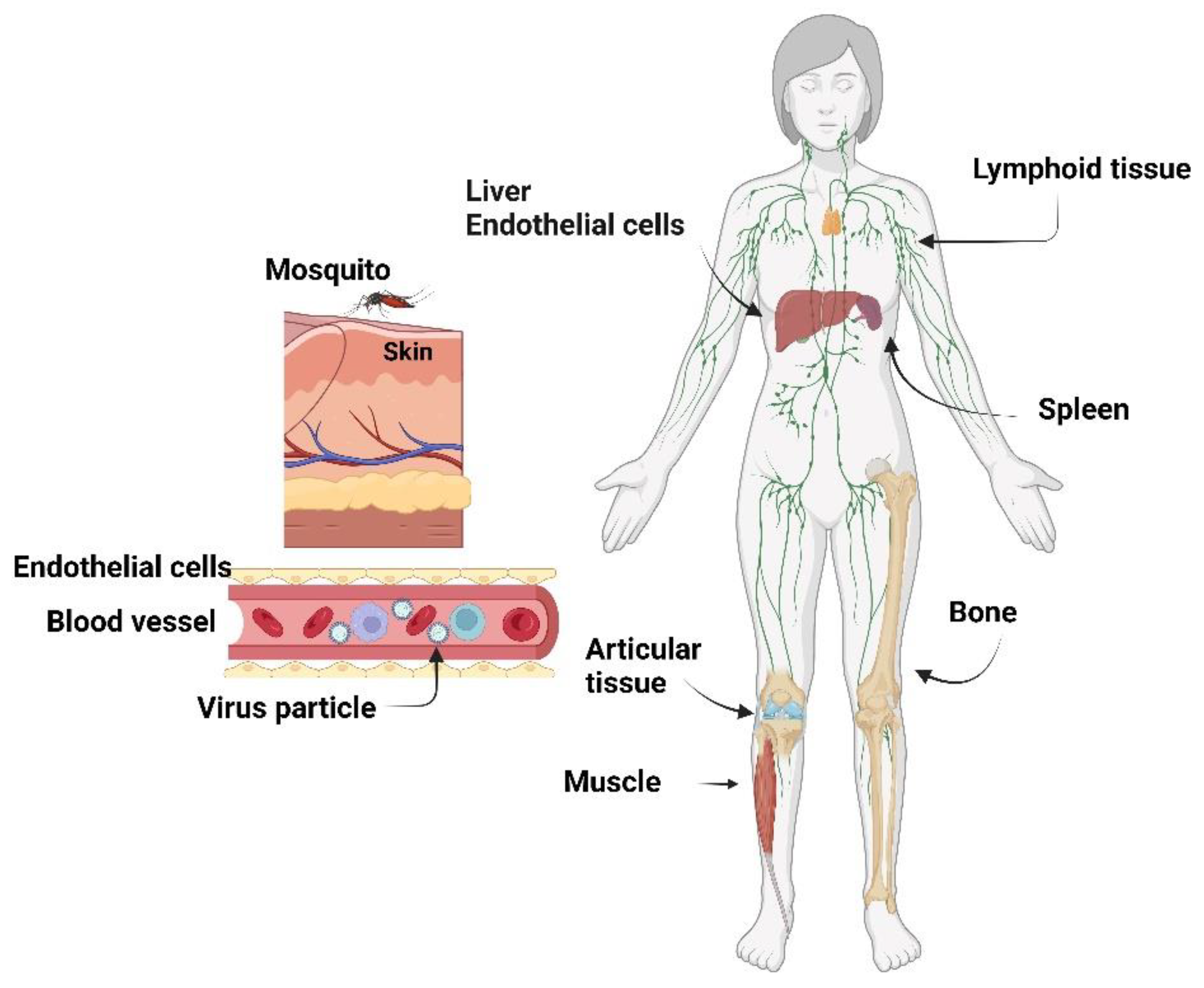
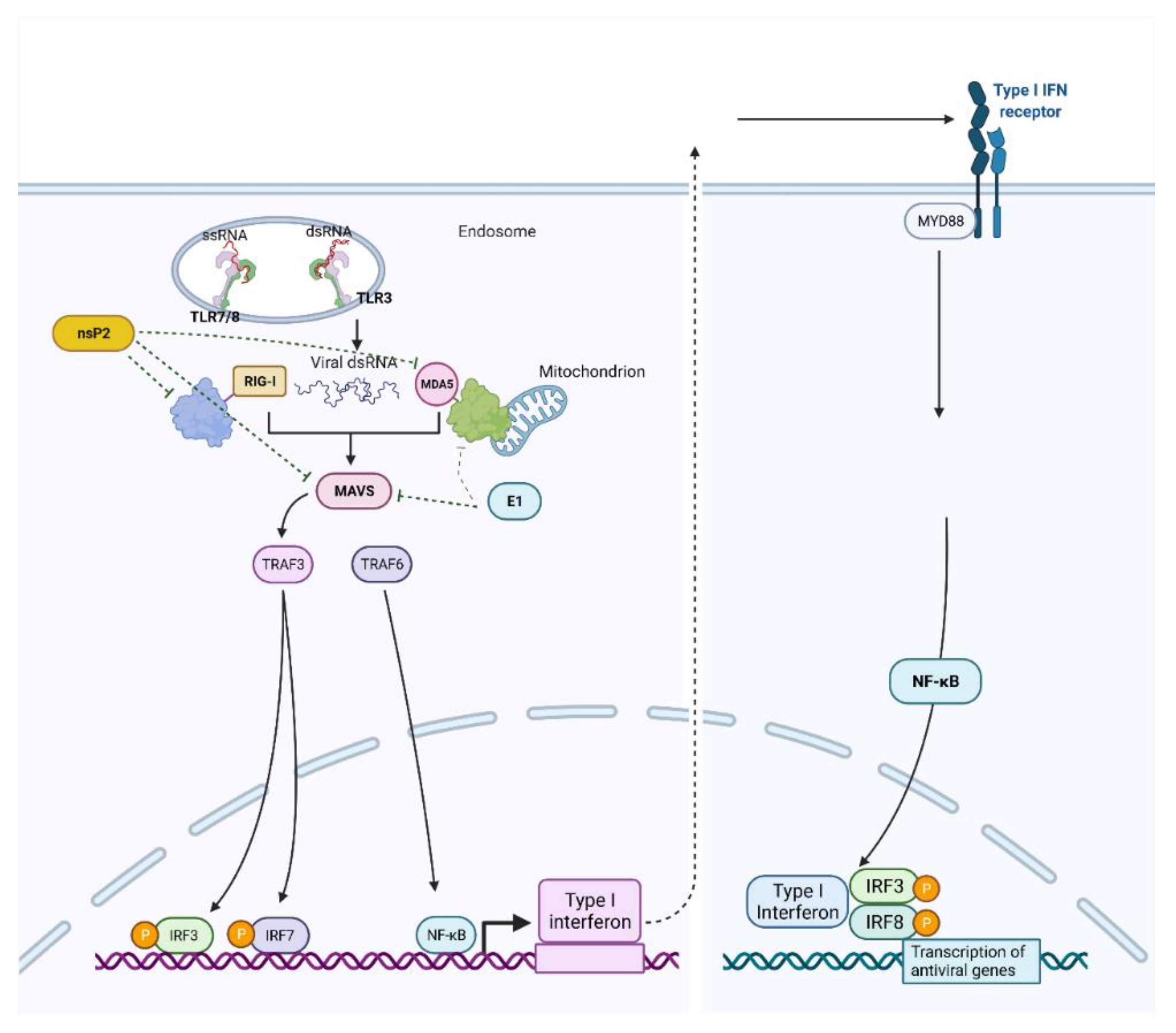
3.1. Chikungunya Virus
3.2. Mayaro Virus
4. Omic’s Study across Members of Bunyavirales Order—Orthobunyavirus and Phlebovirus
4.1. Rift Valley Fever Virus
4.2. Oropouche Virus
5. Remarks and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Young, P.R. Arboviruses: A Family on the Move. Adv. Exp. Med. Biol. 2018, 1062, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Slon-Campos, J.L.; Mongkolsapaya, J.; Screaton, G.R. The immune response against flaviviruses. Nat. Immunol. 2018, 19, 1189–1198. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.-J.S.; Higgs, S.; Vanlandingham, D.L. Emergence and re-emergence of mosquito-borne arboviruses. Curr. Opin. Virol. 2019, 34, 104–109. [Google Scholar] [CrossRef]
- Hasin, Y.; Seldin, M.; Lusis, A. Multi-omics approaches to disease. Genome Biol. 2017, 18, 83. [Google Scholar] [CrossRef] [PubMed]
- Mardis, E.R. Next-Generation DNA Sequencing Methods. Annu. Rev. Genom. Hum. Genet. 2008, 9, 387–402. [Google Scholar] [CrossRef]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, C.; Gillet, L.; Rosenberger, G.; Amon, S.; Collins, B.C.; Aebersold, R. Data-independent acquisition-based SWATH—MS for quantitative proteomics: A tutorial. Mol. Syst. Biol. 2018, 14, e8126. [Google Scholar] [CrossRef]
- Zhang, Z.; Wu, S.; Stenoien, D.L.; Pasa-Tolic, L. High-Throughput Proteomics. Annu. Rev. Anal. Chem. 2014, 7, 427–454. [Google Scholar] [CrossRef]
- Emwas, A.-H.; Roy, R.; McKay, R.T.; Tenori, L.; Saccenti, E.; Gowda, G.A.N.; Raftery, D.; Alahmari, F.; Jaremko, L.; Jaremko, M.; et al. NMR Spectroscopy for Metabolomics Research. Metabolites 2019, 9, 123. [Google Scholar] [CrossRef]
- Dettmer, K.; Aronov, P.A.; Hammock, B.D. Mass Spectrometry-based metabolomics. Mass. Spectrom. Rev. 2007, 26, 51–78. [Google Scholar] [CrossRef]
- Pinu, F.R.; Beale, D.J.; Paten, A.M.; Kouremenos, K.; Swarup, S.; Schirra, H.J.; Wishart, D. Systems Biology and Multi-Omics Integration: Viewpoints from the Metabolomics Research Community. Metabolites 2019, 9, 76. [Google Scholar] [CrossRef]
- Lopes, N.; Linhares, R.E.C.; Nozawa, C. Características gerais e epidemiologia dos arbovírus emergentes no Brasil. Rev. Pan-Amaz. Saúde 2014, 5, 55–64. [Google Scholar] [CrossRef]
- Santos, N.S.O.; Romanos, M.T.V.; Wigg, M.D. Introdução à Virologia Humana, 2nd ed.; Guanabara Koogan, Campo Grande: Rio de Janeiro, Brasil, 2008; p. 546. [Google Scholar]
- Chen, R.; Mukhopadhyay, S.; Merits, A.; Bolling, B.; Nasar, F.; Coffey, L.L.; Powers, A.; Weaver, S. ICTV Report Consortium ICTV Virus Taxonomy Profile: Togaviridae. J. Gen. Virol. 2018, 99, 761–762. [Google Scholar] [CrossRef] [PubMed]
- Cheng, R.H.; Kuhn, R.J.; Olson, N.H.; Choi, M.G.R.-K.; Smith, T.J.; Baker, T.S. Nucleocapsid and glycoprotein organization in an enveloped virus. Cell 1995, 80, 621–630. [Google Scholar] [CrossRef]
- Griffin, D. Alphaviruses. In Fields Virology, 6th ed.; Knipe, D.M., Howley, P.M., Eds.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2013; Volume 1, pp. 651–686. [Google Scholar]
- Hefti, E.; Bishop, D.H.; Dubin, D.T.; Stollar, V. 5’ nucleotide sequence of sindbis viral RNA. J. Virol. 1976, 17, 149–159. [Google Scholar] [CrossRef]
- Lavergne, A.; de Thoisy, B.; Lacoste, V.; Pascalis, H.; Pouliquen, J.F.; Mercier, V.; Tolou, H.; Dussart, P.; Morvan, J.; Talarmin, A.; et al. Mayaro virus: Complete nucleotide sequence and phylogenetic relationships with other alphaviruses. Virus Res. 2006, 117, 283–290. [Google Scholar] [CrossRef]
- Rupp, J.C.; Sokoloski, K.J.; Gebhart, N.N.; Hardy, R.W. Alphavirus RNA synthesis and non-structural protein functions. J. Gen. Virol. 2015, 96, 2483–2500. [Google Scholar] [CrossRef]
- Hyde, J.L.; Chen, R.; Trobaugh, D.W.; Diamond, M.S.; Weaver, S.C.; Klimstra, W.B.; Wilusz, J. The 5′ and 3′ ends of alphavirus RNAs—Non-coding is not non-functional. Virus Res. 2015, 206, 99–107. [Google Scholar] [CrossRef]
- Melancon, P.; Garoff, H. Processing of the Semliki Forest virus structural polyprotein: Role of the capsid protease. J. Virol. 1987, 61, 1301–1309. [Google Scholar] [CrossRef]
- Mota, M.T.D.O.; Ribeiro, M.R.; Vedovello, D.; Nogueira, M.L. Mayaro virus: A neglected arbovirus of the Americas. Futur. Virol. 2015, 10, 1109–1122. [Google Scholar] [CrossRef]
- Omar, A.; Koblet, H. Semliki forest virus particles containing only the E1 envelope glycoprotein are infectious and can induce cell-cell fusion. Virology 1988, 166, 17–23. [Google Scholar] [CrossRef]
- Götte, B.; Liu, L.; McInerney, G.M. The enigmatic alphavirus non-structural protein 3 (nsP3) revealing its secrets at last. Viruses 2018, 10, 105. [Google Scholar] [CrossRef] [PubMed]
- Wintachai, P.; Wikan, N.; Kuadkitkan, A.; Jaimipuk, T.; Ubol, S.; Pulmanausahakul, R.; Auewarakul, P.; Kasinrerk, W.; Weng, W.-Y.; Panyasrivanit, M.; et al. Identification of prohibitin as a Chikungunya virus receptor protein. J. Med. Virol. 2012, 84, 1757–1770. [Google Scholar] [CrossRef] [PubMed]
