Emerging Phage Resistance in Pseudomonas aeruginosa PAO1 Is Accompanied by an Enhanced Heterogeneity and Reduced Virulence
Abstract
:1. Introduction
2. Materials and Methods
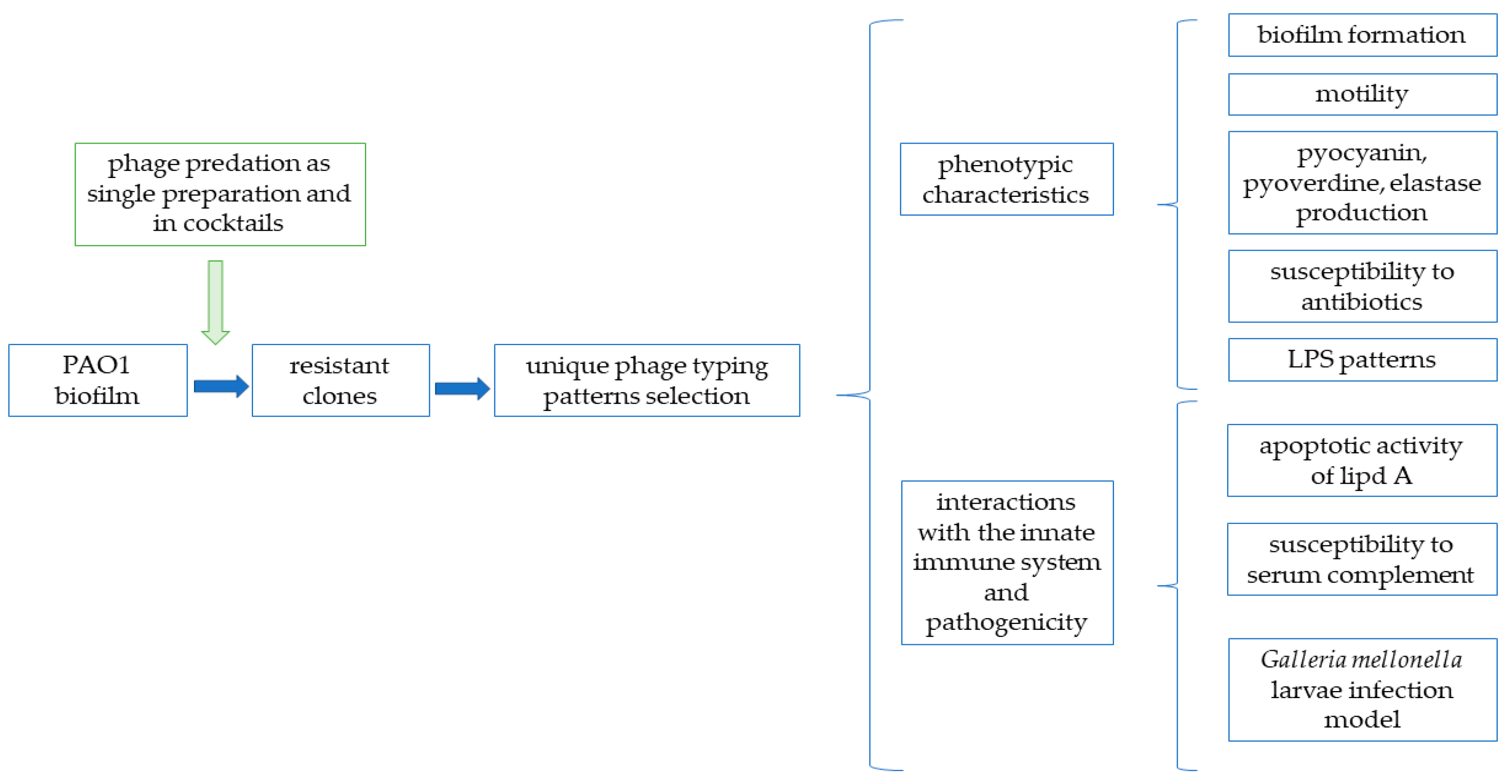
2.1. Experimental Pipeline
2.2. Bacterial Strains and Bacteriophages
2.3. Isolation of Phage-Resistant Clones
2.4. Phage Typing by the Double-Layer Plate Method
2.5. Motility Assays
2.6. Pyocyanin and Pyoverdine Production
2.7. Elastase Production
2.8. Antibiotics Susceptibility Testing
2.9. Biofilm Formation Measurement
2.10. Susceptibility of Bacterial Isolates to Serum Complement Activity
2.11. Lipopolysaccharide Isolation and Pattern Analysis
2.12. Apoptotic Activity of the LPS Isolated from Phage-Resistant PAO1 Mutants
2.13. Galleria mellonella Larvae Infection Model
3. Results
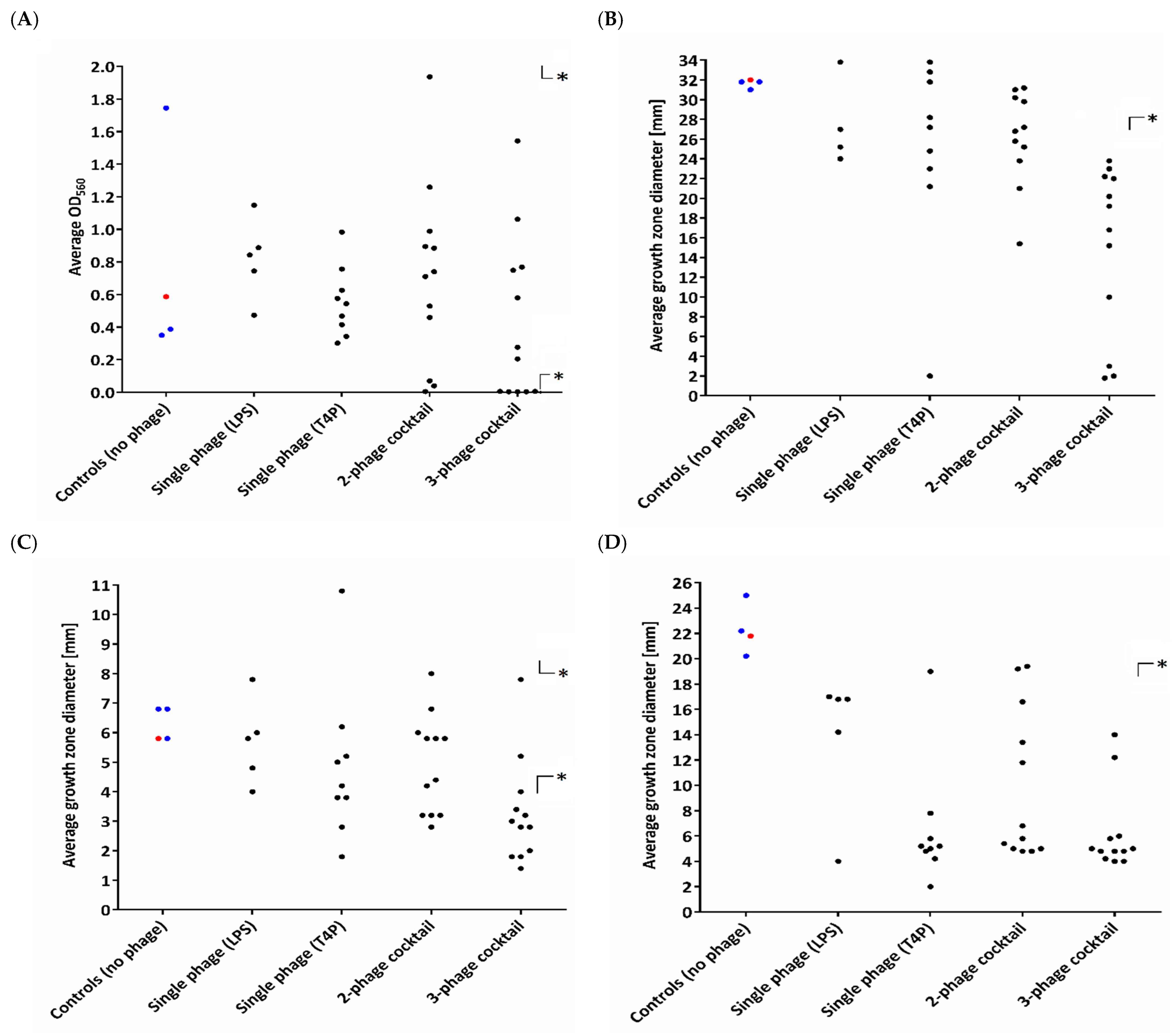
3.1. Heterogeneity in Phenotypic Patterns
3.2. Diversity in Extracellular Virulence Factor Production
3.3. Interactions with the Innate Immune System and Pathogenicity
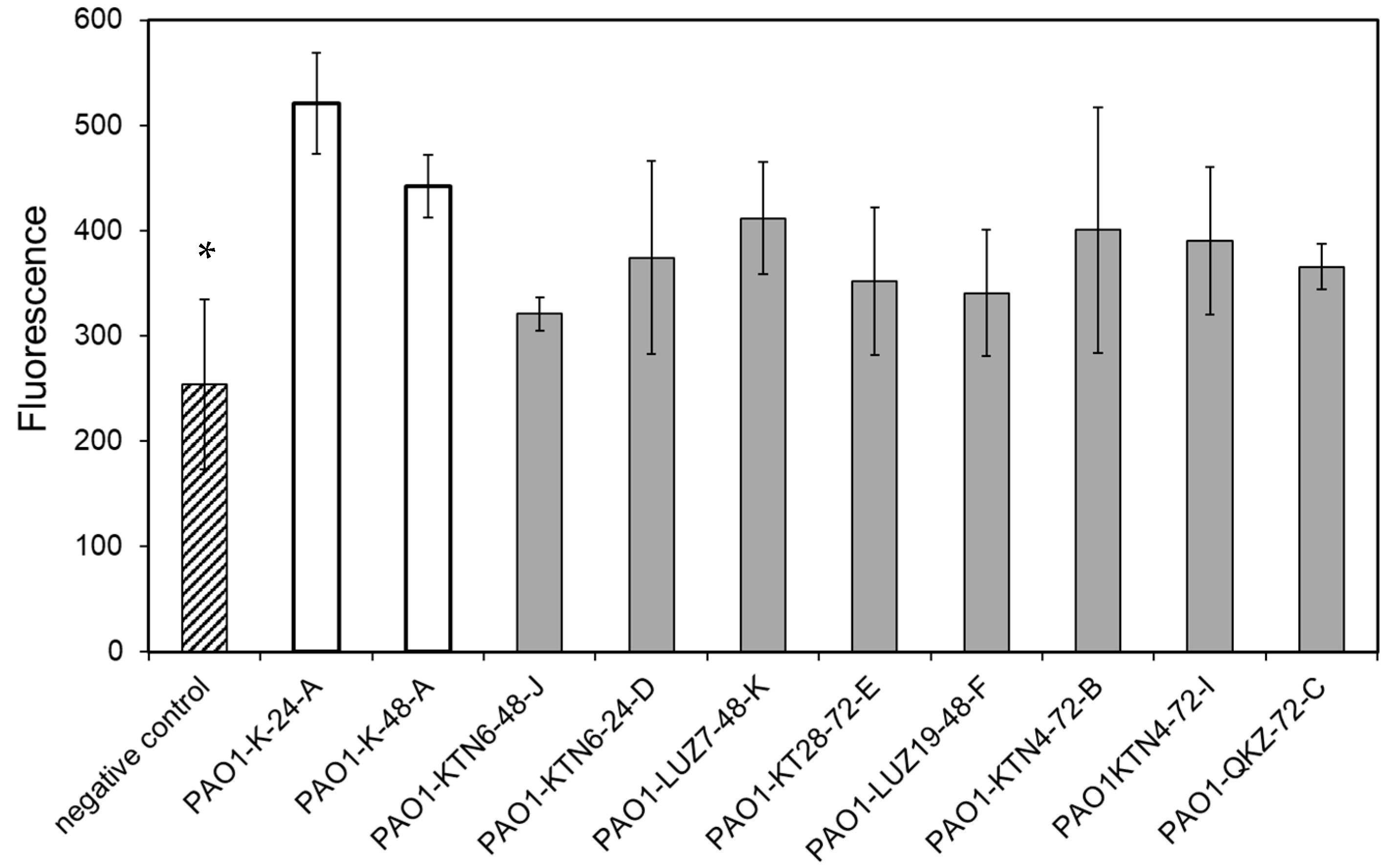
3.3.1. Phage Resistance Does Not Impact the Apoptotic Activity of LPS
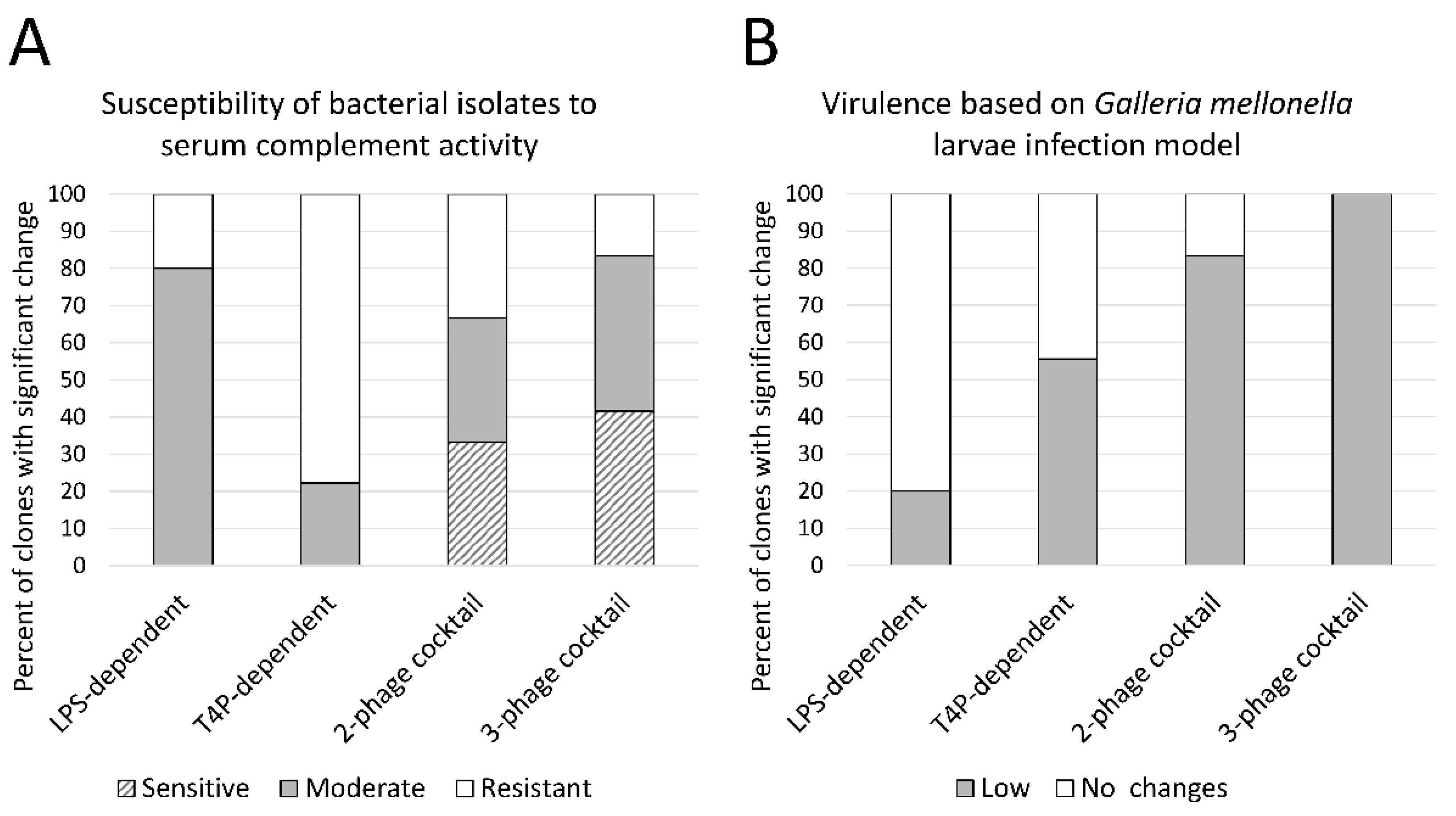
3.3.2. LPS-Dependent Phage Selection Sensitizes Bacteria to Serum Complement
3.3.3. Phage Cocktail Application Results with a Significantly Reduced Virulence in the G. mellonella Infection Model
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Behzadi, P.; Baráth, Z.; Gajdács, M. It’s not easy being green: A narrative review on the microbiology, virulence and therapeutic prospects of multidrug-resistant Pseudomonas aeruginosa. Antibiotics 2021, 10, 42. [Google Scholar] [CrossRef]
