Transcriptome and Proteomic Analysis Reveals Up-Regulation of Innate Immunity-Related Genes Expression in Caprine Herpesvirus 1 Infected Madin Darby Bovine Kidney Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture, Virus Infection and Sample Preparation
2.2. RNA Extraction, Library Preparation and RNA-Seq
2.3. Read Quality Control and Mapping
2.4. Differential Expression Genes (DEGs) Analysis, Functional Enrichment, and Protein–Protein Interaction (PPI) Analysis of DEGs
2.5. Quantitative Real-Time RT-PCR (qRT-PCR)
2.6. Total Protein Extraction
2.7. Protein Quality Test
2.8. iTRAQ Labeling of Peptides
2.9. Separation of Fractions
2.10. LC-MS/MS Analysis
2.11. The Identification and Quantitation of Protein
2.12. The Functional Analysis of Protein
2.13. Western Blot
2.14. Statistical Analysis
3. Results
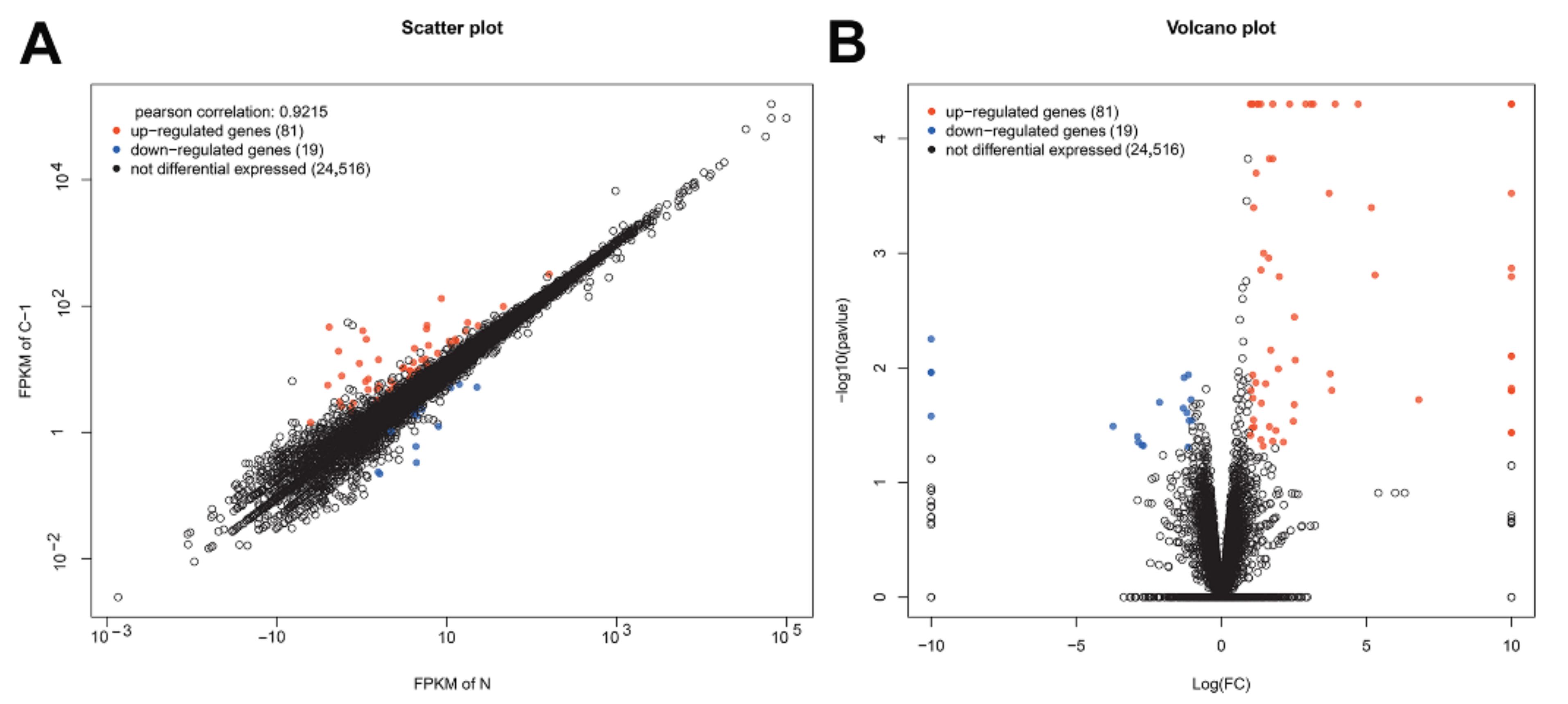
3.1. Transcriptome Quantification
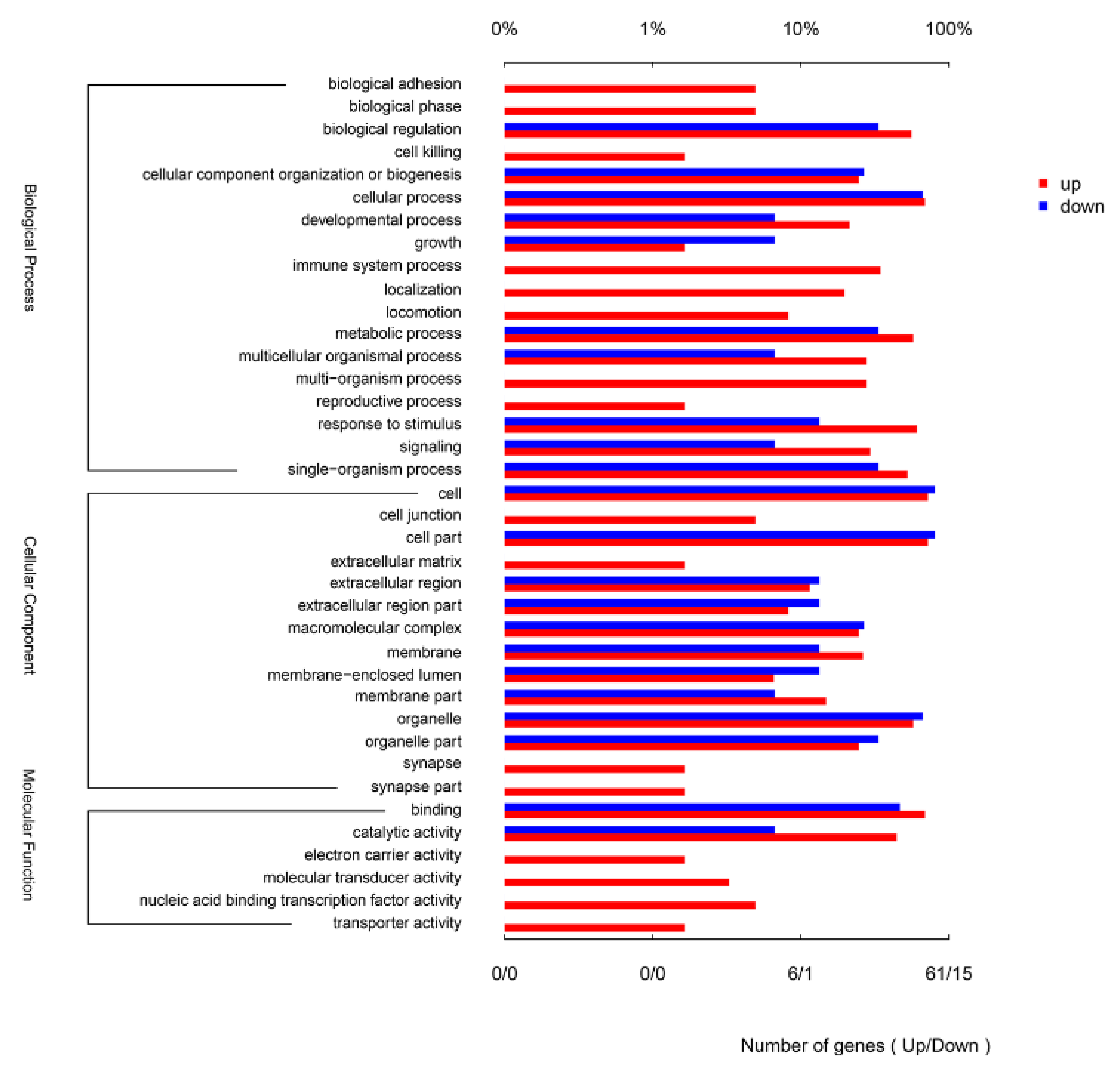
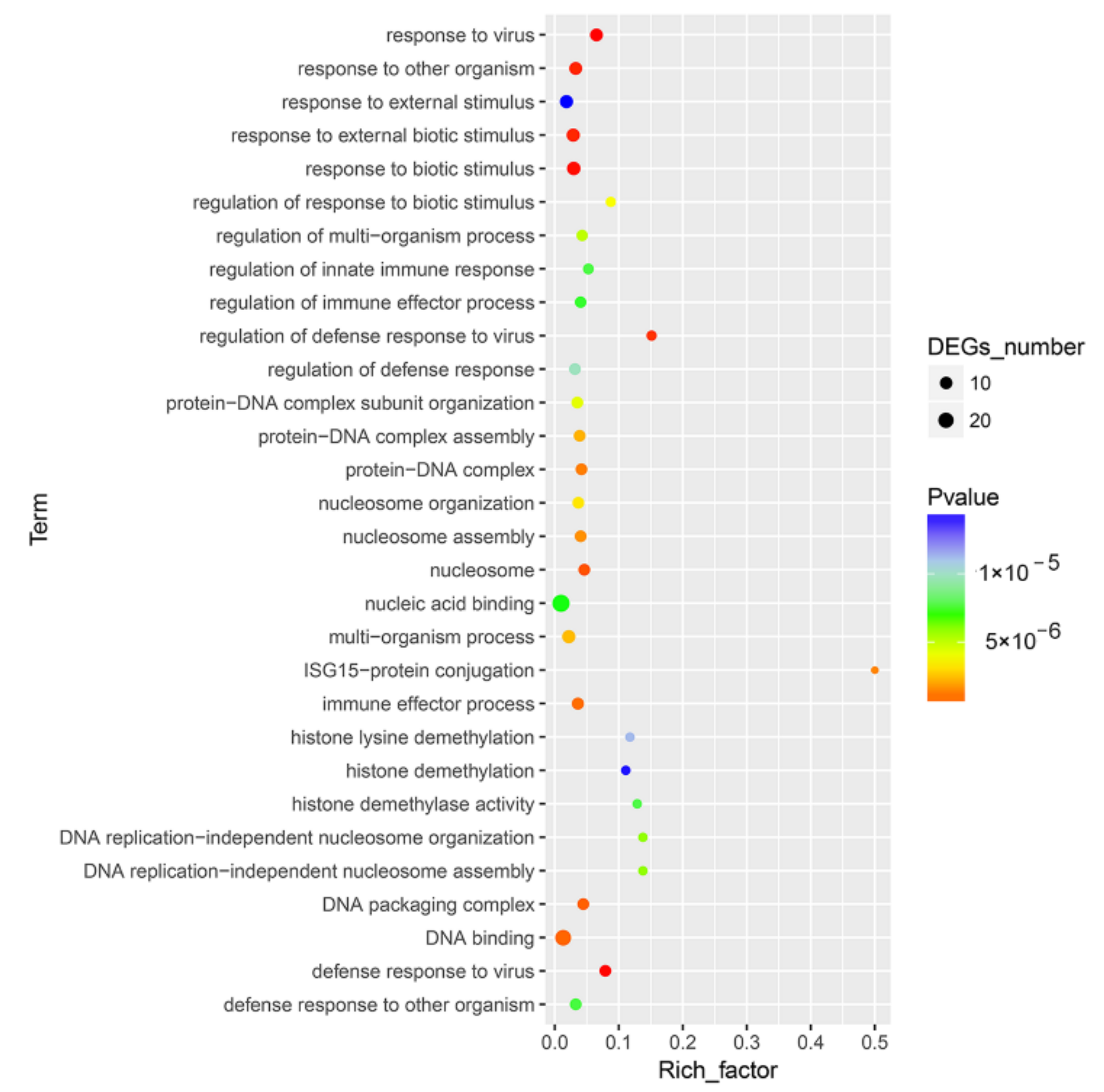
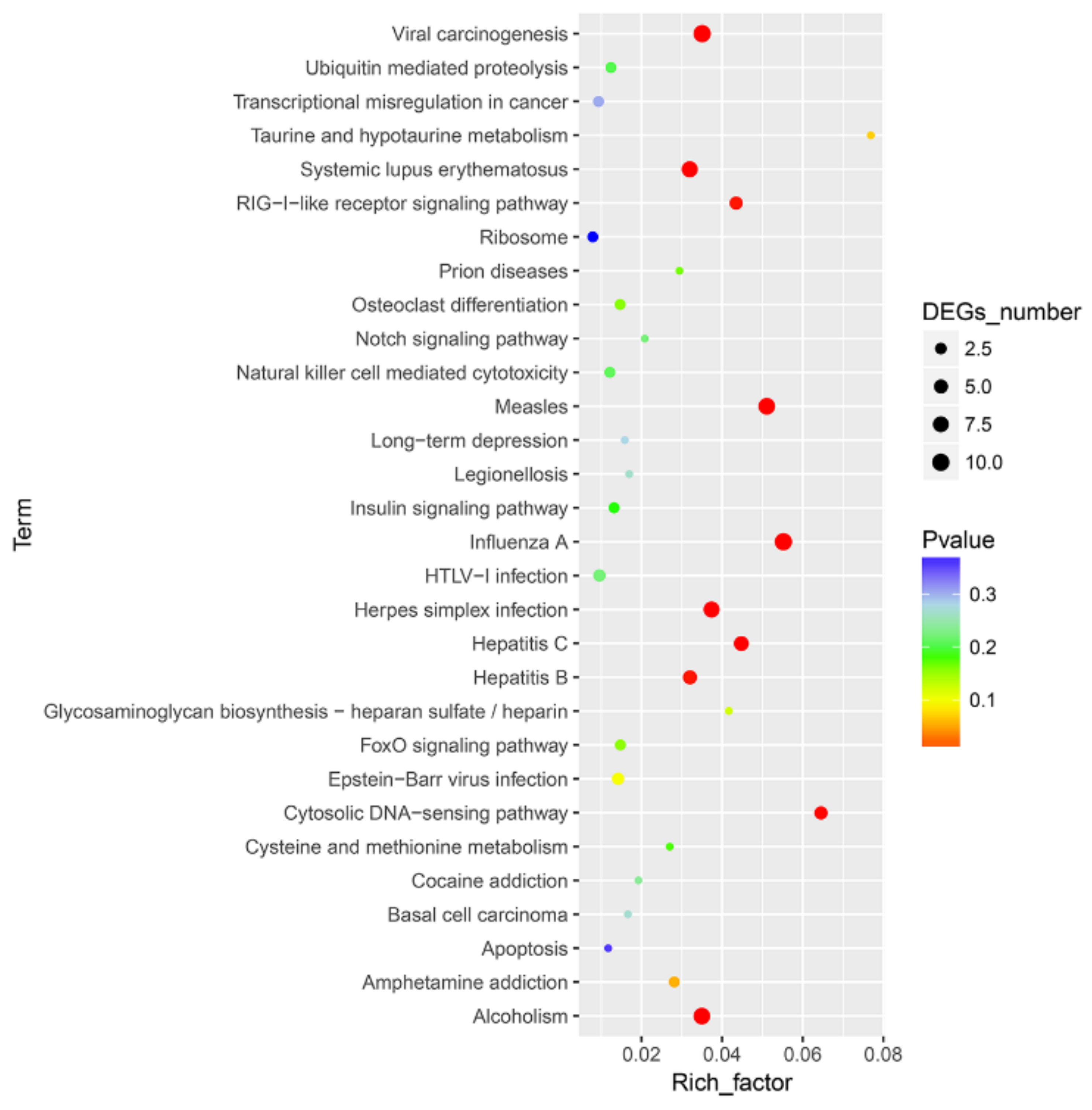
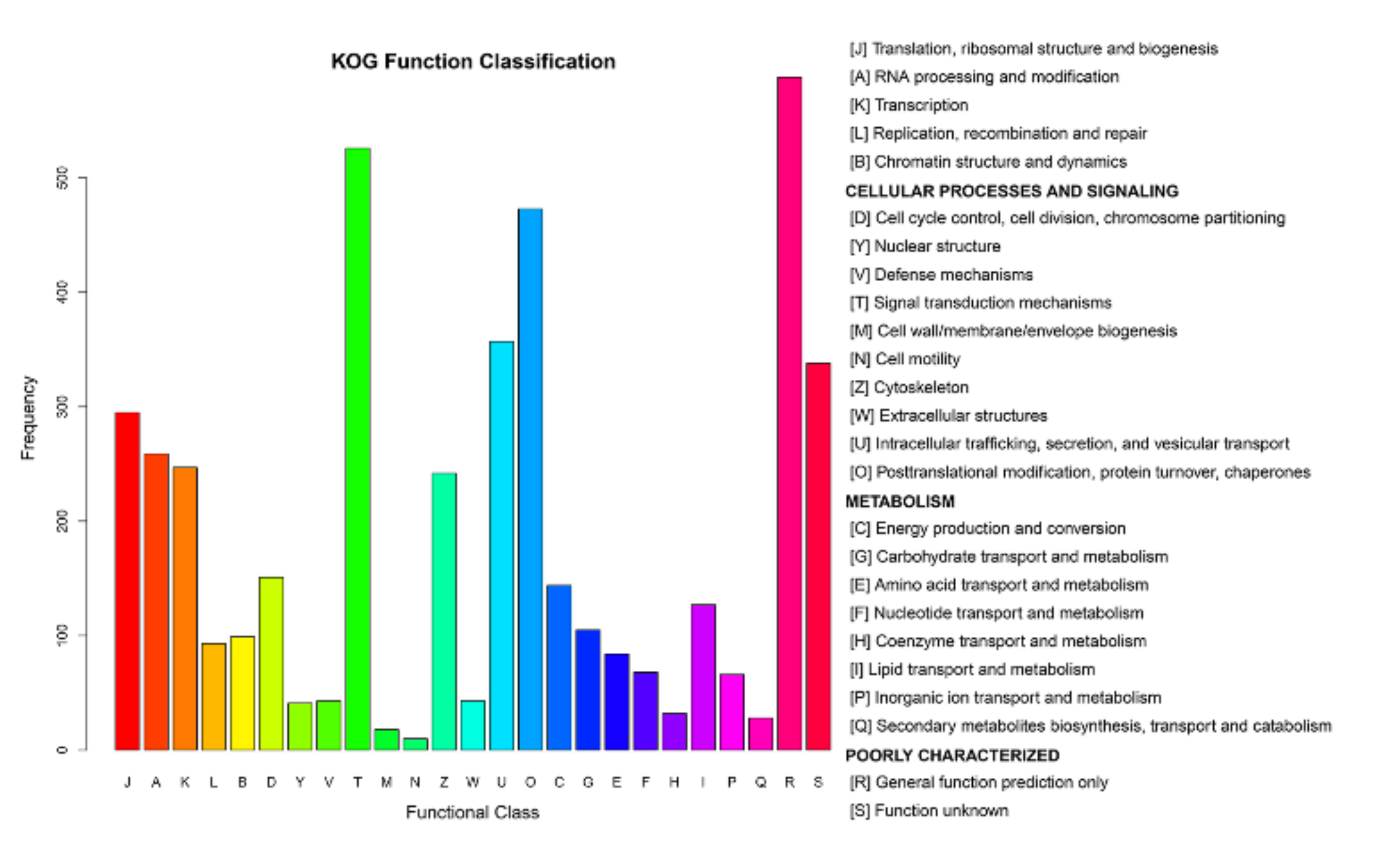
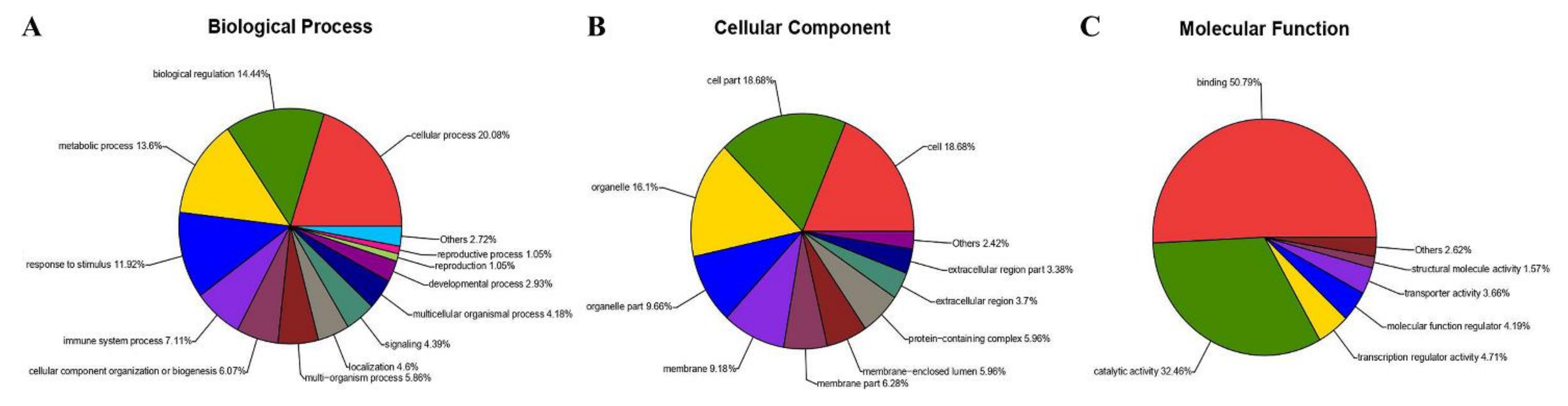
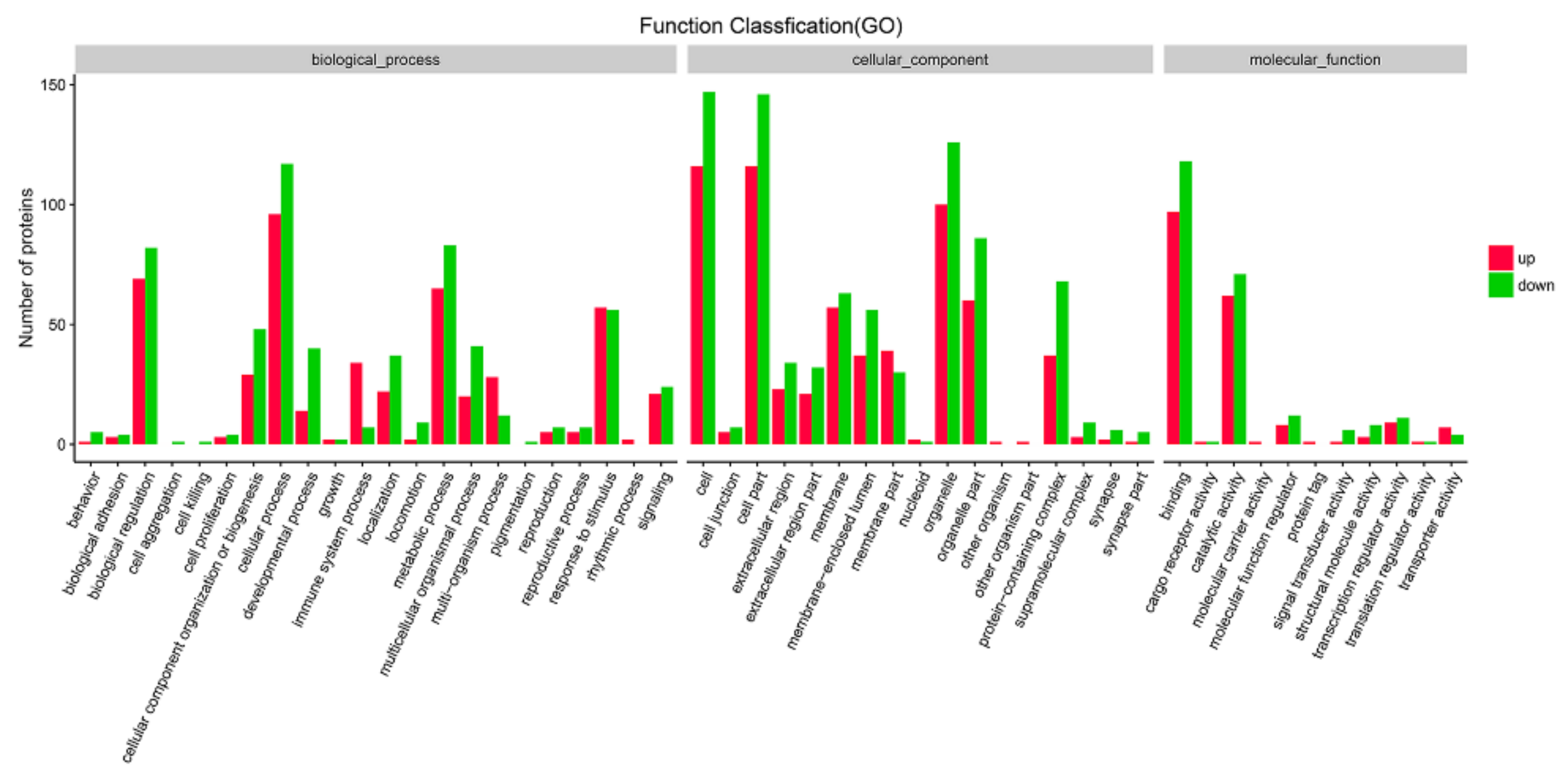
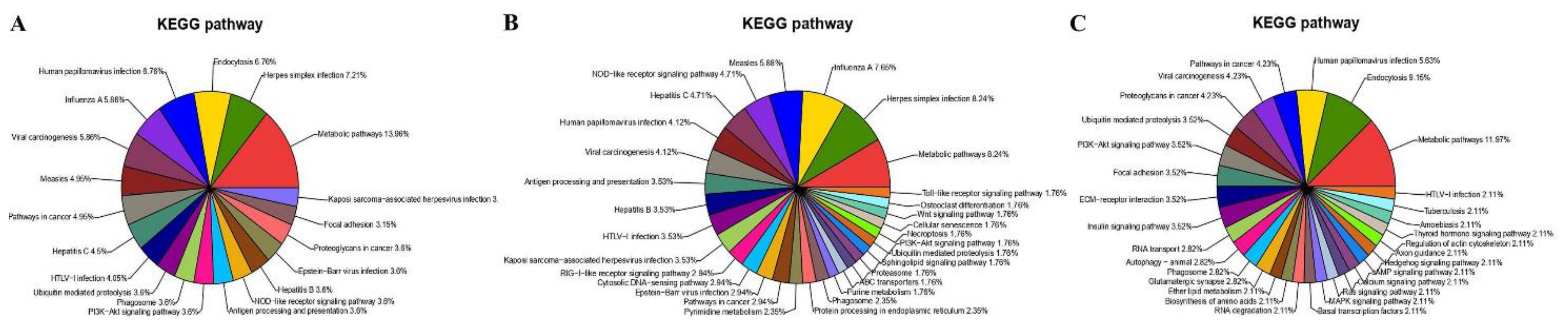
3.2. DEG Analysis and Functional Annotation
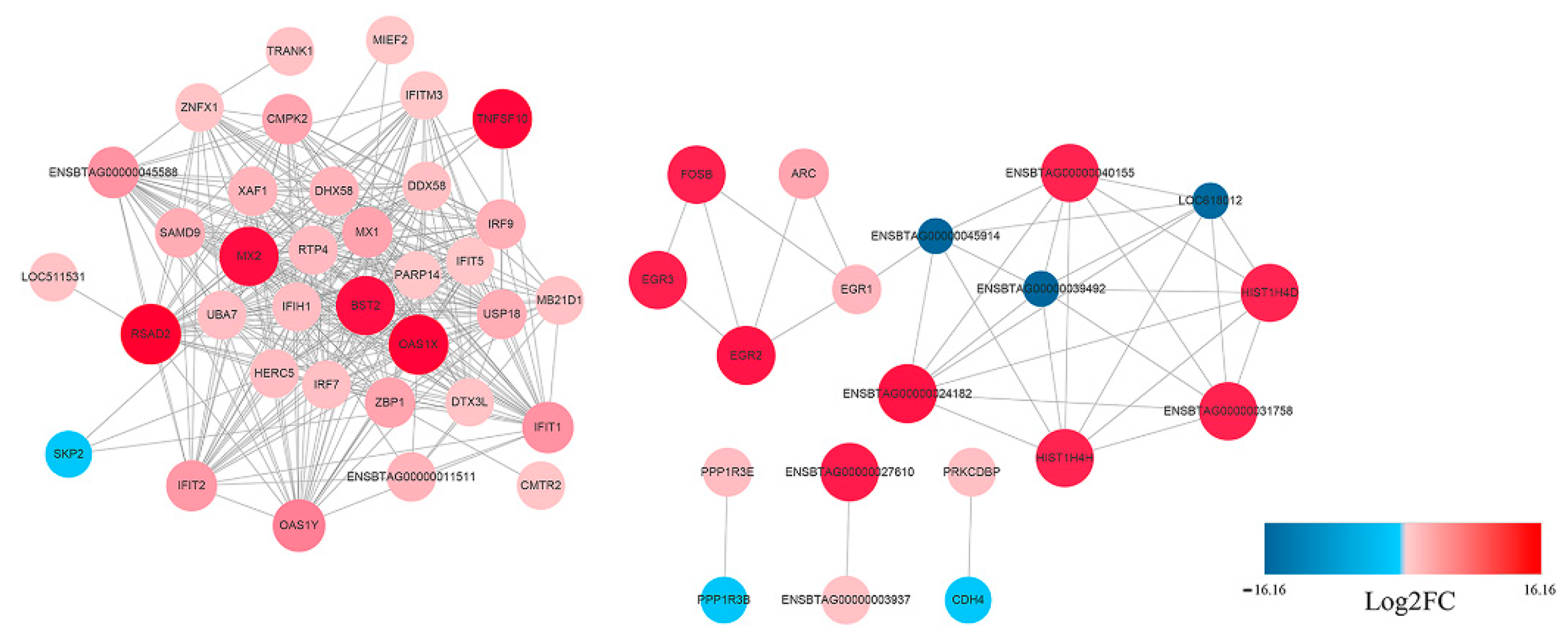
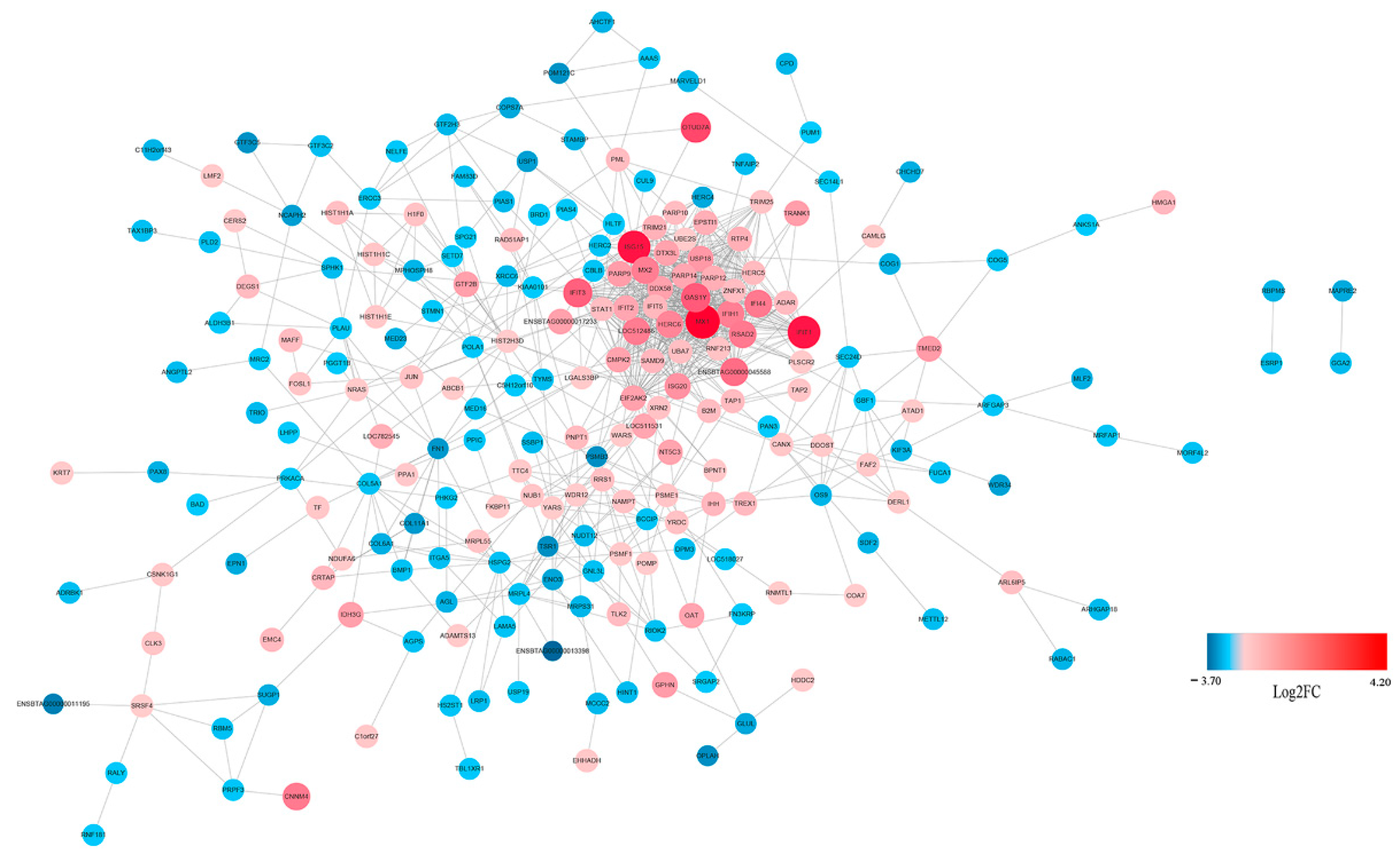
3.3. Functional and PPI Analysis of DEGs
3.4. Protein Profiling of MDBK Cells
3.5. Functional Analysis of Protein Responses to CpHV-1 Infection in MDBK Cells
3.6. Partial Validation of RNA-Seq and iTRAQ Data
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chayavichitsilp, P.; Buckwalter, J.V.; Krakowski, A.C.; Friedlander, S.F. Herpes simplex. Pediatrics Rev. 2009, 30, 119–129. [Google Scholar] [CrossRef]
- Patel, J.R.; Didlick, S. Epidemiology, disease and control of infections in ruminants by herpesviruses––An overview. J. S. Afr. Vet. Assoc. 2008, 79, 8–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berrios, P.E.; McKercher, D.G.; Knight, H.D. Pathogenicity of a caprine herpesvirus. Am. J. Vet. Res. 1975, 36, 1763–1769. [Google Scholar]
- Camero, M.; Larocca, V.; Losurdo, M.; Lorusso, E.; Patruno, G.; Staffa, V.N.; Martella, V.; Buonavoglia, C.; Tempesta, M. Goats are susceptible to Bubaline alphaherpesvirus 1 infection: Results of an experimental study. Comp. Immunol. Microbiol. Infect. Dis. 2017, 50, 97–100. [Google Scholar] [CrossRef]
- Chenier, S.; Montpetit, C.; Helie, P. Caprine herpesvirus-1 abortion storm in a goat herd in Quebec. Can. Vet. J. 2004, 45, 241–243. [Google Scholar] [PubMed]
