Identification of a Ruminant Origin Group B Rotavirus Associated with Diarrhea Outbreaks in Foals
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Fecal Sample Collection and Viral Metagenomic Sequencing
2.3. Genome Sequencing and Analysis
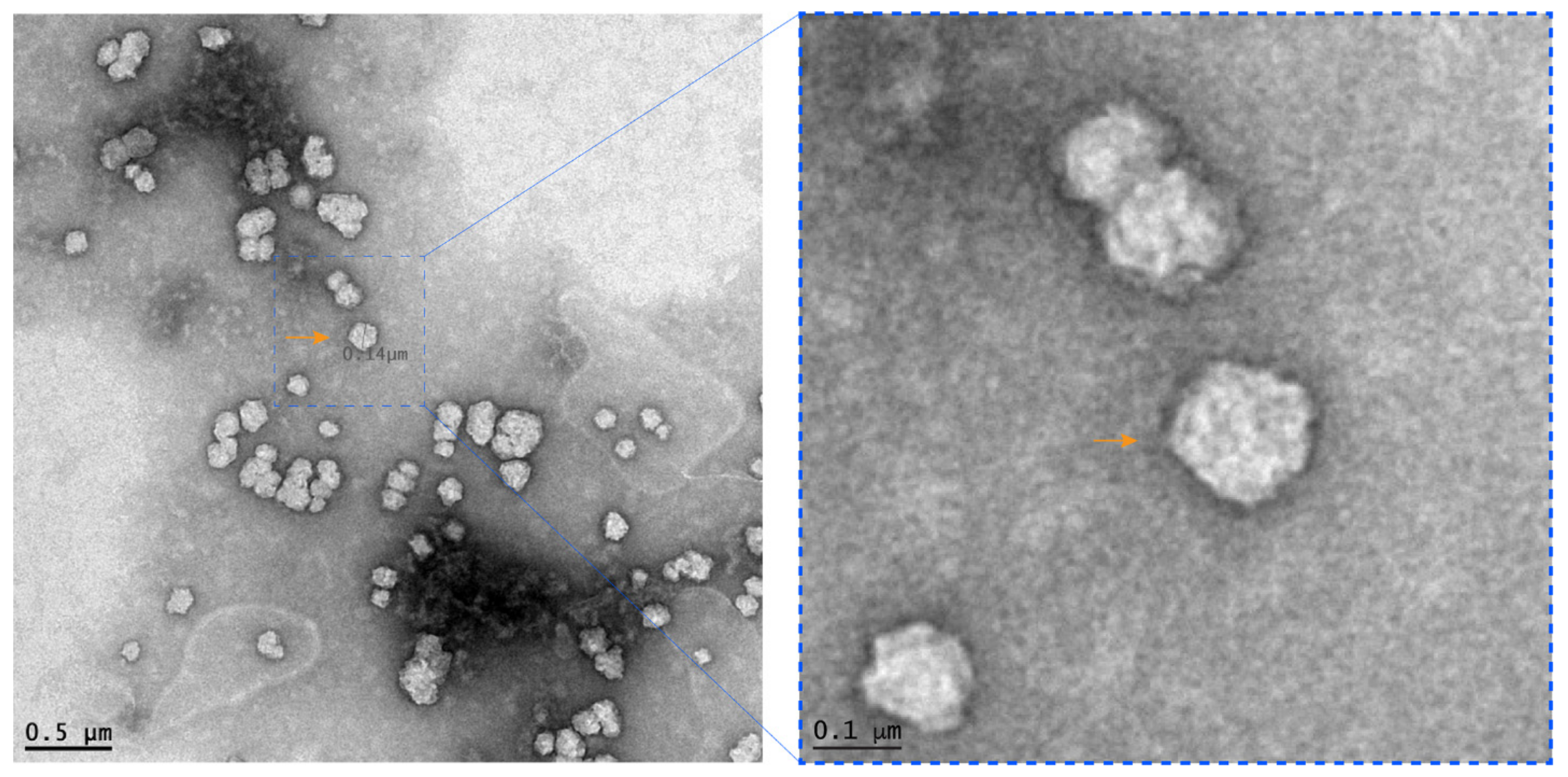
2.4. Transmission Electron Microscopy
2.5. Detection of Equine Rotavirus Group B Genome in Clinical Samples
3. Results
3.1. Foal Diarrhea Outbreak
3.2. Identification of a Novel Group B Rotavirus by Next-Generation Sequencing
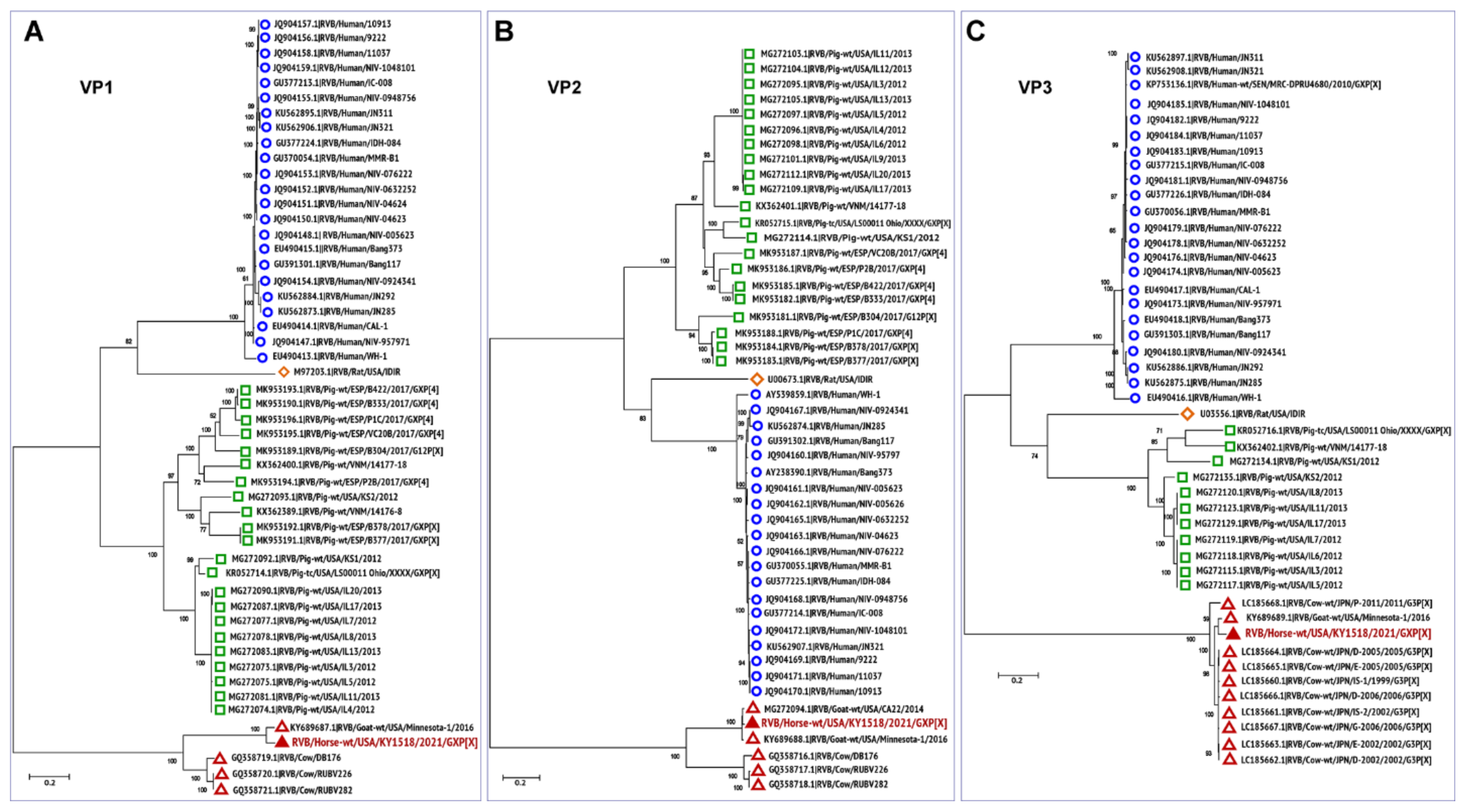
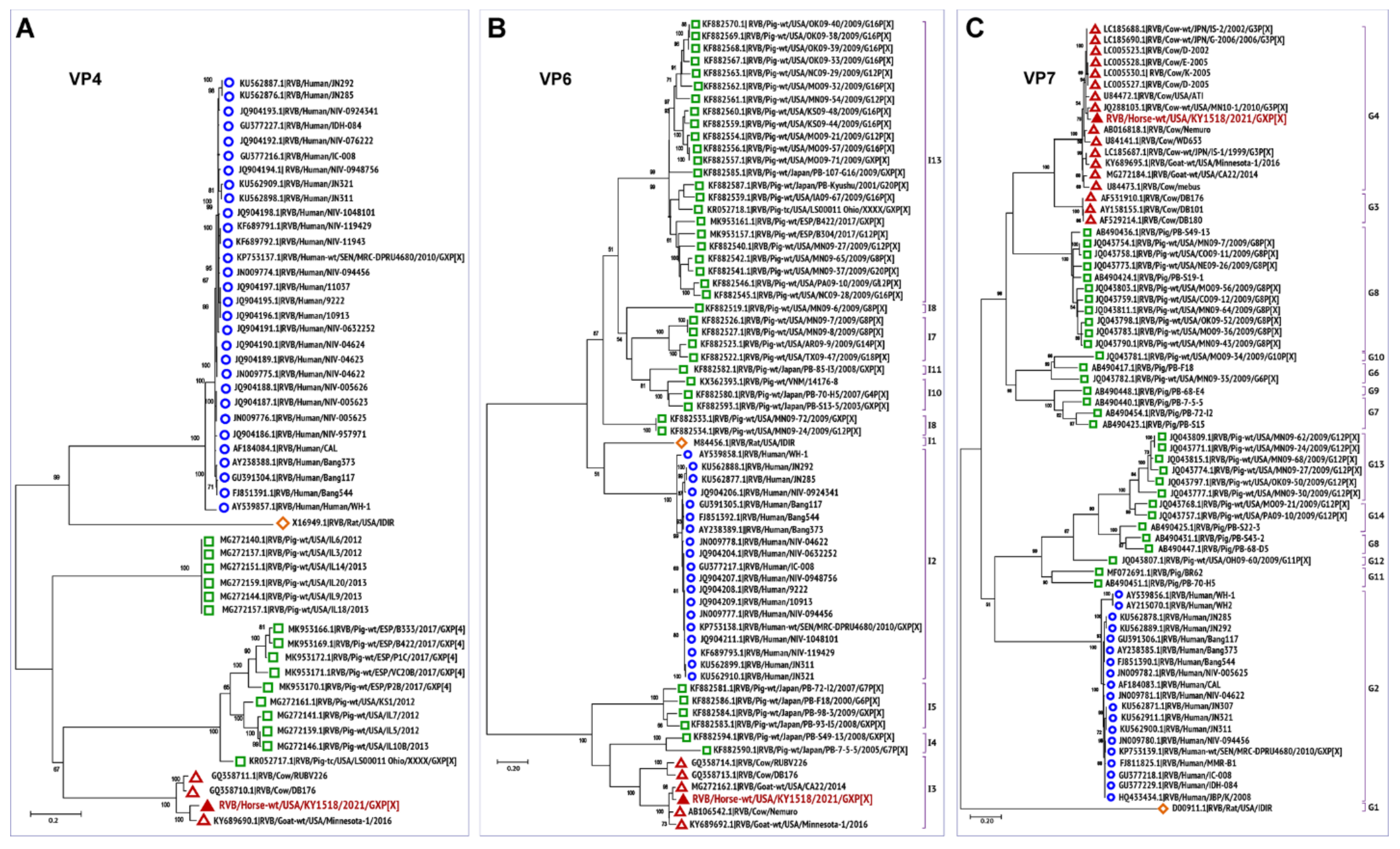
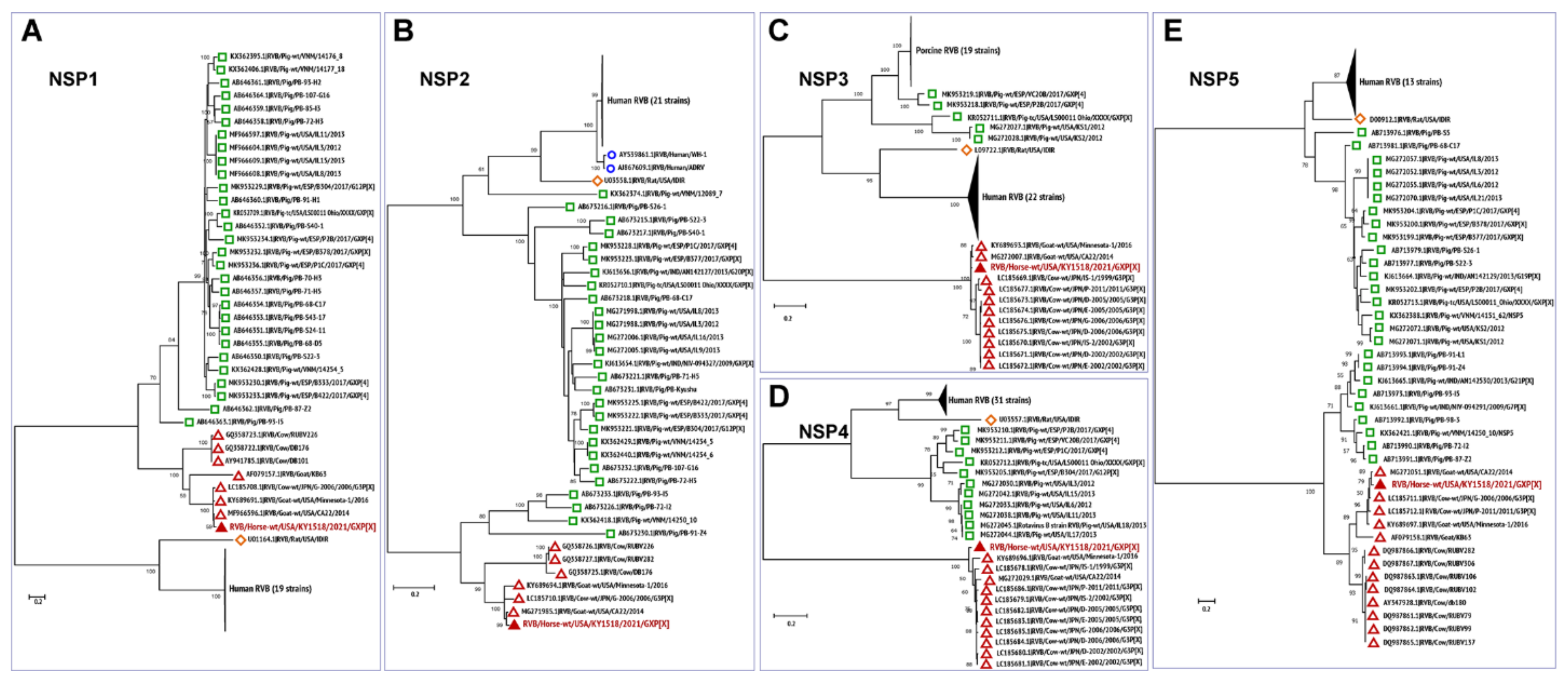
3.3. Phylogenetic Analysis
3.4. Detection of Equine Rotavirus Group B by RT-qPCR
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Troeger, C.; Khalil, I.A.; Rao, P.C.; Cao, S.; Blacker, B.F.; Ahmed, T.; Armah, G.; Bines, J.E.; Brewer, T.G.; Colombara, D.V.; et al. Rotavirus Vaccination and the Global Burden of Rotavirus Diarrhea Among Children Younger Than 5 Years. JAMA Pediatr. 2018, 172, 958–965. [Google Scholar] [CrossRef]
- Dhama, K.; Chauhan, R.S.; Mahendran, M.; Malik, S.V. Rotavirus diarrhea in bovines and other domestic animals. Vet. Res. Commun. 2009, 33, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Anderson, E.J.; Weber, S.G. Rotavirus infection in adults. Lancet Infect. Dis. 2004, 4, 91–99. [Google Scholar] [CrossRef]
- Crawford, S.E.; Ramani, S.; Tate, J.E.; Parashar, U.D.; Svensson, L.; Hagbom, M.; Franco, M.A.; Greenberg, H.B.; O’Ryan, M.; Kang, G.; et al. Rotavirus infection. Nat. Rev. Dis. Primers 2017, 3, 17083. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Crawford, S.E.; Hyser, J.M.; Estes, M.K.; Prasad, B.V. Rotavirus non-structural proteins: Structure and function. Curr. Opin. Virol. 2012, 2, 380–388. [Google Scholar] [CrossRef] [PubMed]
- Trojnar, E.; Otto, P.; Roth, B.; Reetz, J.; Johne, R. The genome segments of a group D rotavirus possess group A-like conserved termini but encode group-specific proteins. J. Virol. 2010, 84, 10254–10265. [Google Scholar] [CrossRef]
- Zhang, X.; Settembre, E.; Xu, C.; Dormitzer, P.R.; Bellamy, R.; Harrison, S.C.; Grigorieff, N. Near-atomic resolution using electron cryomicroscopy and single-particle reconstruction. Proc. Natl. Acad. Sci. USA 2008, 105, 1867–1872. [Google Scholar] [CrossRef]
- Tang, B.; Gilbert, J.M.; Matsui, S.M.; Greenberg, H.B. Comparison of the rotavirus gene 6 from different species by sequence analysis and localization of subgroup-specific epitopes using site-directed mutagenesis. Virology 1997, 237, 89–96. [Google Scholar] [CrossRef][Green Version]
- Matthijnssens, J.; Otto, P.H.; Ciarlet, M.; Desselberger, U.; Van Ranst, M.; Johne, R. VP6-sequence-based cutoff values as a criterion for rotavirus species demarcation. Arch. Virol. 2012, 157, 1177–1182. [Google Scholar] [CrossRef] [PubMed]
- Walker, P.J.; Siddell, S.G.; Lefkowitz, E.J.; Mushegian, A.R.; Dempsey, D.M.; Dutilh, B.E.; Harrach, B.; Harrison, R.L.; Hendrickson, R.C.; Junglen, S.; et al. Changes to virus taxonomy and the International Code of Virus Classification and Nomenclature ratified by the International Committee on Taxonomy of Viruses. Arch. Virol. 2019, 164, 2417–2429. [Google Scholar] [CrossRef]