- Pires, D.P.; Dötsch, A.; Anderson, E.M.; Hao, Y.; Khursigara, C.M.; Lam, J.S.; Sillankorva, S.; Azeredo, J. A genotypic analysis of five P. aeruginosa strains after biofilm infection by phages targeting different cell surface receptors. Front. Microbiol. 2017, 8, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Oliveira, D.M.P.; Forde, B.M.; Kidd, T.J.; Harris, P.N.A.; Schembri, M.A.; Beatson, S.A.; Paterson, D.L.; Walker, M.J. Antimicrobial resistance in ESKAPE pathogens. Clin. Microbiol. Rev. 2020, 33. [Google Scholar] [CrossRef]
- Bonomo, R.A.; Szabo, D. Mechanisms of multidrug resistance in Acinetobacter species and Pseudomonas aeruginosa. Clin. Infect. Dis. 2006, 43, S49–S56. [Google Scholar] [CrossRef] [Green Version]
- Folkesson, A.; Jelsbak, L.; Yang, L.; Johansen, H.K.; Ciofu, O.; Høiby, N.; Molin, S. Adaptation of Pseudomonas aeruginosa to the cystic fibrosis airway: An evolutionary perspective. Nat. Rev. Microbiol. 2012, 10, 841–851. [Google Scholar] [CrossRef]
- Clark, S.T.; Diaz Caballero, J.; Cheang, M.; Coburn, B.; Wang, P.W.; Donaldson, S.L.; Zhang, Y.; Liu, M.; Keshavjee, S.; Yau, Y.C.W.; et al. Phenotypic diversity within a Pseudomonas aeruginosa population infecting an adult with cystic fibrosis. Sci. Rep. 2015, 5, 10932. [Google Scholar] [CrossRef] [PubMed]
- Moradali, M.F.; Ghods, S.; Rehm, B.H.A. Pseudomonas aeruginosa lifestyle: A paradigm for adaptation, survival, and persistence. Front. Cell. Infect. Microbiol. 2017, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allison, K.R.; Brynildsen, M.P.; Collins, J.J. Heterogeneous bacterial persisters and engineering approaches to eliminate them. Curr. Opin. Microbiol. 2011, 14, 593–598. [Google Scholar] [CrossRef] [Green Version]
- Lewis, K. Persister cells. Annu. Rev. Microbiol. 2010, 64, 357–372. [Google Scholar] [CrossRef]