- Gonzalez, J.; Passantino, G.; Esnal, A.; Cuesta, N.; Garcia Vera, J.A.; Mechelli, L.; Saez, A.; Garcia Marin, J.F.; Tempesta, M. Abortion in goats by Caprine alphaherpesvirus 1 in Spain. Reprod. Domest. Anim. 2017, 52, 1093–1096. [Google Scholar] [CrossRef]
- Suavet, F.; Champion, J.L.; Bartolini, L.; Bernou, M.; Alzieu, J.P.; Brugidou, R.; Darnatigues, S.; Reynaud, G.; Perrin, C.; Adam, G.; et al. First Description of Infection of Caprine Herpesvirus 1 (CpHV-1) in Goats in Mainland France. Pathogens 2016, 5, 17. [Google Scholar] [CrossRef] [Green Version]
- Hao, F.; Mao, L.; Li, W.; Li, J.; Yang, L.; Zhang, W.; Jiang, J.; Sun, M.; Xie, X.; Liu, M. Epidemiological investigation and genomic characterization of Caprine herpesvirus 1 from goats in China. Infect. Genet. Evol. 2020, 79, 104168. [Google Scholar] [CrossRef] [PubMed]
- Tempesta, M.; Camero, M.; Greco, G.; Pratelli, A.; Martella, V.; Buonavoglia, C. A classical inactivated vaccine induces protection against caprine herpesvirus 1 infection in goats. Vaccine 2001, 19, 3860–3864. [Google Scholar] [CrossRef]
- Tempesta, M.; Camero, M.; Sciorsci, R.L.; Greco, G.; Minoia, R.; Martella, V.; Pratelli, A.; Buonavoglia, C. Experimental infection of goats at different stages of pregnancy with caprine herpesvirus 1. Comp. Immunol. Microbiol. Infect. Dis. 2004, 27, 25–32. [Google Scholar] [CrossRef]
- Grewal, A.S.; Wells, R. Vulvovaginitis of goats due to a herpesvirus. Aust. Vet. J. 1986, 63, 79–82. [Google Scholar] [CrossRef]
- Horner, G.W.; Hunter, R.; Day, A.M. An outbreak of vulvovaginitis in goats caused by a caprine herpesvirus. N. Zeal. Vet. J. 1982, 30, 150–152. [Google Scholar]
- Tarigan, S.; Webb, R.F.; Kirkland, D. Caprine herpesvirus from balanoposthitis. Aust. Vet. J. 1987, 64, 321. [Google Scholar] [CrossRef]
- Pagnini, U.; Montagnaro, S.; Sanfelice di Monteforte, E.; Pacelli, F.; De Martino, L.; Roperto, S.; Florio, S.; Iovane, G. Caprine herpesvirus-1 (CapHV-1) induces apoptosis in goat peripheral blood mononuclear cells. Vet. Immunol. Immunopathol. 2005, 103, 283–293. [Google Scholar] [CrossRef]
- Longo, M.; Fiorito, F.; Marfe, G.; Montagnaro, S.; Pisanelli, G.; De Martino, L.; Iovane, G.; Pagnini, U. Analysis of apoptosis induced by Caprine Herpesvirus 1 in vitro. Virus Res. 2009, 145, 227–235. [Google Scholar] [CrossRef]