- Moller-Tank, S.; Kondratowicz, A.S.; Davey, R.; Rennert, P.D.; Maury, W. Role of the Phosphatidylserine Receptor TIM-1 in Enveloped-Virus Entry. J. Virol. 2013, 87, 8327–8341. [Google Scholar] [CrossRef]
- Silva, L.A.; Khomandiak, S.; Ashbrook, A.W.; Weller, R.; Heise, M.T.; Morrison, T.E.; Dermody, T.S. A Single-Amino-Acid Polymorphism in Chikungunya Virus E2 Glycoprotein Influences Glycosaminoglycan Utilization. J. Virol. 2014, 88, 2385–2397. [Google Scholar] [CrossRef] [PubMed]
- Van Duijl-Richter, M.K.S.; Hoornweg, T.E.; Rodenhuis-Zybert, I.A.; Smit, J.M. Early events in chikungunya virus infec-tion—From virus cell binding to membrane fusion. Viruses 2015, 7, 3647–3674. [Google Scholar] [CrossRef]
- Burt, F.J.; Chen, W.; Miner, J.J.; Lenschow, D.J.; Merits, A.; Schnettler, E.; Kohl, A.; Rudd, P.A.; Taylor, A.; Herrero, L.; et al. Chikungunya virus: An update on the biology and pathogenesis of this emerging pathogen. Lancet Infect. Dis. 2017, 17, e107–e117. [Google Scholar] [CrossRef]
- Strauss, E.G.; Rice, C.M.; Strauss, J.H. Sequence coding for the alphavirus nonstructural proteins is interrupted by an opal termination codon. Proc. Natl. Acad. Sci. USA 1983, 80, 5271–5275. [Google Scholar] [CrossRef]
- Shirako, Y.; Strauss, J.H. Regulation of Sindbis virus RNA replication: Uncleaved P123 and nsP4 function in minus-strand RNA synthesis, whereas cleaved products from P123 are required for efficient plus-strand RNA synthesis. J. Virol. 1994, 68, 1874–1885. [Google Scholar] [CrossRef]
- Garoff, H.; Sjöberg, M.; Cheng, R.H. Budding of alphaviruses. Virus Res. 2004, 106, 103–116. [Google Scholar] [CrossRef]
- Lemm, J.; Rümenapf, T.; Strauss, E.; Strauss, J.; Rice, C. Polypeptide requirements for assembly of functional Sindbis virus replication complexes: A model for the temporal regulation of minus- and plus-strand RNA synthesis. EMBO J. 1994, 13, 2925–2934. [Google Scholar] [CrossRef] [PubMed]
- Schmaljohn, A.L.; Mcclain, D. Alphaviruses (togaviridae) and flaviviruses (Flaviviridae). In Medical Microbiology, 4th ed.; Baron, S., Ed.; University of Texas Medical Branch at Galveston: Gavelston, TX, USA, 1996. [Google Scholar]
- Strauss, J.H.; Strauss, E.G. The alphaviruses: Gene expression, replication, and evolution. Microbiol. Rev. 1994, 58, 491–562. [Google Scholar] [CrossRef] [PubMed]
- Assuncao-Miranda, I.; Cruz-Oliveira, C.; Da Poian, A.T. Molecular mechanisms involved in the pathogenesis of alpha-virus-induced arthritis. Biomed Res. Int. 2013, 2013, 973516. [Google Scholar] [CrossRef] [PubMed]
- Dupuis-Maguiraga, L.; Noret, M.; Brun, S.; Le Grand, R.; Gras, G.; Roques, P. Chikungunya Disease: Infection-Associated Markers from the Acute to the Chronic Phase of Arbovirus-Induced Arthralgia. PLOS Negl. Trop. Dis. 2012, 6, e1446. [Google Scholar] [CrossRef] [PubMed]
- Morrison, T.E.; Whitmore, A.C.; Shabman, R.S.; Lidbury, B.A.; Mahalingam, S.; Heise, M.T. Characterization of Ross River Virus Tropism and Virus-Induced Inflammation in a Mouse Model of Viral Arthritis and Myositis. J. Virol. 2006, 52, 601–623. [Google Scholar] [CrossRef]
- Gottipati, K.; Woodson, M.; Choi, K.H. Membrane binding and rearrangement by chikungunya virus capping enzyme nsP1. Physiol. Behav. 2020, 544, 31–41. [Google Scholar] [CrossRef]
- Laakkonen, P.; Auvinen, P.; Kujala, P.; Kääriäinen, L. Alphavirus Replicase Protein NSP1 Induces Filopodia and Rear-rangement of Actin Filaments. J. Virol. 1998, 72, 10265–10269. [Google Scholar] [CrossRef]
- Saisawang, C.; Sillapee, P.; Sinsirimongkol, K.; Ubol, S.; Smith, D.R.; Ketterman, A.J. Full length and protease domain activity of chikungunya virus nsP2 differ from other alphavirus nsP2 proteases in recognition of small peptide substrates. Biosci. Rep. 2015, 35, e00196. [Google Scholar] [CrossRef]
- Rikkonen, M.; Peränen, J.; Kääriäinen, L. Nuclear targeting of Semliki Forest virus nsP2. Arch. Virol. Suplemmenta 1994, 9, 369–377. [Google Scholar]
- Garmashova, N.; Gorchakov, R.; Frolova, E.; Frolov, I. Sindbis Virus Nonstructural Protein nsP2 Is Cytotoxic and Inhibits Cellular Transcription. J. Virol. 2006, 80, 5686–5696. [Google Scholar] [CrossRef]
- Bae, S.; Lee, J.Y.; Myoung, J. Chikungunya virus-encoded NSP2, E2 and E1 strongly antagonize the interferon-β signaling pathway. J. Microbiol. Biotechnol. 2019, 29, 1852–1859. [Google Scholar] [CrossRef] [PubMed]
- Park, E.; Griffin, D.E. The nsP3 macro domain is important for Sindbis virus replication in neurons and neurovirulence in mice. Virology 2009, 388, 305–314. [Google Scholar] [CrossRef] [PubMed]
- Egloff, M.-P.; Malet, H.; Putics, A.; Heinonen, M.; Dutartre, H.; Frangeul, A.; Gruez, A.; Campanacci, V.; Cambillau, C.; Ziebuhr, J.; et al. Structural and Functional Basis for ADP-Ribose and Poly(ADP-Ribose) Binding by Viral Macro Domains. J. Virol. 2006, 80, 8493–8502. [Google Scholar] [CrossRef] [PubMed]
- Malet, H.; Coutard, B.; Jamal, S.; Dutartre, H.; Papageorgiou, N.; Neuvonen, M.; Ahola, T.; Forrester, N.; Gould, E.A.; Lafitte, D.; et al. The Crystal Structures of Chikungunya and Venezuelan Equine Encephalitis Virus nsP3 Macro Domains Define a Conserved Adenosine Binding Pocket. J. Virol. 2009, 83, 6534–6545. [Google Scholar] [CrossRef] [PubMed]