- Hoshino, Y.; Kapikian, A.Z. Classification of rotavirus VP4 and VP7 serotypes. Arch. Virol. Suppl. 1996, 12, 99–111. [Google Scholar] [CrossRef]
- van der Heide, R.; Koopmans, M.P.; Shekary, N.; Houwers, D.J.; van Duynhoven, Y.T.; van der Poel, W.H. Molecular characterizations of human and animal group a rotaviruses in the Netherlands. J. Clin. Microbiol. 2005, 43, 669–675. [Google Scholar] [CrossRef]
- Steele, A.; Geyer, A.; Gerdes, G.; Coetzer, J.; Tustin, R. Rotavirus infections. In Infectious Diseases of Livestock; Coetzer, J.A.W., Tustin, R.C., Eds.; Oxford University Press: Cape Town, South Africa, 2004; pp. 1256–1264. [Google Scholar]
- Trovao, N.S.; Shepherd, F.K.; Herzberg, K.; Jarvis, M.C.; Lam, H.C.; Rovira, A.; Culhane, M.R.; Nelson, M.I.; Marthaler, D.G. Evolution of rotavirus C in humans and several domestic animal species. Zoonoses Public Health 2019, 66, 546–557. [Google Scholar] [CrossRef]
- Deol, P.; Kattoor, J.J.; Sircar, S.; Ghosh, S.; Banyai, K.; Dhama, K.; Malik, Y.S. Avian Group D Rotaviruses: Structure, Epidemiology, Diagnosis, and Perspectives on Future Research Challenges. Pathogens 2017, 6, 53. [Google Scholar] [CrossRef]
- Sanekata, T.; Ahmed, M.U.; Kader, A.; Taniguchi, K.; Kobayashi, N. Human group B rotavirus infections cause severe diarrhea in children and adults in Bangladesh. J. Clin. Microbiol. 2003, 41, 2187–2190. [Google Scholar] [CrossRef] [PubMed]
- Tao, H.; Changan, W.; Zhaoying, F.; Zinyi, C.; Xuejian, C.; Xiaoquang, L.; Guangmu, C.; Henli, Y.; Tungxin, C.; Weiwe, Y.; et al. Waterborne outbreak of rotavirus diarrhoea in adults in China caused by a novel rotavirus. Lancet 1984, 323, 1139–1142. [Google Scholar] [CrossRef]
- Chen, G.-M.; Hung, T.; Bridger, J.C.; McCrae, M.A. Chinese adult rotavirus is A group B rotavirus. Lancet 1985, 326, 1123–1124. [Google Scholar] [CrossRef]
- Chang, K.O.; Parwani, A.V.; Smith, D.; Saif, L.J. Detection of group B rotaviruses in fecal samples from diarrheic calves and adult cows and characterization of their VP7 genes. J. Clin. Microbiol. 1997, 35, 2107–2110. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.A.; Khamrin, P.; Trinh, Q.D.; Phan, T.G.; le Duc, P.; le Phuc, H.; Hoang, K.T.; Yagyu, F.; Okitsu, S.; Ushijima, H. Sequence analysis of Vietnamese P(6) rotavirus strains suggests evidence of interspecies transmission. J. Med. Virol. 2007, 79, 1959–1965. [Google Scholar] [CrossRef]
- Cook, N.; Bridger, J.; Kendall, K.; Gomara, M.I.; El-Attar, L.; Gray, J. The zoonotic potential of rotavirus. J. Infect. 2004, 48, 289–302. [Google Scholar] [CrossRef] [PubMed]
- Muller, H.; Johne, R. Rotaviruses: Diversity and zoonotic potential—A brief review. Berl. Munch Tierarztl. Wochenschr. 2007, 120, 108–112. [Google Scholar] [PubMed]
- Martella, V.; Banyai, K.; Matthijnssens, J.; Buonavoglia, C.; Ciarlet, M. Zoonotic aspects of rotaviruses. Vet. Microbiol. 2010, 140, 246–255. [Google Scholar] [CrossRef]