- Sanz-García, F.; Hernando-Amado, S.; Martínez, J.L. Mutation-driven evolution of Pseudomonas aeruginosa in the presence of either ceftazidime or ceftazidime-avibactam. Antimicrob. Agents Chemother. 2018, 62, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Koskella, B.; Brockhurst, M.A. Bacteria-phage coevolution as a driver of ecological and evolutionary processes in microbial communities. FEMS Microbiol. Rev. 2014, 38, 916–931. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oechslin, F.; Piccardi, P.; Mancini, S.; Gabard, J.; Moreillon, P.; Entenza, J.M.; Resch, G.; Que, Y.-A. Synergistic interaction between phage therapy and antibiotics clears Pseudomonas aeruginosa infection in endocarditis and reduces virulence. J. Infect. Dis. 2017, 215, 703–712. [Google Scholar] [CrossRef] [Green Version]
- Roach, D.R.; Leung, C.Y.; Henry, M.; Morello, E.; Singh, D.; Di Santo, J.P.; Weitz, J.S.; Debarbieux, L. Synergy between the host immune system and bacteriophage is essential for successful phage therapy against an acute respiratory pathogen. Cell Host Microbe 2017, 22, 38–47.e4. [Google Scholar] [CrossRef] [PubMed]
- Labrie, S.J.; Samson, J.E.; Moineau, S. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 2010, 8, 317–327. [Google Scholar] [CrossRef]
- De Smet, J.; Hendrix, H.; Blasdel, B.G.; Danis-Wlodarczyk, K.; Lavigne, R. Pseudomonas predators: Understanding and exploiting phage-host interactions. Nat. Rev. Microbiol. 2017, 15, 517–530. [Google Scholar] [CrossRef]
- Majkowska-Skrobek, G.; Markwitz, P.; Sosnowska, E.; Lood, C.; Lavigne, R.; Drulis-Kawa, Z. The evolutionary trade-offs in phage-resistant Klebsiella pneumoniae entail cross-phage sensitization and loss of multidrug resistance. Environ. Microbiol. 2021, 1462–2920. [Google Scholar] [CrossRef]