- Montagnaro, S.; Damiano, S.; Ciarcia, R.; Puzio, M.V.; Ferrara, G.; Iovane, V.; Forte, I.M.; Giordano, A.; Pagnini, U. Caprine herpesvirus 1 (CpHV-1) as a potential candidate for oncolytic virotherapy. Cancer Biol. Ther. 2019, 20, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Camero, M.; Lanave, G.; Lucente, M.S.; Losurdo, M.; Di Paola, G.; Lorusso, E.; Martella, V.; Buonavoglia, C.; Tempesta, M. Bubaline alphaherpesvirus 1 induces a latent/reactivable infection in goats. Comp. Immunol. Microbiol. Infect. Dis. 2019, 62, 54–57. [Google Scholar] [CrossRef]
- Martins, B.; Ebling, R.C.; Martins, M.; Diel, D.G.; Weiblen, R.; Flores, E.F. Antigenic relationships between Caprine alphaherpesvirus 1 (CpHV-1) and Bovine alphaherpesvirus 1 (BoHV-1) and experimental CpHV-1 infection of kids and calves. Microb. Pathog. 2019, 136, 103663. [Google Scholar] [CrossRef]
- Li, W.; Mao, L.; Shu, X.; Liu, R.; Hao, F.; Li, J.; Liu, M.; Yang, L.; Zhang, W.; Sun, M.; et al. Transcriptome analysis reveals differential immune related genes expression in bovine viral diarrhea virus-2 infected goat peripheral blood mononuclear cells (PBMCs). BMC Genom. 2019, 20, 516. [Google Scholar] [CrossRef]
- Unwin, R.D.; Griffiths, J.R.; Whetton, A.D. Simultaneous analysis of relative protein expression levels across multiple samples using iTRAQ isobaric tags with 2D nano LC–MS/MS. Nat. Protoc. 2010, 5, 1574–1582. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Roizman, B.; Whitley, R.J. The nine ages of herpes simplex virus. Herpes J. IHMF 2001, 8, 23–27. [Google Scholar]
- Whitley, R.J.; Roizman, B. Herpes simplex virus infections. Lancet 2001, 357, 1513–1518. [Google Scholar] [CrossRef]
- Zerboni, L.; Sen, N.; Oliver, S.L.; Arvin, A.M. Molecular mechanisms of varicella zoster virus pathogenesis. Nat. Rev. Microbiol. 2014, 12, 197–210. [Google Scholar] [CrossRef] [Green Version]
- Engel, E.A.; Song, R.; Koyuncu, O.O.; Enquist, L.W. Investigating the biology of alpha herpesviruses with MS-based proteomics. Proteomics 2015, 15, 1943–1956. [Google Scholar] [CrossRef] [Green Version]
- Miettinen, J.J.; Matikainen, S.; Nyman, T.A. Global secretome characterization of herpes simplex virus 1-infected human primary macrophages. J. Virol. 2012, 86, 12770–12778. [Google Scholar] [CrossRef] [Green Version]
- Hogue, I.B.; Bosse, J.B.; Hu, J.R.; Thiberge, S.Y.; Enquist, L.W. Cellular mechanisms of alpha herpesvirus egress: Live cell fluorescence microscopy of pseudorabies virus exocytosis. PLoS Pathog. 2014, 10, e1004535. [Google Scholar] [CrossRef] [PubMed]