- Mukherjee, A.; Mullick, S.; Deb, A.K.; Panda, S.; Chawla-Sarkar, M. First report of human rotavirus G8P(4) gastroenteritis in India: Evidence of ruminants-to-human zoonotic transmission. J. Med. Virol. 2013, 85, 537–545. [Google Scholar] [CrossRef]
- Mukherjee, A.; Dutta, D.; Ghosh, S.; Bagchi, P.; Chattopadhyay, S.; Nagashima, S.; Kobayashi, N.; Dutta, P.; Krishnan, T.; Naik, T.N.; et al. Full genomic analysis of a human group A rotavirus G9P(6) strain from Eastern India provides evidence for porcine-to-human interspecies transmission. Arch. Virol. 2009, 154, 733–746. [Google Scholar] [CrossRef]
- Komoto, S.; Tacharoenmuang, R.; Guntapong, R.; Ide, T.; Tsuji, T.; Yoshikawa, T.; Tharmaphornpilas, P.; Sangkitporn, S.; Taniguchi, K. Reassortment of Human and Animal Rotavirus Gene Segments in Emerging DS-1-Like G1P(8) Rotavirus Strains. PLoS ONE 2016, 11, e0148416. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Knutson, T.P.; Ciarlet, M.; Sturos, M.; Marthaler, D.G. Complete genome characterization of a rotavirus B (RVB) strain identified in Alpine goat kids with enteritis reveals inter-species transmission with RVB bovine strains. J. Gen. Virol. 2018, 99, 457–463. [Google Scholar] [CrossRef]
- Miño, S.; Adúriz, M.; Barrandeguy, M.; Parreño, V. Molecular Characterization of Equine Rotavirus Group A Detected in Argentinean Foals During 2009–2014. J. Equine Vet. Sci. 2017, 59, 64–70. [Google Scholar] [CrossRef]
- Hoshino, Y.; Wyatt, R.G.; Greenberg, H.B.; Flores, J.; Kapikian, A.Z. Serotypic similarity and diversity of rotaviruses of mammalian and avian origin as studied by plaque-reduction neutralization. J. Infect. Dis. 1984, 149, 694–702. [Google Scholar] [CrossRef]
- Browning, G.F.; Fitzgerald, T.A.; Chalmers, R.M.; Snodgrass, D.R. A novel group A rotavirus G serotype: Serological and genomic characterization of equine isolate FI23. J. Clin. Microbiol. 1991, 29, 2043–2046. [Google Scholar] [CrossRef] [PubMed]
- Imagawa, H.; Tanaka, T.; Sekiguchi, K.; Fukunaga, Y.; Anzai, T.; Minamoto, N.; Kamada, M. Electropherotypes, serotypes, and subgroups of equine rotaviruses isolated in Japan. Arch. Virol. 1993, 131, 169–176. [Google Scholar] [CrossRef]
- Isa, P.; Wood, A.R.; Netherwood, T.; Ciarlet, M.; Imagawa, H.; Snodgrass, D.R. Survey of equine rotaviruses shows conservation of one P genotype in background of two G genotypes. Arch. Virol. 1996, 141, 1601–1612. [Google Scholar] [CrossRef] [PubMed]
- Imagawa, H.; Ishida, S.; Uesugi, S.; Masanobu, K.; Fukunaga, Y.; Nakagomi, O. Genetic analysis of equine rotavirus by RNA-RNA hybridization. J. Clin. Microbiol. 1994, 32, 2009–2012. [Google Scholar] [CrossRef]
- Browning, G.F.; Chalmers, R.M.; Fitzgerald, T.A.; Snodgrass, D.R. Serological and genomic characterization of L338, a novel equine group A rotavirus G serotype. J. Gen. Virol. 1991, 72 Pt 5, 1059–1064. [Google Scholar] [CrossRef] [PubMed]
- Hardy, M.E.; Gorziglia, M.; Woode, G.N. The outer capsid protein VP4 of equine rotavirus strain H-2 represents a unique VP4 type by amino acid sequence analysis. Virology 1993, 193, 492–497. [Google Scholar] [CrossRef]