- Oberste, M.S.; Parker, M.D.; Smith, J.F. Complete sequence of Venezuelan equine encephalitis virus subtype IE reveals con-served and hypervariable domains within the C terminus of nsP3. Virology 1996, 219, 314–320. [Google Scholar] [CrossRef]
- Shin, G.; Yost, S.A.; Miller, M.T.; Elrod, E.J.; Grakoui, A.; Marcotrigiano, J. Structural and functional insights into alphavirus polyprotein processing and pathogenesis. Proc. Natl. Acad. Sci. USA 2012, 109, 16534–16539. [Google Scholar] [CrossRef]
- Scholte, F.E.M.; Tas, A.; Albulescu, I.C.; Žusinaite, E.; Merits, A.; Snijder, E.J.; van Hemert, M.J. Stress Granule Components G3BP1 and G3BP2 Play a Proviral Role Early in Chikungunya Virus Replication. J. Virol. 2015, 89, 4457–4469. [Google Scholar] [CrossRef]
- Tuittila, M.; Hinkkanen, A.E. Amino acid mutations in the replicase protein nsP3 of Semliki Forest virus cumulatively affect neurovirulence. J. Gen. Virol. 2003, 84, 1525–1533. [Google Scholar] [CrossRef]
- Atkins, G.J.; Sheahan, B.J. Molecular determinants of alphavirus neuropathogenesis in mice. J. Gen. Virol. 2016, 97, 1283–1296. [Google Scholar] [CrossRef]
- Pietilä, M.K.; Hellström, K.; Ahola, T. Alphavirus polymerase and RNA replication. Virus Res. 2017, 234, 44–57. [Google Scholar] [CrossRef]
- de Groot, R.J.; Rümenapf, T.; Kuhn, R.J.; Strauss, E.G.; Strauss, J.H. Sindbis virus RNA polymerase is degraded by the N-end rule pathway. Proc. Natl. Acad. Sci. USA 1991, 88, 8967–8971. [Google Scholar] [CrossRef] [PubMed]
- Tomar, S.; Hardy, R.W.; Smith, J.L.; Kuhn, R.J. Catalytic Core of Alphavirus Nonstructural Protein nsP4 Possesses Terminal Adenylyltransferase Activity. J. Virol. 2006, 80, 9962–9969. [Google Scholar] [CrossRef] [PubMed]
- Lello, L.S.; Bartholomeeusen, K.; Wang, S.; Coppens, S.; Fragkoudis, R.; Alphey, L.; Ariën, K.K.; Merits, A.; Utt, A. nsP4 Is a Major Determinant of Alphavirus Replicase Activity and Template Selectivity. J. Virol. 2021, 95, JVI0035521. [Google Scholar] [CrossRef] [PubMed]
- Kääriäinen, L.; Ahola, T. Functions of alphavirus nonstructural proteins in RNA replication. Prog. Nucleic Acid Res. Mol. Biol. 2002, 71, 187–222. [Google Scholar] [PubMed]
- Gaudreault, N.N.; Indran, S.V.; Balaraman, V.; Wilson, W.C.; Richt, J.A. Molecular aspects of Rift Valley fever virus and the emergence of reassortants. Virus Genes 2019, 55, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Horne, K.M.; VanLandingham, D.L. Bunyavirus-Vector Interactions. Viruses 2014, 6, 4373–4397. [Google Scholar] [CrossRef] [PubMed]
- Walter, C.T.; Barr, J.N. Recent advances in the molecular and cellular biology of bunyaviruses. J. Gen. Virol. 2011, 92, 2467–2484. [Google Scholar] [CrossRef] [PubMed]
- Sakkas, H.; Bozidis, P.; Franks, A.; Papadopoulou, C. Oropouche Fever: A Review. Viruses 2018, 10, 175. [Google Scholar] [CrossRef]
- Romero-Alvarez, D.; Escobar, L.E. Oropouche fever, an emergent disease from the Americas. Microbes Infect. 2018, 20, 135–146. [Google Scholar] [CrossRef]
- Da Rosa, J.F.T.; De Souza, W.M.; De Paula Pinheiro, F.; Figueiredo, M.L.; Cardoso, J.F.; Acrani, G.O.; Nunes, M.R.T. Oropouche Virus: Clinical, Epidemiological, and Molecular Aspects of a Neglected Orthobunyavirus. Am. J. Trop. Med. Hyg. 2017, 96, 1019–1030. [Google Scholar] [CrossRef]
- Tauro, L.B.; de Souza, W.M.; Rivarola, M.E.; de Oliveira, R.; Konigheim, B.; Silva, S.P.; Lima, C.; Oliveira, L.; Vasconcelos, J.M.; Cardoso, J.F.; et al. Genomic characterization of orthobunyavirus of veterinary importance in America. Infect. Genet. Evol. 2019, 73, 205–209. [Google Scholar] [CrossRef] [PubMed]
- Ferron, F.; Weber, F.; De la Torre, J.C.; Reguera, J. Transcription and replication mechanisms of Bunyaviridae and Are-naviridae L proteins. Virus Res. 2017, 234, 118–134. [Google Scholar] [CrossRef] [PubMed]
- Walter, C.T.; Bento, D.F.C.; Alonso, A.G.; Barr, J.N. Amino acid changes within the Bunyamwera virus nucleocapsid protein differentially affect the mRNA transcription and RNA replication activities of assembled ribonucleoprotein templates. J. Gen. Virol. 2011, 92, 80–84. [Google Scholar] [CrossRef] [PubMed]
- Schmaljohn, C.S.; Elliot, R.M. Bunyaviridae. In Fields Virology, 6th ed.; Knipe, D.M., Howley, P.M., Eds.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2013; Volume 1, pp. 1244–1282. [Google Scholar]
- Liu, L.; Celma, C.C.P.; Roy, P. Rift Valley fever virus structural proteins: Expression, characterization and assembly of re-combinant proteins. Virol. J. 2008, 5, 82. [Google Scholar] [CrossRef] [PubMed]
- Gerrard, S.R.; Bird, B.H.; Albariño, C.G.; Nichol, S.T. The NSm proteins of Rift Valley fever virus are dispensable for matu-ration, replication and infection. Virology 2007, 359, 459–465. [Google Scholar] [CrossRef]
- Billecocq, A.; Spiegel, M.; Vialat, P.; Kohl, A.; Weber, F.; Bouloy, M.; Haller, O. NSs Protein of Rift Valley Fever Virus Blocks Interferon Production by Inhibiting Host Gene Transcription. J. Virol. 2004, 78, 9798–9806. [Google Scholar] [CrossRef]