- Olszak, T.; Shneider, M.M.; Latka, A.; Maciejewska, B.; Browning, C.; Sycheva, L.V.; Cornelissen, A.; Danis-Wlodarczyk, K.; Senchenkova, S.N.; Shashkov, A.S.; et al. The O-specific polysaccharide lyase from the phage LKA1 tailspike reduces Pseudomonas virulence. Sci. Rep. 2017, 7, 16302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ha, D.-G.; Kuchma, S.L.; O’Toole, G.A. Plate-based assay for swimming motility in Pseudomonas aeruginosa. In Pseudomonas Methods and Protocols; Filloux, A., Ramos, J.L., Eds.; Humana Press Inc.: New York, NY, USA, 2014; pp. 59–66. [Google Scholar]
- Turnbull, L.; Whitchurch, C.B. Motility assay: Twitching motility. In Pseudomonas Methods and Protocols; Filloux, A., Ramos, J.-L., Eds.; Humana Press Inc.: New York, NY, USA, 2014; pp. 73–86. [Google Scholar]
- Ha, D.-G.; Kuchma, S.L.; O’Toole, G.A. Plate-based assay for swarming motility in Pseudomonas aeruginosa. In Pseudomonas Methods and Protocols; Filloux, A., Ramos, J.L., Eds.; Humana Press Inc.: New York, NY, USA, 2014; pp. 73–86. [Google Scholar]
- Das, T.; Manefield, M. Pyocyanin promotes extracellular DNA release in Pseudomonas aeruginosa. PLoS ONE 2012, 7, e46718. [Google Scholar] [CrossRef] [Green Version]
- Rust, L.; Messing, C.R.; Iglewski, B.H. Elastase assays. Methods Enzymol. 1994, 235, 554–562. [Google Scholar]
- O’Toole, G.A. Microtiter dish biofilm formation assay. J. Vis. Exp. 2011. [CrossRef]
- Khan, F.; Lee, J.-W.; Javaid, A.; Park, S.-K.; Kim, Y.-M. Inhibition of biofilm and virulence properties of Pseudomonas aeruginosa by sub-inhibitory concentrations of aminoglycosides. Microb. Pathog. 2020, 146, 104249. [Google Scholar] [CrossRef]
- Westphal, O.; Jann, K. Methods in Carbohydrate Chemistry. In Methods in Carbohydrate Chemistry Volume V; Whistler, L.R., Ed.; Academic Press: New York, NY, USA, 1965; p. 83. [Google Scholar]