- Lam, N.; Letchworth, G.J. Bovine herpesvirus 1 U(L)3.5 interacts with bovine herpesvirus 1 alpha-transinducing factor. J. Virol. 2000, 74, 2876–2884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, M.; Anderson, B.D.; Wang, T.; Chen, Y.; Zhang, D.; Gray, G.C.; Lu, J. Serological Evidence and Risk Factors for Swine Influenza Infections among Chinese Swine Workers in Guangdong Province. PLoS ONE 2015, 10, e0128479. [Google Scholar] [CrossRef] [Green Version]
- Camero, M.; Buonavoglia, D.; Lucente, M.S.; Losurdo, M.; Crescenzo, G.; Trerotoli, P.; Casalino, E.; Martella, V.; Elia, G.; Tempesta, M. Enhancement of the antiviral activity against caprine herpesvirus type 1 of Acyclovir in association with Mizoribine. Res. Vet. Sci. 2017, 111, 120–123. [Google Scholar] [CrossRef] [PubMed]
- Camero, M.; Lanave, G.; Catella, C.; Capozza, P.; Gentile, A.; Fracchiolla, G.; Britti, D.; Martella, V.; Buonavoglia, C.; Tempesta, M. Virucidal activity of ginger essential oil against caprine alphaherpesvirus-1. Vet. Microbiol. 2019, 230, 150–155. [Google Scholar] [CrossRef] [PubMed]
- Lanave, G.; Lucente, M.S.; Siciliano, P.; Zizzadoro, C.; Trerotoli, P.; Martella, V.; Buonavoglia, C.; Tempesta, M.; Camero, M. Antiviral activity of PHA767491 on Caprine alphaherpesvirus-1 in vitro. Res. Vet. Sci. 2019, 126, 113–117. [Google Scholar] [CrossRef] [PubMed]
- Bivens, N.J.; Zhou, M. RNA-Seq Library Construction Methods for Transcriptome Analysis. Curr. Protoc. Plant Biol. 2016, 1, 197–215. [Google Scholar] [CrossRef]
- Hrdlickova, R.; Toloue, M.; Tian, B. RNA-Seq methods for transcriptome analysis. Wiley Interdiscip. Rev. RNA 2017, 8, e1364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crosse, K.M.; Monson, E.A.; Beard, M.R.; Helbig, K.J. Interferon-Stimulated Genes as Enhancers of Antiviral Innate Immune Signaling. J. Innate Immun. 2018, 10, 85–93. [Google Scholar] [CrossRef]
- Gurtler, C.; Bowie, A.G. Innate immune detection of microbial nucleic acids. Trends Microbiol. 2013, 21, 413–420. [Google Scholar] [CrossRef] [Green Version]
- Iwasaki, A. A virological view of innate immune recognition. Annu. Rev. Microbiol. 2012, 66, 177–196. [Google Scholar] [CrossRef] [Green Version]
- Severa, M.; Coccia, E.M.; Fitzgerald, K.A. Toll-like receptor-dependent and -independent viperin gene expression and counter-regulation by PRDI-binding factor-1/BLIMP1. J. Biol. Chem. 2006, 281, 26188–26195. [Google Scholar] [CrossRef] [Green Version]
- Saitoh, T.; Satoh, T.; Yamamoto, N.; Uematsu, S.; Takeuchi, O.; Kawai, T.; Akira, S. Antiviral protein Viperin promotes Toll-like receptor 7- and Toll-like receptor 9-mediated type I interferon production in plasmacytoid dendritic cells. Immunity 2011, 34, 352–363. [Google Scholar] [CrossRef] [Green Version]