- Pepin, M.; Bouloy, M.; Bird, B.H.; Kemp, A.; Paweska, J. Rift Valley fever virus (Bunyaviridae: Phlebovirus): An update on pathogenesis, molecular epidemiology, vectors, diagnostics and prevention. Vet. Res. 2010, 41, 61. [Google Scholar] [CrossRef]
- Guu, T.S.Y.; Zheng, W.; Tao, Y.J. Bunyavirus: Structure and Replication. In Advances in Experimental Medicine and Biology: Viral Molecular Machines; Rossmann, M.G., Rao, V.B., Eds.; Springer: Boston, MA, USA, 2012; pp. 245–266. [Google Scholar]
- Fontana, J.; López-Montero, N.; Elliott, R.M.; Fernández, J.J.; Risco, C. The unique architecture of Bunyamwera virus factories around the Golgi complex. Cell. Microbiol. 2012, 10, 2012–2028. [Google Scholar] [CrossRef]
- Ikegami, T.; Makino, S. The Pathogenesis of Rift Valley Fever. Viruses 2011, 3, 493–519. [Google Scholar] [CrossRef]
- Robinson, M.C. An epidemic of virus disease in Southern Province, Tanganyika territory, in 1952–1953. Trans. R. Soc. Trop. Med. Hyg. 1955, 49, 28–32. [Google Scholar] [CrossRef]
- Staples, J.E.; Breiman, R.F.; Powers, A.M. Chikungunya Fever: An Epidemiological Review of a Re-Emerging Infectious Disease. Clin. Infect. Dis. 2009, 49, 942–948. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, O.; Albert, M.L. Biology and pathogenesis of chikungunya virus. Nat. Rev. Microbiol. 2010, 8, 491–500. [Google Scholar] [CrossRef] [PubMed]
- Vu, D.M.; Jungkind, D.; Labeaud, A.D. Chikungunya virus. Clin. Lab. Med. 2017, 37, 371–382. [Google Scholar] [CrossRef] [PubMed]
- Filomatori, C.V.; Bardossy, E.S.; Merwaiss, F.; Suzuki, Y.; Henrion, A.; Saleh, M.C.; Alvarez, D.E. RNA recombination at Chikungunya virus 3’UTR as an evolutionary mechanism that provides adaptability. PLOS Pathog. 2019, 15, e1007706. [Google Scholar] [CrossRef]
- Schneider, A.D.B.; Ochsenreiter, R.; Hostager, R.; Hofacker, I.L.; Janies, D.; Wolfinger, M.T. Updated Phylogeny of Chikungunya Virus Suggests Lineage-Specific RNA Architecture. Viruses 2019, 11, 798. [Google Scholar] [CrossRef]
- Pichlmair, A.; Sousa, C.R. Innate recognition of viruses. Immunity 2007, 27, 370–383. [Google Scholar] [CrossRef]
- McCartney, S.A.; Colonna, M. Viral sensors: Diversity in pathogen recognition. Immunol. Rev. 2009, 227, 87–94. [Google Scholar] [CrossRef]
- Her, Z.; Malleret, B.; Chan, M.; Ong, E.K.S.; Wong, S.-C.; Kwek, D.J.C.; Tolou, H.; Lin, R.T.P.; Tambyah, P.A.; Rénia, L.; et al. Active Infection of Human Blood Monocytes by Chikungunya Virus Triggers an Innate Immune Response. J. Immunol. 2010, 184, 5903–5913. [Google Scholar] [CrossRef]
- Akhrymuk, I.; Kulemzin, S.V.; Frolova, E.I. Evasion of the Innate Immune Response: The Old World Alphavirus nsP2 Protein Induces Rapid Degradation of Rpb1, a Catalytic Subunit of RNA Polymerase II. J. Virol. 2012, 86, 7180–7191. [Google Scholar] [CrossRef]
- Sourisseau, M.; Schilte, C.; Casartelli, N.; Trouillet, C.; Guivel-Benhassine, F.; Rudnicka, D.; Sol-Foulon, N.; Roux, K.L.; Prevost, M.C.; Fisihi, H.; et al. Char-acterization of reemerging chikungunya virus. PLoS Pathog. 2007, 3, e89. [Google Scholar] [CrossRef]
- Caglioti, C.; Lalle, E.; Castilletti, C.; Carletti, F.; Capobianchi, M.R.; Bordi, L. Chikungunya virus infection: An overview. New Microbiol. 2013, 36, 211–227. [Google Scholar] [PubMed]
- Jungfleisch, J.; Böttcher, R.; Talló-Parra, M.; Pérez-Vilaró, G.; Merits, A.; Novoa, E.M.; Díez, J. CHIKV infection repro-grams codon optimality to favor viral RNA translation by altering the tRNA epitranscriptome. Nat. Commun. 2022, 13, 4725. [Google Scholar] [CrossRef] [PubMed]
- Levi, L.I.; Rezelj, V.V.; Henrion-Lacritick, A.; Erazo, D.; Boussier, J.; Vallet, T.; Bernhauerová, V.; Suzuki, Y.; Carrau, L.; Weger-Lucarelli, J.; et al. Defective viral genomes from chikungunya virus are broad-spectrum antivirals and prevent virus dissemination in mosquitoes. PLOS Pathog. 2021, 17, e1009110. [Google Scholar] [CrossRef] [PubMed]
- Valdés-López, J.F.; Fernandez, G.J.; Urcuqui-Inchima, S. Interleukin 27 as an inducer of antiviral response against chikungunya virus infection in human macrophages. Cell. Immunol. 2021, 367, 104411. [Google Scholar] [CrossRef] [PubMed]
- Dhanwani, R.; Khan, M.; Alam, S.I.; Rao, P.V.L.; Parida, M. Differential proteome analysis of Chikungunya virus- infected new-born mice tissues reveal implication of stress, inflammatory and apoptotic pathways in disease pathogenesis. Proteomics 2011, 11, 1936–1951. [Google Scholar] [CrossRef] [PubMed]
- Thio, C.L.; Yusof, R.; Abdul-rahman, P.S.A.; Karsani, A. Differential Proteome Analysis of Chikungunya Virus Infection on Host Cells. PLoS ONE 2013, 8, e61444. [Google Scholar] [CrossRef] [PubMed]
- Treffers, E.E.; Tas, A.; Scholte, F.E.; Van, M.N.; Heemskerk, M.T.; de Ru, A.H.; Snijder, E.J.; van Hemert, M.J.; van Veelen, P.A. Temporal SILAC-based quantitative proteomics identifies host factors involved in chikungunya virus replication. Proteomics 2015, 15, 2267–2280. [Google Scholar] [CrossRef]
- Issac, T.H.K.; Tan, E.L.; Chu, J.J.H. Proteomic profiling of chikungunya virus-infected human muscle cells: Reveal the role of cytoskeleton network in CHIKV replication. J. Proteom. 2014, 108, 445–464. [Google Scholar] [CrossRef]