- Marolda, C.L.; Welsh, J.; Dafoe, L.; Valvano, M.A. Genetic analysis of the O7-polysaccharide biosynthesis region from the Escherichia coli O7:K1 strain VW187. J. Bacteriol. 1990, 172, 3590–3599. [Google Scholar] [CrossRef] [Green Version]
- Lee, C.H.; Tsai, C.M. Quantification of bacterial lipopolysaccharides by the purpald assay: Measuring formaldehyde generated from 2-keto-3-deoxyoctonate and heptose at the inner core by periodate oxidation. Anal. Biochem. 1999, 267, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.M.; Frasch, C.E. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal. Biochem. 1982, 119, 115–119. [Google Scholar] [CrossRef]
- Rand, J.H.; Wu, X.-X.; Lin, E.Y.; Griffel, A.; Gialanella, P.; McKitrick, J.C. Annexin A5 binds to lipopolysaccharide and reduces its endotoxin activity. MBio 2012, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Courcot, E.; Leclerc, J.; Lafitte, J.-J.; Mensier, E.; Jaillard, S.; Gosset, P.; Shirali, P.; Pottier, N.; Broly, F.; Lo-Guidice, J.-M. Xenobiotic metabolism and disposition in human lung cell models: Comparison with in vivo expression profiles. Drug Metab. Dispos. 2012, 40, 1953–1965. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gałczyńska, K.; Ciepluch, K.; Madej, Ł.; Kurdziel, K.; Maciejewska, B.; Drulis-Kawa, Z.; Węgierek-Ciuk, A.; Lankoff, A.; Arabski, M. Selective cytotoxicity and antifungal properties of copper(II) and cobalt(II) complexes with imidazole-4-acetate anion or 1-allylimidazole. Sci. Rep. 2019, 9, 9777. [Google Scholar] [CrossRef]