- Haller, O.; Staeheli, P.; Schwemmle, M.; Kochs, G. Mx GTPases: Dynamin-like antiviral machines of innate immunity. Trends Microbiol. 2015, 23, 154–163. [Google Scholar] [CrossRef]
- Ghosh, S.; Marsh, E.N.G. Viperin: An ancient radical SAM enzyme finds its place in modern cellular metabolism and innate immunity. J. Biol. Chem. 2020, 295, 11513–11528. [Google Scholar] [CrossRef]
- Li, M.; Liao, Z.; Xu, Z.; Zou, X.; Wang, Y.; Peng, H.; Li, Y.; Ou, X.; Deng, Y.; Guo, Y.; et al. The Interaction Mechanism Between Herpes Simplex Virus 1 Glycoprotein D and Host Antiviral Protein Viperin. Front. Immunol. 2019, 10, 2810. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nasr, N.; Maddocks, S.; Turville, S.G.; Harman, A.N.; Woolger, N.; Helbig, K.J.; Wilkinson, J.; Bye, C.R.; Wright, T.K.; Rambukwelle, D.; et al. HIV-1 infection of human macrophages directly induces viperin which inhibits viral production. Blood 2012, 120, 778–788. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wu, X.; Pan, T.; Song, W.; Wang, Y.; Zhang, F.; Yuan, Z. Viperin inhibits hepatitis C virus replication by interfering with binding of NS5A to host protein hVAP-33. J. Gen. Virol. 2012, 93 Pt 1, 83–92. [Google Scholar] [CrossRef]
- Wang, X.; Hinson, E.R.; Cresswell, P. The interferon-inducible protein viperin inhibits influenza virus release by perturbing lipid rafts. Cell Host Microbe 2007, 2, 96–105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fitzgerald, K.A. The interferon inducible gene: Viperin. J. Interferon Cytokine Res. 2011, 31, 131–135. [Google Scholar] [CrossRef]
- Sun, Y.; Hu, B.; Fan, C.; Jia, L.; Zhang, Y.; Du, A.; Zheng, X.; Zhou, J. iTRAQ-based quantitative subcellular proteomic analysis of Avibirnavirus-infected cells. Electrophoresis 2015, 36, 1596–1611. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.K.; Chai, F.; Li, H.Y.; Xiao, G.; Guo, L. Identification of host proteins involved in Japanese encephalitis virus infection by quantitative proteomics analysis. J. Proteome Res. 2013, 12, 2666–2678. [Google Scholar] [CrossRef]
- Lu, Q.; Bai, J.; Zhang, L.; Liu, J.; Jiang, Z.; Michal, J.J.; He, Q.; Jiang, P. Two-dimensional liquid chromatography-tandem mass spectrometry coupled with isobaric tags for relative and absolute quantification (iTRAQ) labeling approach revealed first proteome profiles of pulmonary alveolar macrophages infected with porcine reproductive and respiratory syndrome virus. J. Proteome Res. 2012, 11, 2890–2903. [Google Scholar]