- Cui, Y.; Liu, P.; Mooney, B.P.; Franz, A.W.E. Quantitative Proteomic Analysis of Chikungunya Virus-Infected Aedes aegypti Reveals Proteome Modulations Indicative of Persistent Infection. J. Proteome Res. 2020, 19, 2443–2456. [Google Scholar] [CrossRef]
- Chowdhury, A.; Modahl, C.M.; Missé, D.; Kini, R.M.; Pompon, J. High resolution proteomics of Aedes aegypti salivary glands infected with either dengue, Zika or chikungunya viruses identify new virus specific and broad antiviral factors. Sci. Rep. 2021, 11, 23696. [Google Scholar] [CrossRef]
- Vasconcellos, A.F.; Melo, R.M.; Mandacaru, S.C.; de Oliveira, L.S.; de Oliveira, A.S.; Moraes, E.C.d.S.; Trugilho, M.R.D.O.; Ricart, C.A.O.; Báo, S.N.; Resende, R.O.; et al. Aedes aegypti Aag-2 Cell Proteome Modulation in Response to Chikungunya Virus Infection. Front. Cell. Infect. Microbiol. 2022, 12, 920425. [Google Scholar] [CrossRef] [PubMed]
- Sukkaew, A.; Suksatu, A.; Roytrakul, S.; Smith, D.R.; Ubol, S. Proteomic analysis of CHIKV-infected human fibro-blast-like synoviocytes: Identification of host factors potentially associated with CHIKV replication and cellular pathogenesis. Microbiol. Immunol. 2020, 64, 445–457. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.M.; Ham, H.-J.V.D.; Oduber, M.; Martina, E.; Zaaraoui-Boutahar, F.; Roose, J.M.; van Ijcken, W.; Osterhaus, A.D.M.E.; Andeweg, A.C.; Koraka, P.; et al. Transcriptomic Analyses Reveal Differential Gene Expression of Immune and Cell Death Pathways in the Brains of Mice Infected with West Nile Virus and Chikungunya Virus. Front. Microbiol. 2017, 8, 1556. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Alto, B.W.; Jiang, Y.; Yu, F.; Zhang, Y. Transcriptomic Analysis of Aedes aegypti Innate Immune System in Response to Ingestion of Chikungunya Virus. Int. J. Mol. Sci. 2019, 20, 3133. [Google Scholar] [CrossRef]
- Campbell, C.L.; Lehmann, C.J.; Gill, S.S.; Dunn, W.A.; James, A.A.; Foy, B.D. A role for endosomal proteins in alphavirus dissemination in mosquitoes. Insect Mol. Biol. 2011, 20, 429–436. [Google Scholar] [CrossRef][Green Version]
- Liu, Y.; Zhang, F.; Liu, J.; Xiao, X.; Zhang, S.; Qin, C.; Xiang, Y.; Wang, P.; Cheng, G. Transmission-Blocking Antibodies against Mosquito C-Type Lectins for Dengue Prevention. PLOS Pathog. 2014, 10, e1003931. [Google Scholar] [CrossRef]
- Dong, S.; Kantor, A.M.; Lin, J.; Passarelli, A.L.; Clem, R.J.; Franz, A.W.E. Infection pattern and transmission potential of chikungunya virus in two New World laboratory-adapted Aedes aegypti strains. Sci. Rep. 2004, 41, 467–475. [Google Scholar] [CrossRef]
- Romoser, W.S.; Wasieloski, L.P., Jr.; Pushko, P.; Kondig, J.P.; Lerdthusnee, K.; Neira, M.; Ludwig, G.V. Evidence for arbovirus dissemination conduits from the mosquito (Diptera: Culicidae) midgut. J. Med. Entomol. 2004, 41, 467–475. [Google Scholar] [CrossRef]
- Dong, S.; Behura, S.K.; Franz, A.W.E. The midgut transcriptome of Aedes aegypti fed with saline or protein meals containing chikungunya virus reveals genes potentially involved in viral midgut escape. BMC Genom. 2017, 18, 382. [Google Scholar] [CrossRef]
- Vedururu, R.K.; Neave, M.J.; Tachedjian, M.; Klein, M.J.; Gorry, P.R.; Duchemin, J.-B.; Paradkar, P.N. RNASeq Analysis of Aedes albopictus Mosquito Midguts after Chikungunya Virus Infection. Viruses 2019, 11, 513. [Google Scholar] [CrossRef]
- Vedururu, R.K.; Neave, M.J.; Sundaramoorthy, V.; Green, D.; Harper, J.A.; Gorry, P.R.; Duchemin, J.-B.; Paradkar, P.N. Whole Transcriptome Analysis of Aedes albopictus Mosquito Head and Thorax Post-Chikungunya Virus Infection. Pathogens 2019, 8, 132. [Google Scholar] [CrossRef] [PubMed]
- Byers, N.M.; Fleshman, A.C.; Perera, R.; Molins, C.R. Metabolomic insights into human arboviral infections: Dengue, chikungunya, and zika viruses. Viruses 2019, 11, 225. [Google Scholar] [CrossRef] [PubMed]
- Shrinet, J.; Shastri, J.S.; Gaind, R.; Bhavesh, N.S.; Sunil, S. Serum metabolomics analysis of patients with chikungunya and dengue mono/co-infections reveals distinct metabolite signatures in the three disease conditions. Sci. Rep. 2016, 6, 36833. [Google Scholar] [CrossRef]
- Pego, P.N.; Gomes, L.P.; Provance, D.W., Jr.; De Simone, S.G. Mayaro virus disease. J. Hum. Virol. Retrovirology 2014, 1, 1–11. [Google Scholar]
- Acosta-Ampudia, Y.; Monsalve, D.M.; Rodríguez, Y.; Pacheco, Y.; Anaya, J.M.; Ramírez-Santana, C. Mayaro: An emerging viral threat? Emerg. Microbes Infect. 2018, 7, 163. [Google Scholar] [CrossRef]
- Sun, J.; Wu, D. Mayaro virus, a regional or global threat? Travel Med. Infect. Dis. 2019, 32, 101462. [Google Scholar] [CrossRef] [PubMed]
- Mavian, C.; Rife, B.D.; Dollar, J.J.; Cella, E.; Ciccozzi, M.; Prosperi, M.C.F.; Lednicky, J.; Morris, J.G.; Capua, I.; Salemi, M. Emergence of recombinant Mayaro virus strains from the Amazon basin. Sci. Rep. 2017, 7, 8718. [Google Scholar] [CrossRef]
- Dalle-Donne, I.; Rossi, R.; Colombo, R.; Giustarini, D.; Milzani, A.D.G. Biomarkers of Oxidative Damage in Human Disease. Clin. Chem. 2006, 52, 601–623. [Google Scholar] [CrossRef]