- Cullen, L.; Weiser, R.; Olszak, T.; Maldonado, R.; Moreira, A.; Slachmuylders, L.; Brackman, G.; Paunova-Krasteva, T.; Zarnowiec, P.; Czerwonka, G.; et al. Phenotypic characterization of an international Pseudomonas aeruginosa reference panel: Strains of cystic fibrosis (CF) origin show less in vivo virulence than non-CF strains. Microbiology 2015, 161, 1961–1977. [Google Scholar] [CrossRef] [PubMed]
- Lorè, N.I.; Cigana, C.; De Fino, I.; Riva, C.; Juhas, M.; Schwager, S.; Eberl, L.; Bragonzi, A. Cystic fibrosis-niche adaptation of Pseudomonas aeruginosa reduces virulence in multiple infection hosts. PLoS ONE 2012, 7, e35648. [Google Scholar] [CrossRef]
- Danis-Wlodarczyk, K.; Olszak, T.; Arabski, M.; Wasik, S.; Majkowska-Skrobek, G.; Augustyniak, D.; Gula, G.; Briers, Y.; Jang, H.B.; Vandenheuvel, D.; et al. Characterization of the newly isolated lytic bacteriophages KTN6 and KT28 and their efficacy against Pseudomonas aeruginosa biofilm. PLoS ONE 2015, 10, e0127603. [Google Scholar] [CrossRef]
- Forti, F.; Roach, D.R.; Cafora, M.; Pasini, M.E.; Horner, D.S.; Fiscarelli, E.V.; Rossitto, M.; Cariani, L.; Briani, F.; Debarbieux, L.; et al. Design of a broad-range bacteriophage cocktail that reduces Pseudomonas aeruginosa biofilms and treats acute infections in two animal models. Antimicrob. Agents Chemother. 2018, 62, e02573-17. [Google Scholar] [CrossRef] [Green Version]
- Latz, S.; Krüttgen, A.; Häfner, H.; Buhl, E.; Ritter, K.; Horz, H.-P. Differential effect of newly isolated phages belonging to PB1-Like, phiKZ-Like and LUZ24-Like viruses against multi-drug resistant Pseudomonas aeruginosa under varying growth conditions. Viruses 2017, 9, 315. [Google Scholar] [CrossRef] [Green Version]
- Latino, L.; Midoux, C.; Hauck, Y.; Vergnaud, G.; Pourcel, C. Pseudolysogeny and sequential mutations build multiresistance to virulent bacteriophages in Pseudomonas aeruginosa. Microbiology 2016, 162, 748–763. [Google Scholar] [CrossRef]