- Diamond, M.S.; Farzan, M. The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nat. Rev. Immunol. 2013, 13, 46–57. [Google Scholar] [CrossRef] [PubMed]
- Pichlmair, A.; Lassnig, C.; Eberle, C.A.; Gorna, M.W.; Baumann, C.L.; Burkard, T.R.; Burckstummer, T.; Stefanovic, A.; Krieger, S.; Bennett, K.L.; et al. IFIT1 is an antiviral protein that recognizes 5′–triphosphate RNA. Nat. Immunol. 2011, 12, 624–630. [Google Scholar] [CrossRef] [PubMed]
- Raychoudhuri, A.; Shrivastava, S.; Steele, R.; Kim, H.; Ray, R.; Ray, R.B. ISG56 and IFITM1 proteins inhibit hepatitis C virus replication. J. Virol. 2011, 85, 12881–12889. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meroni, G.; Diez–Roux, G. TRIM/RBCC, a novel class of “single protein RING finger” E3 ubiquitin ligases. BioEssays 2005, 27, 1147–1157. [Google Scholar] [CrossRef] [PubMed]
- Carthagena, L.; Bergamaschi, A.; Luna, J.M.; David, A.; Uchil, P.D.; Margottin–Goguet, F.; Mothes, W.; Hazan, U.; Transy, C.; Pancino, G.; et al. Human TRIM gene expression in response to interferons. PLoS ONE 2009, 4, e4894. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rajsbaum, R.; Garcia-Sastre, A.; Versteeg, G.A. TRIMmunity: The roles of the TRIM E3-ubiquitin ligase family in innate antiviral immunity. J. Mol. Biol. 2014, 426, 1265–1284. [Google Scholar] [CrossRef] [Green Version]
- Liu, B.; Li, N.L.; Shen, Y.; Bao, X.; Fabrizio, T.; Elbahesh, H.; Wehbby, R.J.; Li, K. The C-Terminal Tail of TRIM56 Dictates Antiviral Restriction of Influenza A and B Viruses by Impeding Viral RNA Synthesis. J. Virol. 2016, 90, 4369–4382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Merkulova, T.; Dehaupas, M.; Nevers, M.C.; Creminon, C.; Alameddine, H.; Keller, A. Differential modulation of alpha, beta and gamma enolase isoforms in regenerating mouse skeletal muscle. Eur. J. Biochem. 2000, 267, 3735–3743. [Google Scholar] [CrossRef]
- Ho, J.A.; Chang, H.C.; Shih, N.Y.; Wu, L.C.; Chang, Y.F.; Chen, C.C.; Chou, C. Diagnostic detection of human lung cancer–associated antigen using a gold nanoparticle-based electrochemical immunosensor. Anal. Chem. 2010, 82, 5944–5950. [Google Scholar] [CrossRef]
- Keller, A.; Peltzer, J.; Carpentier, G.; Horvath, I.; Olah, J.; Duchesnay, A.; Orosz, F.; Ovadi, J. Interactions of enolase isoforms with tubulin and microtubules during myogenesis. Biochim. Biophys. Acta 2007, 1770, 919–926. [Google Scholar] [CrossRef]