- Isa, P.; Snodgrass, D.R. Serological and genomic characterization of equine rotavirus VP4 proteins identifies three different P serotypes. Virology 1994, 201, 364–372. [Google Scholar] [CrossRef]
- Collins, P.J.; Cullinane, A.; Martella, V.; O’Shea, H. Molecular characterization of equine rotavirus in Ireland. J. Clin. Microbiol. 2008, 46, 3346–3354. [Google Scholar] [CrossRef] [PubMed]
- Carossino, M.; Barrandeguy, M.E.; Li, Y.; Parreno, V.; Janes, J.; Loynachan, A.T.; Balasuriya, U.B.R. Detection, molecular characterization and phylogenetic analysis of G3P(12) and G14P(12) equine rotavirus strains co-circulating in central Kentucky. Virus Res. 2018, 255, 39–54. [Google Scholar] [CrossRef]
- Athiyyah, A.F.; Utsumi, T.; Wahyuni, R.M.; Dinana, Z.; Yamani, L.N. Molecular Epidemiology and Clinical Features of Rotavirus Infection Among Pediatric Patients in East Java, Indonesia During 2015–2018: Dynamic Changes in Rotavirus Genotypes from Equine-Like G3 to Typical Human G1/G3. Front. Microbiol. 2019, 10, 940. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Shintani, T.; Kobayashi, N. Evidence for the porcine origin of equine rotavirus strain H-1. Vet. Microbiol. 2012, 158, 410–414. [Google Scholar] [CrossRef] [PubMed]
- Mino, S.; Matthijnssens, J.; Badaracco, A.; Garaicoechea, L.; Zeller, M.; Heylen, E.; Van Ranst, M.; Barrandeguy, M.; Parreno, V. Equine G3P(3) rotavirus strain E3198 related to simian RRV and feline/canine-like rotaviruses based on complete genome analyses. Vet. Microbiol. 2013, 161, 239–246. [Google Scholar] [CrossRef]
- Bailey, K.E.; Gilkerson, J.R.; Browning, G.F. Equine rotaviruses—Current understanding and continuing challenges. Vet. Microbiol. 2013, 167, 135–144. [Google Scholar] [CrossRef] [PubMed]
- Otto, P.H.; Rosenhain, S.; Elschner, M.C.; Hotzel, H.; Machnowska, P.; Trojnar, E.; Hoffmann, K.; Johne, R. Detection of rotavirus species A, B and C in domestic mammalian animals with diarrhoea and genotyping of bovine species A rotavirus strains. Vet. Microbiol. 2015, 179, 168–176. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Shepherd, F.K.; Herrera-Ibata, D.M.; Porter, E.; Homwong, N.; Hesse, R.; Bai, J.; Marthaler, D.G. Whole Genome Classification and Phylogenetic Analyses of Rotavirus B strains from the United States. Pathogens 2018, 7, 44. [Google Scholar] [CrossRef]
- Lu, X.; McDonald, S.M.; Tortorici, M.A.; Tao, Y.J.; Vasquez-Del Carpio, R.; Nibert, M.L.; Patton, J.T.; Harrison, S.C. Mechanism for coordinated RNA packaging and genome replication by rotavirus polymerase VP1. Structure 2008, 16, 1678–1688. [Google Scholar] [CrossRef] [PubMed]
- McDonald, S.M.; Patton, J.T. Rotavirus VP2 core shell regions critical for viral polymerase activation. J. Virol. 2011, 85, 3095–3105. [Google Scholar] [CrossRef]
- Patton, J.T.; Chen, D. RNA-binding and capping activities of proteins in rotavirus open cores. J. Virol. 1999, 73, 1382–1391. [Google Scholar] [CrossRef]