- Shen, M.; Zhang, H.; Shen, W.; Zou, Z.; Lu, S.; Li, G.; He, X.; Agnello, M.; Shi, W.; Hu, F.; et al. Pseudomonas aeruginosa MutL promotes large chromosomal deletions through non-homologous end joining to prevent bacteriophage predation. Nucleic Acids Res. 2018, 46, 4505–4514. [Google Scholar] [CrossRef] [Green Version]
- Wright, R.C.T.; Friman, V.-P.; Smith, M.C.M.; Brockhurst, M.A. Cross-resistance is modular in bacteria–phage interactions. PLoS Biol. 2018, 16, e2006057. [Google Scholar] [CrossRef] [PubMed]
- Ripp, S.; Miller, R.V. Dynamics of the pseudolysogenic response in slowly growing cells of Pseudomonas aeruginosa. Microbiology 1998. [CrossRef] [Green Version]
- Hosseinidoust, Z.; Tufenkji, N.; van de Ven, T.G.M. Predation in homogeneous and heterogeneous phage environments affects virulence determinants of Pseudomonas aeruginosa. Appl. Environ. Microbiol. 2013, 79, 2862–2871. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Merz, A.J.; So, M.; Sheetz, M.P. Pilus retraction powers bacterial twitching motility. Nature 2000, 407, 98–102. [Google Scholar] [CrossRef] [PubMed]
- Mattick, J.S. Type IV pili and twitching motility. Annu. Rev. Microbiol. 2002, 56, 289–314. [Google Scholar] [CrossRef]
- Burns, S.M.; Hull, S.I. Comparison of loss of serum resistance by defined lipopolysaccharide mutants and an acapsular mutant of uropathogenic Escherichia coli O75:K5. Infect. Immun. 1998, 66, 4244–4253. [Google Scholar] [CrossRef]
- Grossman, N.; Leive, L. Complement activation via the alternative pathway by purified Salmonella lipopolysaccharide is affected by its structure but not its O-antigen length. J. Immunol. 1984, 132, 376–385. [Google Scholar] [PubMed]