- Avilán, L.; Gualdrón–López, M.; Quiñones, W.; González-González, L.; Hannaert, V.; Michels, P.A.; Concepción, J.L. Enolase: A key player in the metabolism and a probable virulence factor of trypanosomatid parasites–perspectives for its use as a therapeutic target. Enzym. Res. 2011, 2011, 932549. [Google Scholar] [CrossRef] [Green Version]
- Shanda, S.K.; Wilson, D.W. UL36p is required for efficient transport of membrane-associated herpes simplex virus type 1 along microtubules. J. Virol. 2008, 82, 7388–7394. [Google Scholar] [CrossRef] [Green Version]
- Pasdeloup, D.; McElwee, M.; Beilstein, F.; Labetoulle, M.; Rixon, F.J. Herpesvirus tegument protein pUL37 interacts with dystonin/BPAG1 to promote capsid transport on microtubules during egress. J. Virol. 2013, 87, 2857–2867. [Google Scholar] [CrossRef] [Green Version]
- Ding, X.-C.; Yang, X.-Q.; Yan, T.-T.; Lu, Z.-H.; Liu, X.-Y.; Luo, X.; Hu, Y.-C.; Yang, Y.-X.; Ma, L.-N. Alpha–enolase regulates hepatitis B virus replication through suppression of the interferon signalling pathway. J. Viral Hepat. 2018, 25, 289–295. [Google Scholar]
- Perez–Riverol, Y.; Csordas, A.; Bai, J.; Bernal–Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef]

| Gene | Accession No. | Primer | Target Position | Product Length (bp) |
|---|---|---|---|---|
| IRF7 | NM_001105040.1 | F: 5′-GCTCCACTACACCGAGAAGC-3′ | 1367-1386 | 196 |
| R: 5′-GAAGTCAAAGATGGGCGTGT-3′ | 1562-1543 | |||
| IRF9 | NM_001024506.1 | F: 5′-AAGGCCTGGGCGATATACAA-3′ | 694-713 | 125 |
| R: 5′-CCGATCTCAGGAACCTCCTC-3′ | 818-799 | |||
| IFIT1 | XM_015469501.1 | F: 5′-GGAACGTGCTGTGCAACTAA-3′ | 1305-1324 | 134 |
| R: 5′-TGTCGAGTGCTTTCATGCAG-3′ | 1438-1419 | |||
| IFIT2 | XM_001787823.5 | F: 5′-GGCAGCAAAGCTGTATCGAA-3′ | 975-994 | 137 |
| R: 5′-CTTCCAGGACTTTGGCCCTA-3′ | 1111-1092 | |||
| IFIT5 | NM_001075698.1 | F: 5′-CGTGGAGCGAGACTCTATGT-3′ | 1160-1179 | 162 |
| R: 5′-CGGCCATAGTGGTAGTGGAT-3′ | 1321-1302 | |||
| IFIH1 | XM_010802053.1 | F: 5′-ATTCTGAGGCAGACGGGAAA-3′ | 896-915 | 74 |
| R: 5′-TCTGTACTGCCTTCACAGCA-3′ | 969-950 | |||
| IFITM3 | NM_001078141.2 | F: 5′-TGGTCCCTGTTCAACACCAT-3′ | 231-250 | 91 |
| R: 5′-CCATCTTCCGGTCCCTAGAC-3′ | 321-302 | |||
| OAS1X | NM_178108.2 | F: 5′-AGCACTGGTACCAACTGTGT-3′ | 671-690 | 132 |
| R: 5′-GAAATCCCTGAGCTGTGCTG-3′ | 802-783 | |||
| OASIY | NM_001040606.1 | F: 5′-CTCAGCTTTGTGCTGAGGTC-3′ | 453-472 | 131 |
| R: 5′-TGGATGAGCCGGACATAGAC-3′ | 583-564 | |||
| MX1 | JQ766265.1 | F: 5′-TTTTTCAACCTCCACCGAAC-3′ | 1450-1469 | 132 |
| R: 5′-GTACACCTGGTCCTGGCAGT-3′ | 1581-1562 | |||
| RSAD2 | NM_001045941.1 | F: 5′-TTCAACGTGGACGAGGATATG-3′ | 734-754 | 98 |
| R: 5′-CCAGAGTTCTCACCCTCAATTAT-3′ | 831-809 | |||