- Camini, F.C.; Caetano, C.C.D.S.; Almeida, L.T.; Guerra, J.F.D.C.; Silva, B.D.M.; Silva, S.D.Q.; de Magalhães, J.C.; Magalhães, C.L.D.B. Oxidative stress in Mayaro virus infection. Virus Res. 2017, 236, 1–8. [Google Scholar] [CrossRef]
- Caetano, C.C.D.S.; Camini, F.C.; Almeida, L.T.; Ferraz, A.C.; Da Silva, T.F.; Lima, R.L.S.; Carvalho, M.M.D.F.; Castro, T.D.F.; Carneiro, C.M.; Silva, B.D.M.; et al. Mayaro Virus Induction of Oxidative Stress is Associated With Liver Pathology in a Non-Lethal Mouse Model. Sci. Rep. 2019, 9, 15289. [Google Scholar] [CrossRef]
- Hoque, H.; Islam, R.; Ghosh, S.; Rahaman, M.; Jewel, N.A.; Miah, A. Implementation of in silico methods to predict common epitopes for vaccine development against Chikungunya and Mayaro viruses. Heliyon 2021, 7, e06396. [Google Scholar] [CrossRef] [PubMed]
- Vasconcellos, A.F.; Mandacaru, S.; De Oliveira, A.S.; Fontes, W.; Melo, R.M.; De Sousa, M.V.; Resende, R.O.; Charneau, S. Dynamic proteomic analysis of Aedes aegypti Aag-2 cells infected with Mayaro virus. Parasites Vectors 2020, 13, 297. [Google Scholar] [CrossRef] [PubMed]
- Henderson, C.; Brustolin, M.; Hedge, S.; Dayama, G.; Lau, N.; Hughes, G.L.; Bergey, C.; Rasgon, J.L. Mayaro Virus infection elicits an innate immune response in Anopheles stephensi. bioRxiv 2021, 2020, 1–37. [Google Scholar]
- Bengue, M.; Ferraris, P.; Barthelemy, J.; Diagne, C.T.; Hamel, R.; Liégeois, F.; Nougairède, A.; de Lamballerie, X.; Simonin, Y.; Pompon, J.; et al. Mayaro Virus Infects Human Brain Cells and Induces a Potent Antiviral Response in Human Astrocytes. Viruses 2021, 13, 465. [Google Scholar] [CrossRef] [PubMed]
- De Castro, C.M.O. Análise Metabolômica de Alterações Induzidas Pelo Vírus Mayaro em Células Vero. Ph.D. Thesis, Faculty of Medicine of Sao Jose do Rio Preto, Sao Paulo, Brazil, September 2015. [Google Scholar]
- Bird, B.H.; Ksiazek, T.G.; Nichol, S.T.; MacLachlan, N.J. Rift Valley fever virus. J. Am. Vet. Med. Assoc. 2009, 234, 883–893. [Google Scholar] [CrossRef]
- Flick, R.; Bouloy, M. Rift Valley fever virus. Curr. Mol. Med. 2005, 5, 827–834. [Google Scholar] [CrossRef]
- Grobbelaar, A.A.; Weyer, J.; Leman, P.A.; Kemp, A.; Paweska, J.T.; Swanepoel, R. Molecular Epidemiology of Rift Valley Fever Virus. Emerg. Infect. Dis. 2011, 17, 2270–2276. [Google Scholar] [CrossRef]
- Juma, J.; Fonseca, V.; Konongoi, S.L.; van Heusden, P.; Roesel, K.; Sang, R.; Bett, B.; Christoffels, A.; de Oliveira, T.; Oyola, S.O. Genomic surveillance of Rift Valley fever virus: From sequencing to lineage assignment. BMC Genom. 2022, 23, 520. [Google Scholar] [CrossRef]
- Arishi, H.M.; Aqeel, A.Y.; Al Hazmi, M.M. Vertical transmission of fatal Rift Valley fever in a newborn. Ann. Trop. Paediatr. 2006, 26, 251–253. [Google Scholar] [CrossRef]
- Raymond, R.; Gibbs, C.J., Jr.; Aulisio, C.G.; Binn, L.N.; Harrison, V.R. The development of a formalin-killed Rift Valley fever virus vaccine for use in man. J. Immunol. 1962, 89, 660–671. [Google Scholar]
- Harmon, J.R.; Barbeau, D.J.; Nichol, S.T.; Spiropoulou, C.F.; McElroy, A.K. Rift Valley fever virus vaccination induces long-lived, antigen-specific human T cell responses. npj Vaccines 2020, 5, 17. [Google Scholar] [CrossRef] [PubMed]
- Ikegami, T.; Makino, S. Rift Valley fever vaccines. Vaccine 2009, 27, D62–D72. [Google Scholar] [CrossRef] [PubMed]
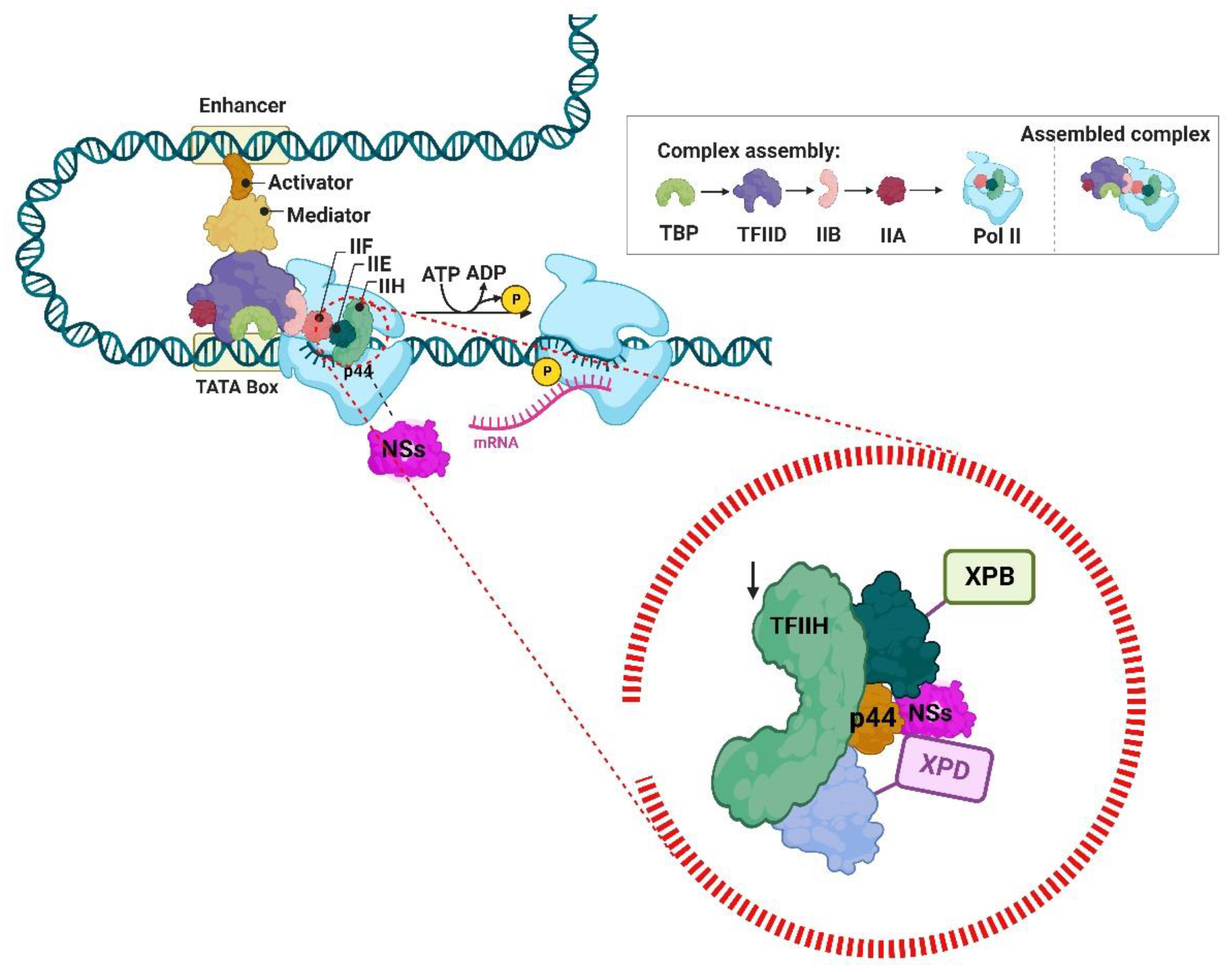
- Le May, N.; Dubaele, S.; De Santis, L.P.; Billecocq, A.; Bouloy, M.; Egly, J.-M. TFIIH Transcription Factor, a Target for the Rift Valley Hemorrhagic Fever Virus. Cell 2004, 116, 541–550. [Google Scholar] [CrossRef]
- Nfon, C.K.; Marszal, P.; Zhang, S.; Weingartl, H.M. Innate Immune Response to Rift Valley Fever Virus in Goats. PLOS Negl. Trop. Dis. 2012, 6, e1623. [Google Scholar] [CrossRef] [PubMed]
- Bouloy, M.; Janzen, C.; Vialat, P.; Khun, H.; Pavlovic, J.; Huerre, M.; Haller, O. Genetic Evidence for an Interferon-Antagonistic Function of Rift Valley Fever Virus Nonstructural Protein NSs. J. Virol. 2001, 75, 1371–1377. [Google Scholar] [CrossRef] [PubMed]
- Ikegami, T.; Narayanan, K.; Won, S.; Kamitani, W.; Peters, C.J.; Makino, S. Rift Valley Fever Virus NSs Protein Promotes Post-Transcriptional Downregulation of Protein Kinase PKR and Inhibits eIF2α Phosphorylation. PLOS Pathog. 2009, 5, e1000287. [Google Scholar] [CrossRef]
- Won, S.; Ikegami, T.; Peters, C.J.; Makino, S. NSm Protein of Rift Valley Fever Virus Suppresses Virus-Induced Apoptosis. J. Virol. 2007, 81, 13335–13345. [Google Scholar] [CrossRef]
- Nuss, J.E.; Kehn-Hall, K.; Benedict, A.; Constantino, J.; Ward, M.; Peyser, B.D.; Retterer, C.J.; Tressler, L.E.; Wanner, L.M.; McGovern, H.F.; et al. Multi-faceted proteomic characterization of host protein complement of rift valley fever virus virions and identification of specific Heat Shock Proteins, including HSP90, as important viral host factors. PLoS ONE 2014, 9, e93483. [Google Scholar] [CrossRef]
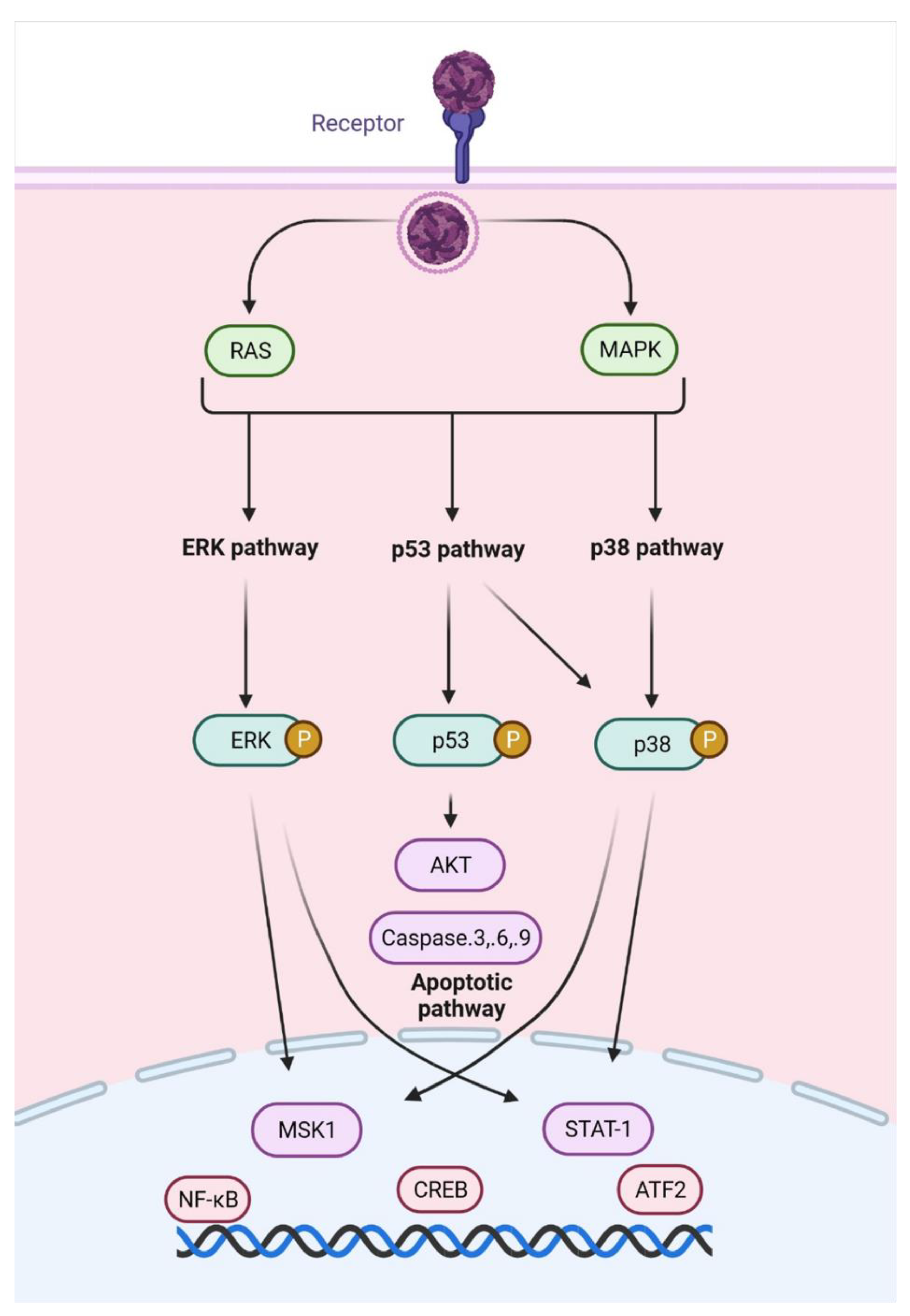