- Díaz-Salinas, M.A.; Romero, P.; Espinosa, R.; Hoshino, Y.; López, S.; Arias, C.F. The Spike Protein VP4 Defines the Endocytic Pathway Used by Rotavirus to Enter MA104 Cells. J. Virol. 2013, 87, 1658–1663. [Google Scholar] [CrossRef]
- Lopez, S.; Arias, C.F. Multistep entry of rotavirus into cells: A Versaillesque dance. Trends Microbiol. 2004, 12, 271–278. [Google Scholar] [CrossRef]
- Greenberg, H.B.; Estes, M.K. Rotaviruses: From pathogenesis to vaccination. Gastroenterology 2009, 136, 1939–1951. [Google Scholar] [CrossRef]
- Marthaler, D.; Suzuki, T.; Rossow, K.; Culhane, M.; Collins, J.; Goyal, S.; Tsunemitsu, H.; Ciarlet, M.; Matthijnssens, J. VP6 genetic diversity, reassortment, intragenic recombination and classification of rotavirus B in American and Japanese pigs. Vet. Microbiol. 2014, 172, 359–366. [Google Scholar] [CrossRef]
- Barro, M.; Patton, J.T. Rotavirus NSP1 inhibits expression of type I interferon by antagonizing the function of interferon regulatory factors IRF3, IRF5, and IRF7. J. Virol. 2007, 81, 4473–4481. [Google Scholar] [CrossRef] [PubMed]
- Eichwald, C.; Rodriguez, J.F.; Burrone, O.R. Characterization of rotavirus NSP2/NSP5 interactions and the dynamics of viroplasm formation. J. Gen. Virol. 2004, 85, 625–634. [Google Scholar] [CrossRef]
- Elschner, M.; Schrader, C.; Hotzel, H.; Prudlo, J.; Sachse, K.; Eichhorn, W.; Herbst, W.; Otto, P. Isolation and molecular characterisation of equine rotaviruses from Germany. Vet. Microbiol. 2005, 105, 123–129. [Google Scholar] [CrossRef]
- Browning, G.F.; Begg, A.P. Prevalence of G and P serotypes among equine rotaviruses in the faeces of diarrhoeic foals. Arch. Virol. 1996, 141, 1077–1089. [Google Scholar] [CrossRef] [PubMed]
- Tsunemitsu, H.; Imagawa, H.; Togo, M.; Shouji, T.; Kawashima, K.; Horino, R.; Imai, K.; Nishimori, T.; Takagi, M.; Higuchi, T. Predominance of G3B and G14 equine group A rotaviruses of a single VP4 serotype in Japan. Arch. Virol. 2001, 146, 1949–1962. [Google Scholar] [CrossRef]
- Eiden, J.J.; Nataro, J.; Vonderfecht, S.; Petric, M. Molecular cloning, sequence analysis, in vitro expression, and immunoprecipitation of the major inner capsid protein of the IDIR strain of group B rotavirus (GBR). Virology 1992, 188, 580–589. [Google Scholar] [CrossRef]
- Miyabe, F.M.; Dall Agnol, A.M.; Leme, R.A.; Oliveira, T.E.S.; Headley, S.A.; Fernandes, T.; de Oliveira, A.G.; Alfieri, A.F.; Alfieri, A.A. Porcine rotavirus B as primary causative agent of diarrhea outbreaks in newborn piglets. Sci. Rep. 2020, 10, 22002. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, M.; Murakami, T.; Kuroda, Y.; Takai, H.; Ide, H.; Awang, A.; Suzuki, T.; Miyazaki, A.; Nagai, M.; Tsunemitsu, H. Reinfection of adult cattle with rotavirus B during repeated outbreaks of epidemic diarrhea. Can. J. Vet. Res. 2016, 80, 189–196. [Google Scholar]
- Theil, K.W.; Grooms, D.L.; McCloskey, C.M.; Redman, D.R. Group B rotavirus associated with an outbreak of neonatal lamb diarrhea. J. Vet. Diagn. Investig. 1995, 7, 148–150. [Google Scholar] [CrossRef]