| Phage | Taxonomy (Family, Genus) | Genome Size | GenBank Accession Number | Recognized Bacterial Receptor |
|---|---|---|---|---|
| LUZ7 * | Schitoviridae,Luzseptimavirus | 74,901 bp | NC_013691 | LPS |
| KTN6 ** | Myoviridae, Pbunavirus | 65,994 bp | KP340288 | LPS |
| KT28 ** | Myoviridae, Pbunavirus | 66,381 bp | KP340287 | LPS |
| LUZ19 * | Autographiviridae, Phikmvvirus | 43,548 bp | NC_010326 | T4P |
| phiKZ * | Myoviridae, Phikzvirus | 280,334 bp | AF399011 | T4P |
| KTN4 ** | Myoviridae, Phikzvirus | 279,593 bp | KU521356 | T4P |
| PA5oct ** | Myoviridae, unclassified | 286,783 bp | MK797984 | LPS/T4P |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Markwitz, P.; Olszak, T.; Gula, G.; Kowalska, M.; Arabski, M.; Drulis-Kawa, Z. Emerging Phage Resistance in Pseudomonas aeruginosa PAO1 Is Accompanied by an Enhanced Heterogeneity and Reduced Virulence. Viruses 2021, 13, 1332. https://doi.org/10.3390/v13071332
Markwitz P, Olszak T, Gula G, Kowalska M, Arabski M, Drulis-Kawa Z. Emerging Phage Resistance in Pseudomonas aeruginosa PAO1 Is Accompanied by an Enhanced Heterogeneity and Reduced Virulence. Viruses. 2021; 13(7):1332. https://doi.org/10.3390/v13071332
Chicago/Turabian StyleMarkwitz, Pawel, Tomasz Olszak, Grzegorz Gula, Magdalena Kowalska, Michal Arabski, and Zuzanna Drulis-Kawa. 2021. "Emerging Phage Resistance in Pseudomonas aeruginosa PAO1 Is Accompanied by an Enhanced Heterogeneity and Reduced Virulence" Viruses 13, no. 7: 1332. https://doi.org/10.3390/v13071332
APA StyleMarkwitz, P., Olszak, T., Gula, G., Kowalska, M., Arabski, M., & Drulis-Kawa, Z. (2021). Emerging Phage Resistance in Pseudomonas aeruginosa PAO1 Is Accompanied by an Enhanced Heterogeneity and Reduced Virulence. Viruses, 13(7), 1332. https://doi.org/10.3390/v13071332