- De la Fuente, C.; Pinkham, C.; Dabbagh, D.; Beitzel, B.; Garrison, A.; Palacios, G.; Hodge, K.A.; Petricoin, E.F.; Schmaljohn, C.; Campbell, C.E.; et al. Phosphoproteomic analysis reveals Smad protein family activation following Rift valley fever virus infection. PLoS ONE 2018, 13, e0191983. [Google Scholar]
- Popova, T.G.; Turell, M.J.; Espina, V.; Kehn-Hall, K.; Kidd, J.; Narayanan, A.; Liotta, L.; Petricoin, E.F., III; Kashanchi, F.; Bailey, C.; et al. Reverse-phase phosphoproteome analysis of signaling pathways induced by rift valley fever virus in human small airway epithelial cells. PLoS ONE 2010, 5, e13805. [Google Scholar] [CrossRef][Green Version]
- Le May, N.; Mansuroglu, Z.; Léger, P.; Josse, T.; Blot, G.; Billecocq, A.; Flick, R.; Jacob, Y.; Bonnefoy, E.; Bouloy, M. A SAP30 Complex Inhibits IFN-β Expression in Rift Valley Fever Virus Infected Cells. PLOS Pathog. 2008, 4, e13. [Google Scholar] [CrossRef] [PubMed]
- Havranek, K.E.; White, L.A.; Lanchy, J.-M.; Lodmell, J.S. Transcriptome profiling in Rift Valley fever virus infected cells reveals modified transcriptional and alternative splicing programs. PLoS ONE 2019, 14, e0217497. [Google Scholar] [CrossRef] [PubMed]
- Wickström, S.A.; Lange, A.; Hess, M.W.; Polleux, J.; Spatz, J.P.; Krüger, M.; Pfaller, K.; Lambacher, A.; Bloch, W.; Mann, M.; et al. Integrin-Linked Kinase Controls Microtubule Dynamics Required for Plasma Membrane Targeting of Caveolae. Dev. Cell 2010, 19, 574–588. [Google Scholar] [CrossRef] [PubMed]
- Harpen, M.; Barik, T.; Musiyenko, A.; Barik, S. Mutational Analysis Reveals a Noncontractile but Interactive Role of Actin and Profilin in Viral RNA-Dependent RNA Synthesis. J. Virol. 2009, 83, 10869–10876. [Google Scholar] [CrossRef]
- Pinkham, C.; Dahal, B.; De La Fuente, C.L.; Bracci, N.; Beitzel, B.; Lindquist, M.; Garrison, A.; Schmaljohn, C.; Palacios, G.; Narayanan, A.; et al. Alterations in the host transcriptome in vitro following Rift Valley fever virus infection. Sci. Rep. 2017, 7, 14385. [Google Scholar] [CrossRef]
- Alem, F.; Olanrewaju, A.A.; Omole, S.; Hobbs, H.E.; Ahsan, N.; Matulis, G.; Brantner, C.A.; Zhou, W.; Petricoin, E.F.; Liotta, L.A.; et al. Exosomes originating from infection with the cyto-plasmic single—Stranded RNA virus Rift Valley fever virus (RVFV) protect recipient cells by inducing RIG—I mediated IFN—B response that leads to activation of autophagy. Cell Biosci. 2021, 11, 1–21. [Google Scholar] [CrossRef]
- Licciardi, S.; Loire, E.; Cardinale, E.; Gislard, M.; Dubois, E.; Cêtre-Sossah, C. In vitro shared transcriptomic re-sponses of Aedes aegypti to arboviral infections: Example of dengue and Rift Valley fever viruses. Parasit. Vectors 2020, 13, 395. [Google Scholar] [CrossRef]
- Núñez, A.I.; Esteve-Codina, A.; Gómez-Garrido, J.; Brustolin, M.; Talavera, S.; Berdugo, M.; Dabad, M.; Alioto, T.; Bensaid, A.; Busquets, N. Alteration in the Culex pipiens transcriptome reveals diverse mechanisms of the mosquito immune system implicated upon Rift Valley fever phlebovirus exposure. PLOS Negl. Trop. Dis. 2020, 14, e0008870. [Google Scholar] [CrossRef]
- Kgosi, G.M.; Vervoort, J.; Anton, P.A.; Prinsloo, G. Metabolomic profile of medicinal plants with anti-RVFV activity. Heliyon 2022, 8, e08936. [Google Scholar]
- Mourão, M.P.G.; Bastos, M.S.; Gimaque, J.B.L.; Mota, B.R.; Souza, G.S.; Grimmer, G.H.N.; Galusso, E.S.; Arruda, E.; Figueiredo, L.T.M. Oropouche fever outbreak, Manaus, Brazil, 2007–2008. Emerg. Infect. Dis. 2009, 15, 2063–2064. [Google Scholar] [CrossRef]
- Anderson, C.R.; Spence, L.; Downs, W.G.; Aitken, T.H. Oropouche virus: A new human disease agent from Trinidad, West Indies. Am. J. Trop. Med. Hyg. 1961, 104, 574–578. [Google Scholar] [CrossRef] [PubMed]
- Nunes, M.R.T.; de Souza, W.M.; Savji, N.; Figueiredo, M.L.; Cardoso, J.F.; da Silva, S.P.; Lima, C.P.D.S.D.; Vasconcelos, H.B.; Rodrigues, S.G.; Lipkin, W.I.; et al. Oropouche orthobunyavirus: Genetic characterization of full-length genomes and development of molecular methods to discriminate natural reassortments. Infect. Genet. Evol. 2019, 68, 16–22. [Google Scholar] [CrossRef] [PubMed]
- Nunes, M.R.T.; Martins, L.C.; Rodrigues, S.G.; Chiang, J.O.; Da Silva, R.D.S.A.; Da Rosa, A.P.T.; da Costa, P.F.V. Oropouche virus isolation, southeast Brazil. Emerg. Infect. Dis. 2005, 11, 1610–1613. [Google Scholar] [CrossRef] [PubMed]
- Tilston-Lunel, N.L.; Hughes, J.; Acrani, G.O.; da Silva, D.E.A.; Azevedo, R.S.S.; Rodrigues, S.G.; Vasconcelos, P.F.C.; Nunes, M.R.T.; Elliot, R.M. Genetic analysis of members of the species Oropouche virus and identification of a novel M segment se-quence. J. Gen. Virol. 2015, 96, 1636–1650. [Google Scholar] [CrossRef] [PubMed]
- Santos, R.I.; Rodrigues, A.H.; Silva, M.L.; Mortara, R.A.; Rossi, M.A.; Jamur, M.C.; Oliver, C.; Arruda, E. Oropouche virus entry into HeLa cells involves clathrin and requires endosomal acidification. Virus Res. 2008, 138, 139–143. [Google Scholar] [CrossRef]
- Honda, K.; Takaoka, A.; Taniguchi, T. Type I inteferon gene induction by the interferon regulatory factor family of tran-scription factors. Immunity 2006, 25, 349–360. [Google Scholar] [CrossRef]