- Joshi, M.S.; Ganorkar, N.N.; Ranshing, S.S.; Basu, A.; Chavan, N.A.; Gopalkrishna, V. Identification of group B rotavirus as an etiological agent in the gastroenteritis outbreak in Maharashtra, India. J. Med. Virol. 2017, 89, 2244–2248. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, T.; Sen, A.; Choudhury, J.S.; Das, S.; Naik, T.N.; Bhattacharya, S.K. Emergence of adult diarrhoea rotavirus in Calcutta, India. Lancet 1999, 353, 380–381. [Google Scholar] [CrossRef]
- Kuga, K.; Miyazaki, A.; Suzuki, T.; Takagi, M.; Hattori, N.; Katsuda, K.; Mase, M.; Sugiyama, M.; Tsunemitsu, H. Genetic diversity and classification of the outer capsid glycoprotein VP7 of porcine group B rotaviruses. Arch. Virol. 2009, 154, 1785–1795. [Google Scholar] [CrossRef] [PubMed]
- Marthaler, D.; Rossow, K.; Gramer, M.; Collins, J.; Goyal, S.; Tsunemitsu, H.; Kuga, K.; Suzuki, T.; Ciarlet, M.; Matthijnssens, J. Detection of substantial porcine group B rotavirus genetic diversity in the United States, resulting in a modified classification proposal for G genotypes. Virology 2012, 433, 85–96. [Google Scholar] [CrossRef]
- Alekseev, K.P.; Penin, A.A.; Mukhin, A.N.; Khametova, K.M.; Grebennikova, T.V.; Yuzhakov, A.G.; Moskvina, A.S.; Musienko, M.I.; Raev, S.A.; Mishin, A.M.; et al. Genome Characterization of a Pathogenic Porcine Rotavirus B Strain Identified in Buryat Republic, Russia in 2015. Pathogens 2018, 7, 46. [Google Scholar] [CrossRef]
- Papp, H.; Malik, Y.S.; Farkas, S.L.; Jakab, F.; Martella, V.; Banyai, K. Rotavirus strains in neglected animal species including lambs, goats and camelids. VirusDisease 2014, 25, 215–222. [Google Scholar] [CrossRef] [PubMed]
- Holland, R.E. Some infectious causes of diarrhea in young farm animals. J. Clin. Microbiol. Rev. 1990, 3, 345–375. [Google Scholar] [CrossRef] [PubMed]
- Magdesian, K.G. Neonatal foal diarrhea. Vet. Clin. N. Am. Equine Pract. 2005, 21, 295–312. [Google Scholar] [CrossRef] [PubMed]
- Arana, A.; Montes, M.; Jere, K.C.; Alkorta, M.; Iturriza-Gomara, M.; Cilla, G. Emergence and spread of G3P(8) rotaviruses possessing an equine-like VP7 and a DS-1-like genetic backbone in the Basque Country (North of Spain), 2015. Infect. Genet. Evol. 2016, 44, 137–144. [Google Scholar] [CrossRef]
- Cowley, D.; Donato, C.M.; Roczo-Farkas, S.; Kirkwood, C.D. Emergence of a novel equine-like G3P(8) inter-genogroup reassortant rotavirus strain associated with gastroenteritis in Australian children. J. Gen. Virol. 2016, 97, 403–410. [Google Scholar] [CrossRef]
- Perkins, C.; Mijatovic-Rustempasic, S.; Ward, M.L.; Cortese, M.M.; Bowen, M.D. Genomic Characterization of the First Equine-Like G3P(8) Rotavirus Strain Detected in the United States. Genome Announc. 2017, 5, e01341-17. [Google Scholar] [CrossRef] [PubMed]
- Martella, V.; Colombrita, D.; Lorusso, E.; Draghin, E.; Fiorentini, S.; De Grazia, S.; Banyai, K.; Ciarlet, M.; Caruso, A.; Buonavoglia, C. Detection of a porcine-like rotavirus in a child with enteritis in Italy. J. Clin. Microbiol. 2008, 46, 3501–3507. [Google Scholar] [CrossRef] [PubMed]