| β-actin | AY141970.1 | F: 5′-ACATCCGCAAGGACCTCTA-3′ | 902-920 | 89 |
| R: 5′-GATCTCTTTCTGCATCCTGTCC-3′ | 990-969 |
| Sample | Total_Raw Reads | Total_Clean Reads | Mapped_Reads | Mapped_Rate | FPKM a > 0 | FPKM > 1 | FPKM > 5 | FPKM > 10 |
|---|---|---|---|---|---|---|---|---|
| C-1 | 30,065,892 | 25,480,936 | 19,704,426 | 77.33% | 13,345 | 10,888 | 8437 | 6426 |
| N | 26,223,456 | 22,774,082 | 17,675,904 | 77.61% | 13,153 | 10,864 | 8426 | 6431 |
| Gene/Protein Name | Fold Change (FC) (CpHV-1-Infected/Mock-Infected) | |||
|---|---|---|---|---|
| RNA-Seq (log2) | qRT-PCR (12 hpi, log2) | qRT-PCR (24 hpi, log2) | iTRAQ | |
| MX1 | 3.93 | 6.86 | 6.6 | 4.20 |
| MX2 | / a | / | / | 2.35 |
| RSAD2 (Viperin) | inf b | 10.59 | 13.06 | 2.42 |
| IFIT1 | 5.3 | 4.85 | 7.67 | 3.80 |
| IFIT2 | 4.71 | 5.34 | 5.16 | 1.76 |
| IFIT3 | / | / | / | 2.78 |
| IFIT5 | 1.09 | 1.35 | 2.42 | 1.5 |
| IFITM3 | 1.008 | 0.61 | 1.6 | / |
| IFI44 | / | / | / | 2.43 |
| OAS1X | inf | 8.87 | 8.93 | 1.76 |
| OAS1Y | 6.81 | 3.21 | 4.39 | 2.86 |
| IRF7 | 1.63 | 0.48 | 0.36 | 1.41 |
| IRF9 | 2.91 | 3 | 2.76 | / |
| IFIH1 (MDA5) | 1.77 | 4.1 | 3.11 | 2.34 |
| DDX58 (RIG-I) | / | / | / | 1.83 |
| TRIM5 | / | / | / | 1.31 |
| TRIM21 | / | / | / | 1.60 |
| TRIM25 | / | / | / | 1.32 |
| ISG15 | / | / | / | 3.81 |
| ISG20 | / | / | / | 2.05 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hao, F.; Xie, X.; Liu, M.; Mao, L.; Li, W.; Na, W. Transcriptome and Proteomic Analysis Reveals Up-Regulation of Innate Immunity-Related Genes Expression in Caprine Herpesvirus 1 Infected Madin Darby Bovine Kidney Cells. Viruses 2021, 13, 1293. https://doi.org/10.3390/v13071293
Hao F, Xie X, Liu M, Mao L, Li W, Na W. Transcriptome and Proteomic Analysis Reveals Up-Regulation of Innate Immunity-Related Genes Expression in Caprine Herpesvirus 1 Infected Madin Darby Bovine Kidney Cells. Viruses. 2021; 13(7):1293. https://doi.org/10.3390/v13071293
Chicago/Turabian StyleHao, Fei, Xing Xie, Maojun Liu, Li Mao, Wenliang Li, and Woonsung Na. 2021. "Transcriptome and Proteomic Analysis Reveals Up-Regulation of Innate Immunity-Related Genes Expression in Caprine Herpesvirus 1 Infected Madin Darby Bovine Kidney Cells" Viruses 13, no. 7: 1293. https://doi.org/10.3390/v13071293
APA StyleHao, F., Xie, X., Liu, M., Mao, L., Li, W., & Na, W. (2021). Transcriptome and Proteomic Analysis Reveals Up-Regulation of Innate Immunity-Related Genes Expression in Caprine Herpesvirus 1 Infected Madin Darby Bovine Kidney Cells. Viruses, 13(7), 1293. https://doi.org/10.3390/v13071293