- Pinheiro, F.P.; Da Rosa, A.P.T.; Da Rosa, J.F.T.; Ishak, R.; Freitas, R.B.; Gomes, M.L.; LeDuc, J.W.; Oliva, O.F. Oropouche virus. I. A review of clinical, epidemiological, and ecological findings. Am. J. Trop. Med. Hyg. 1981, 30, 4720–4737. [Google Scholar] [CrossRef]
- Proenca-Modena, J.L.; Sesti-Costa, R.; Pinto, A.K.; Richner, J.M.; Lazear, H.M.; Lucas, T.; Hyde, J.L.; Diamond, M.S. Oropouche virus infection and pathogenesis are restricted by MAVS, IRF-3, IRF-7, and type I interferon signaling pathways in nonmyeloid cells. J. Virol. 2015, 89, 4720–4737. [Google Scholar] [CrossRef]
- Proenca-Modena, J.L.; Hyde, J.L.; Sesti-Costa, R.; Lucas, T.; Pinto, A.K.; Richner, J.M.; Gorman, M.J.; Lazear, H.M.; Diamond, M.S. Interferon-regulatory factor 5-dependent signaling restricts Orthobunyavirus dissemination to the central nervous sys-tem. J. Virol. 2016, 90, 189–205. [Google Scholar] [CrossRef]
- Geddes, V.E.V.; De Oliveira, A.S.; Tanuri, A.; Arruda, E.; Ribeiro-Alves, M.; Aguiar, R.S. MicroRNA and cellular targets profiling reveal miR-217 and miR-576-3p as proviral factors during Oropouche infection. PLOS Negl. Trop. Dis. 2018, 12, e0006508. [Google Scholar] [CrossRef]
- De-Oliveira-Pinto, L.M.; Gandini, M.; Freitas, L.P.; Siqueira, M.M.; Marinho, C.F.; Setúbal, S.; Kubella, C.F.; Cruz, O.G.; Oliveira, S.A.D. Profile of circulating levels of IL-1Ra, CXCL10/IP-10, CCL4/MIP-1β and CCL2/MCP-1 in dengue fever and parvovirosis. Mem. Inst. Oswaldo Cruz 2012, 107, 48–56. [Google Scholar] [CrossRef] [PubMed][Green Version]
- DE Oliveira, E.; Azevedo, R.D.S.S.; Coelho-Dos-Reis, J.G.; Antonelli, L.R.D.V.; Ferreira, M.S.; Campi-Azevedo, A.C.; Costa-Silva, M.F.; Martins, L.C.; Chiang, J.O.; Teixeira-Carvalho, A.; et al. IFN-α as a time-sensitive biomarker during Oropouche virus infection in early and late seroconverters. Sci. Rep. 2019, 9, 17924. [Google Scholar] [CrossRef] [PubMed]
- Amorim, M.R.; Pontelli, M.C.; De Souza, G.F.; Muraro, S.P.; De Toledo-Teixeira, D.A.; Forato, J.; Bispo-Dos-Santos, K.; Barbosa, N.S.; Martini, M.C.; Parise, P.L.; et al. Oropouche Virus Infects, Persists and Induces IFN Response in Human Peripheral Blood Mononuclear Cells as Identified by RNA PrimeFlow™ and qRT-PCR Assays. Viruses 2020, 12, 785. [Google Scholar] [CrossRef]
- Almeida, G.M.; Souza, J.P.; Mendes, N.D.; Pontelli, M.C.; Pinheiro, N.R.; Nogueira, G.O.; Cardoso, R.S.; Paiva, I.M.; Ferrari, G.D.; Veras, F.P.; et al. Neural Infection by Oropouche Virus in Adult Human Brain Slices Induces an Inflammatory and Toxic Response. Front. Neurosci. 2021, 15, 674576. [Google Scholar] [CrossRef] [PubMed]
- Skalsky, R.L.; Cullen, B.R. Viruses, microRNAs, and host interactions. Annu. Rev. Microbiol. 2010, 64, 123–141. [Google Scholar] [CrossRef]
- Schoen, A.; Weber, F. Orthobunyaviruses and innate immunity induction: AlieNSs vs. PredatoRRs. Eur. J. Cell Biol. 2015, 94, 384–390. [Google Scholar] [CrossRef]
- Gouzil, J.; Fablet, A.; Lara, E.; Caignard, G.; Cochet, M.; Kundlacz, C.; Palmarini, M.; Varela, M.; Breard, E.; Sailleau, C.; et al. Nonstructural Protein NSs of Schmallenberg Virus Is Targeted to the Nucleolus and Induces Nucleolar Disorganization. J. Virol. 2017, 91, e01263-16. [Google Scholar] [CrossRef]
- Hopkins, K.C.; McLane, L.M.; Maqbool, T.; Panda, D.; Gordesky-Gold, B.; Cherry, S. A genome-wide RNAi screen reveals that mRNA deccaping restricts bunyaviral replication by limiting the pools of Cdp2-accessible targets for cap-snatching. Genes Dev. 2013, 27, 1511–1525. [Google Scholar] [CrossRef]
- Geddes, V.E.V.; Brustolini, O.J.B.; Cavalcante, L.T.F.; Guimarães, A.P.C.; Gerber, A.L.; Figueiredo, C.M.; Diniz, L.P.; Neto, E.A.; Tanuri, A.; Souza, R.P.; et al. Common Dysregulation of Innate Immunity Pathways in Human Primary Astrocytes Infected With and Zika Viruses. Front. Cell. Infect. Microbiol. 2021, 11, 641261. [Google Scholar] [CrossRef]
- Adhikari, U.K.; Tayebi, M.; Rahman, M.M. Immunoinformatics Approach for Epitope-Based Peptide Vaccine Design and Active Site Prediction against Polyprotein of Emerging Oropouche Virus. J. Immunol. Res. 2018, 2018, 6718083. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peinado, R.d.S.; Eberle, R.J.; Arni, R.K.; Coronado, M.A. A Review of Omics Studies on Arboviruses: Alphavirus, Orthobunyavirus and Phlebovirus. Viruses 2022, 14, 2194. https://doi.org/10.3390/v14102194
Peinado RdS, Eberle RJ, Arni RK, Coronado MA. A Review of Omics Studies on Arboviruses: Alphavirus, Orthobunyavirus and Phlebovirus. Viruses. 2022; 14(10):2194. https://doi.org/10.3390/v14102194
Chicago/Turabian StylePeinado, Rafaela dos S., Raphael J. Eberle, Raghuvir K. Arni, and Mônika A. Coronado. 2022. "A Review of Omics Studies on Arboviruses: Alphavirus, Orthobunyavirus and Phlebovirus" Viruses 14, no. 10: 2194. https://doi.org/10.3390/v14102194
APA StylePeinado, R. d. S., Eberle, R. J., Arni, R. K., & Coronado, M. A. (2022). A Review of Omics Studies on Arboviruses: Alphavirus, Orthobunyavirus and Phlebovirus. Viruses, 14(10), 2194. https://doi.org/10.3390/v14102194