- Jeong, S.; Than, V.T.; Lim, I.; Kim, W. Whole-genome analysis of a rare human Korean G3P rotavirus strain suggests a complex evolutionary origin potentially involving reassortment events between feline and bovine rotaviruses. PLoS ONE 2014, 9, e97127. [Google Scholar] [CrossRef] [PubMed]
- Dwyer, R.M. Diarrhea among young foals. Equine Dis. Q. 1995, 3, 5–6. Available online: https://gluck.ca.uky.edu/sites/gluck.ca.uky.edu/files/jul95.pdf (accessed on 15 June 2021).
- Vlasova, A.N.; Amimo, J.O.; Saif, L.J. Porcine Rotaviruses: Epidemiology, Immune Responses and Control Strategies. Viruses 2017, 9, 48. [Google Scholar] [CrossRef] [PubMed]
| Genome Segment ¦ Protein ¦ Length(nt) | Best BLASTn Hit Virus ¦ Accession Number ¦ Identity |
|---|---|
| 1 ¦ VP1 ¦ 3503 | RVB/Goat-wt/USA/Minnesota-1/2016 ¦ KY689687.1 ¦ 93.19% |
| 2 ¦ VP2 ¦ 2889 | RVB/Goat-wt/USA/Minnesota-1/2016 ¦ KY689688.1 ¦ 97.82% |
| 3 ¦ VP3 ¦ 2340 | RVB/Goat-wt/USA/Minnesota-1/2016 ¦ KY689689.1 ¦ 95.04% |
| 4 ¦ VP4 ¦ 2324 | RVB/Goat-wt/USA/CA22/2014 ¦ MG272136.1 ¦ 97.40% |
| 5 ¦ NSP1 ¦ 1252 | RVB/Goat-wt/USA/Minnesota-1/2016 ¦ KY689691.1 ¦ 95.75% |
| 6 ¦ VP6 ¦ 1221 | RVB/Goat-wt/USA/CA22/2014 ¦ MG272162.1 ¦ 96.34% |
| 7 ¦ NSP3 ¦ 1007 | RVB/Goat-wt/USA/Minnesota-1/2016 ¦ KY689693.1 ¦ 97.84% |
| 8 ¦ NSP2 ¦ 961 | RVB/Goat-wt/USA/CA22/2014 ¦ MG271985.1 ¦ 97.90% |
| 9 ¦ VP7 ¦ 773 | RVB/Cow-wt/USA/MN10-1/2010/G3P[x] ¦ JQ288103.1 ¦ 96.37% |
| 10 ¦ NSP4 ¦ 697 | RVB/Cow-wt/JPN/IS-1/1999/G3P[x] ¦ LC185678.1 ¦ 94.26% |
| 11 ¦ NSP5 ¦ 764 | RVB/Goat-wt/USA/CA22/2014 ¦ MG272051.1 ¦ 96.41% |
| Genome Segment ¦ Protein ¦ ORF (aa) | Best BLASTp Hit Virus ¦ Accession Number ¦ Identity | Protein Function |
|---|---|---|
| 1 ¦ VP1 ¦ 1158 | RVB/Goat-wt/USA/Minnesota-1/2016 ¦ ASV45167.1 ¦ 97.15% | RNA-dependent RNA polymerase (RdRp) |
| 2 ¦ VP2 ¦ 937 | RVB/Goat-wt/USA/CA22/2014 ¦ AUG44960.1 ¦ 99.15% | Outer core protein; essential for RdRp activity |
| 3 ¦ VP3 ¦ 763 | RVB/Goat-wt/USA/Minnesota-1/2016 ¦ ASV45169.1 ¦ 97.38% | Catalyzes the addition of 5′ cap on vRNA |
| 4 ¦ VP4 ¦ 759 | RVB/Goat-wt/USA/Minnesota-1/2016 ¦ ASV45170.1 ¦ 97.10% | Essential for attachment to host cell |
| 5 ¦ NSP1 ¦ 320 | RVB/Goat-wt/USA/CA22/2014 ¦ AUG44808.1 ¦ 96.25% | Interferon antagonist |
| 6 ¦ VP6 ¦ 391 | RVB/Goat-wt/USA/CA22/2014 ¦ AUG45028.1 ¦ 98.98% | Essential for transcription of double layered particles |
| 7 ¦ NSP3 ¦ 296 | RVB/Goat-wt/USA/Minnesota-1/2016 ¦ ASV45174.1 ¦ 99.66% | Inhibits host mRNA translation; promotes vRNA translation |
| 8 ¦ NSP2 ¦ 300 | RVB/Goat-wt/USA/CA22/2014 ¦ AUG44851.1 ¦ 99.33% | Packaging of vRNA; viroplasm formation |
| 9 ¦ VP7 ¦ 247 | RVB/Cow/Nemuro ¦ BAA78609.1 ¦ 99.60% | Interacts with host cell receptor |
| 10 ¦ NSP4 ¦ 208 | RVB/Cow-wt/JPN/IS-2/2002/G3P[x] ¦ BAW98439.1 ¦ 96.63% | Acts as viroporin; enterotoxin |
| 11 ¦ NSP5 ¦ 166 | RVB/Goat-wt/USA/CA22/2014 ¦ AUG44917.1 ¦ 97.59% | NSP5: interacts with VP2; viroplasm Formation NSP6: interacts with NSP5 |
| Farm | Foal | Ct (Assay I) | Ct (Assay II) | Age (Days) | Clinical Status |
|---|---|---|---|---|---|
| A | A1 | 20.9 | 21.0 | 3 d | Diarrhea |
| A2 | 13.1 | 13.9 | <5 d | Diarrhea | |
| B | B1 | 24.5 | 28.8 | 2 d | Diarrhea |
| B2 | 18 | 19.8 | 2 d | Bloody diarrhea | |
| B3 | 19.1 | 18.6 | 3 d | Diarrhea | |
| B4 | 11.6 | 15.1 | 2 d | Diarrhea | |
| B5 | 23.4 | 28.9 | 2 d | Diarrhea | |
| C | C1 | 15.2 | 13.7 | <7 d | Diarrhea |
| D | D1 | 25.6 | 24.0 | <7 d | Diarrhea |
| E | E1 | ND | ND | <7 d | Diarrhea |
| F | F1 | ND | ND | <7 d | Diarrhea |
| G | G1 | ND | ND | <7 d | Diarrhea |
| H | H1 | 15.3 | 16.5 | 3 d | Diarrhea |
| H2 | ND | ND | 12 d | Diarrhea | |
| I | I1 | 11.3 | 12.7 | <3 d | Diarrhea |
| J | J1 | 17.3 | 17.4 | 3 d | Diarrhea |
| J2 | 17.3 | 17.7 | <2 d | Diarrhea | |
| J3 | 10.6 | 9.6 | <2 d | Diarrhea | |
| J4 | 13 | 11.4 | <3 d | Diarrhea | |
| J5 | 21 | 22.3 | 2 d | Diarrhea | |
| K | K1 | 15 | 18.2 | 2 d | Diarrhea |
| L | L1 | ND | ND | 1 d | Normal |
| L2 | ND | ND | 3 d | Normal | |
| M | M1 | ND | ND | <5 d | Normal |
| M2 | ND | ND | <5 d | Normal | |
| M3 | ND | ND | <5 d | Normal | |
| M4 | ND | ND | <5 d | Normal | |
| M5 | ND | ND | <5 d | Normal | |
| N | N1 | 13 | 11.9 | 1 d | Diarrhea |
| N2 | 20.1 | 19.1 | 3 d | Diarrhea | |
| O | O1 | ND | ND | 12 d | Normal |
| P | P1 | ND | ND | <5 d | Normal |
| Q | Q1 | 15.5 | 15.3 | <7 d | Diarrhea |
| R | R1 | ND | ND | <19 d | Diarrhea |
| S | S1 | ND | ND | 4 d | Diarrhea |
| S2 | ND | ND | 4 d | Diarrhea | |
| S3 | ND | ND | 3 d | Diarrhea | |
| S4 | 30.3 | 28.1 | 4 d | Mucoid diarrhea | |
| T | T1 | 15.8 | 18.7 | 3 d | Bloody diarrhea |
| T2 | 14.2 | 15.2 | <2 d | Diarrhea | |
| U | U1 | ND | ND | <7 d | Diarrhea |
| V | V1 | ND | ND | <2 d | Diarrhea |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Uprety, T.; Sreenivasan, C.C.; Hause, B.M.; Li, G.; Odemuyiwa, S.O.; Locke, S.; Morgan, J.; Zeng, L.; Gilsenan, W.F.; Slovis, N.; et al. Identification of a Ruminant Origin Group B Rotavirus Associated with Diarrhea Outbreaks in Foals. Viruses 2021, 13, 1330. https://doi.org/10.3390/v13071330
Uprety T, Sreenivasan CC, Hause BM, Li G, Odemuyiwa SO, Locke S, Morgan J, Zeng L, Gilsenan WF, Slovis N, et al. Identification of a Ruminant Origin Group B Rotavirus Associated with Diarrhea Outbreaks in Foals. Viruses. 2021; 13(7):1330. https://doi.org/10.3390/v13071330
Chicago/Turabian StyleUprety, Tirth, Chithra C. Sreenivasan, Ben M. Hause, Ganwu Li, Solomon O. Odemuyiwa, Stephan Locke, Jocelynn Morgan, Li Zeng, William F. Gilsenan, Nathan Slovis, and et al. 2021. "Identification of a Ruminant Origin Group B Rotavirus Associated with Diarrhea Outbreaks in Foals" Viruses 13, no. 7: 1330. https://doi.org/10.3390/v13071330
APA StyleUprety, T., Sreenivasan, C. C., Hause, B. M., Li, G., Odemuyiwa, S. O., Locke, S., Morgan, J., Zeng, L., Gilsenan, W. F., Slovis, N., Metcalfe, L., Carter, C. N., Timoney, P., Horohov, D., Wang, D., Erol, E., Adam, E., & Li, F. (2021). Identification of a Ruminant Origin Group B Rotavirus Associated with Diarrhea Outbreaks in Foals. Viruses, 13(7), 1330. https://doi.org/10.3390/v13071330








