Human Defensins Inhibit SARS-CoV-2 Infection by Blocking Viral Entry
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents, Plasmids, and Cell Lines
2.2. Cell Culture
2.3. Production of Pseudotyped Viruses
2.4. Replication-Competent Virus Infection
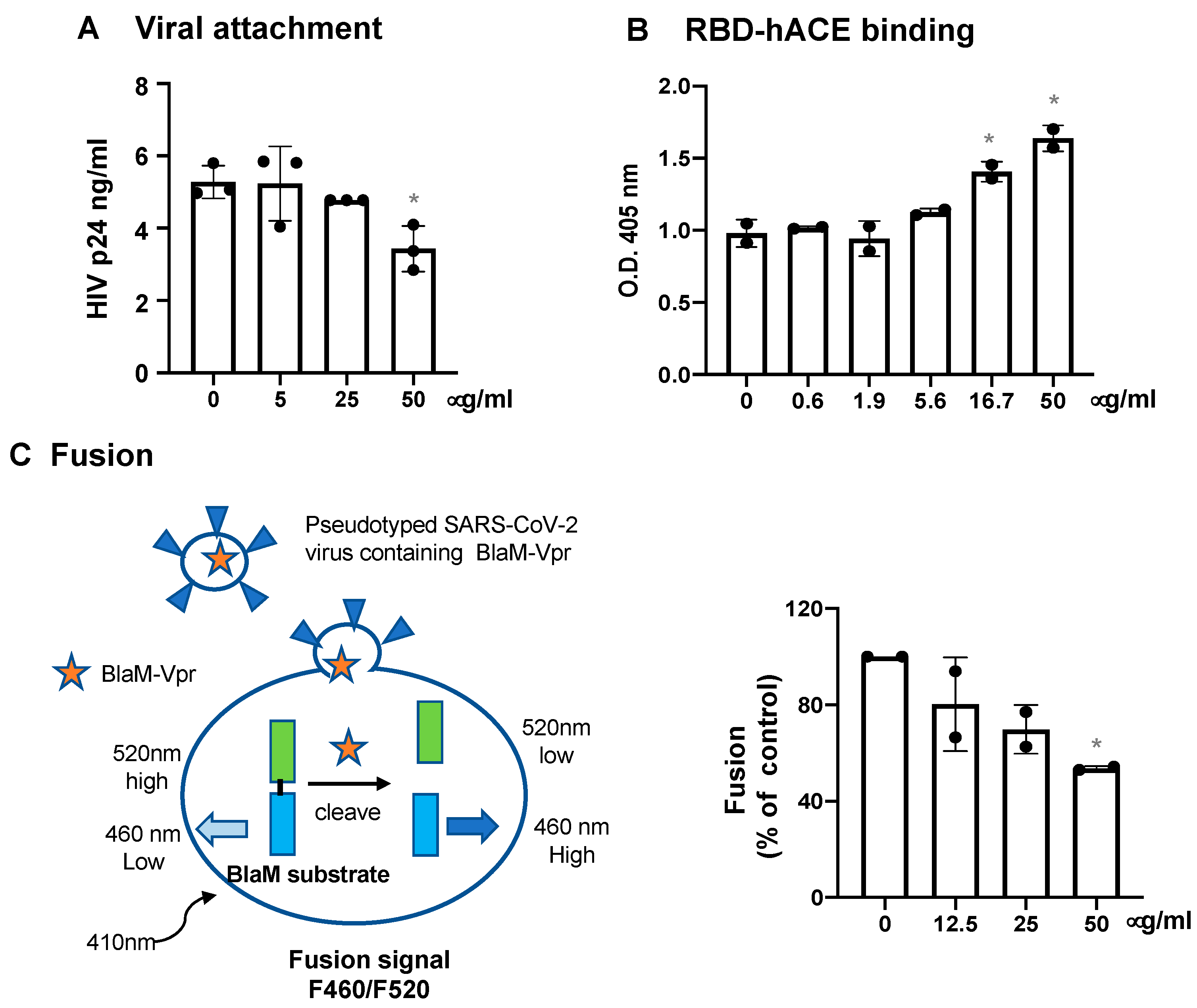
2.5. Viral Attachment Assay
2.6. SARS-CoV-2 RBD and hACE Binding Assay
2.7. Virus-Cell Fusion Assay
2.8. Cytotoxicity Assay
2.9. Statistical Analysis
3. Results
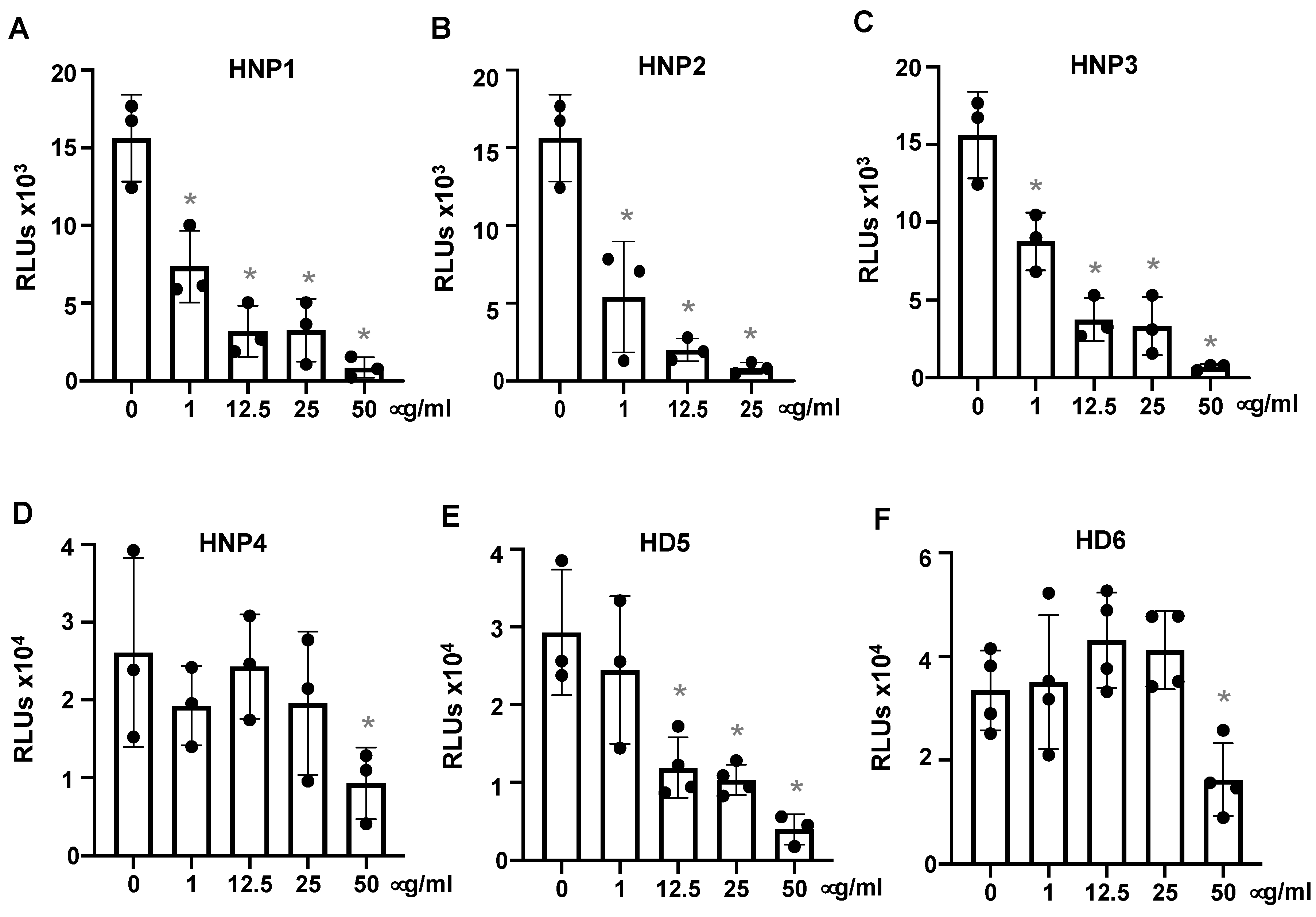
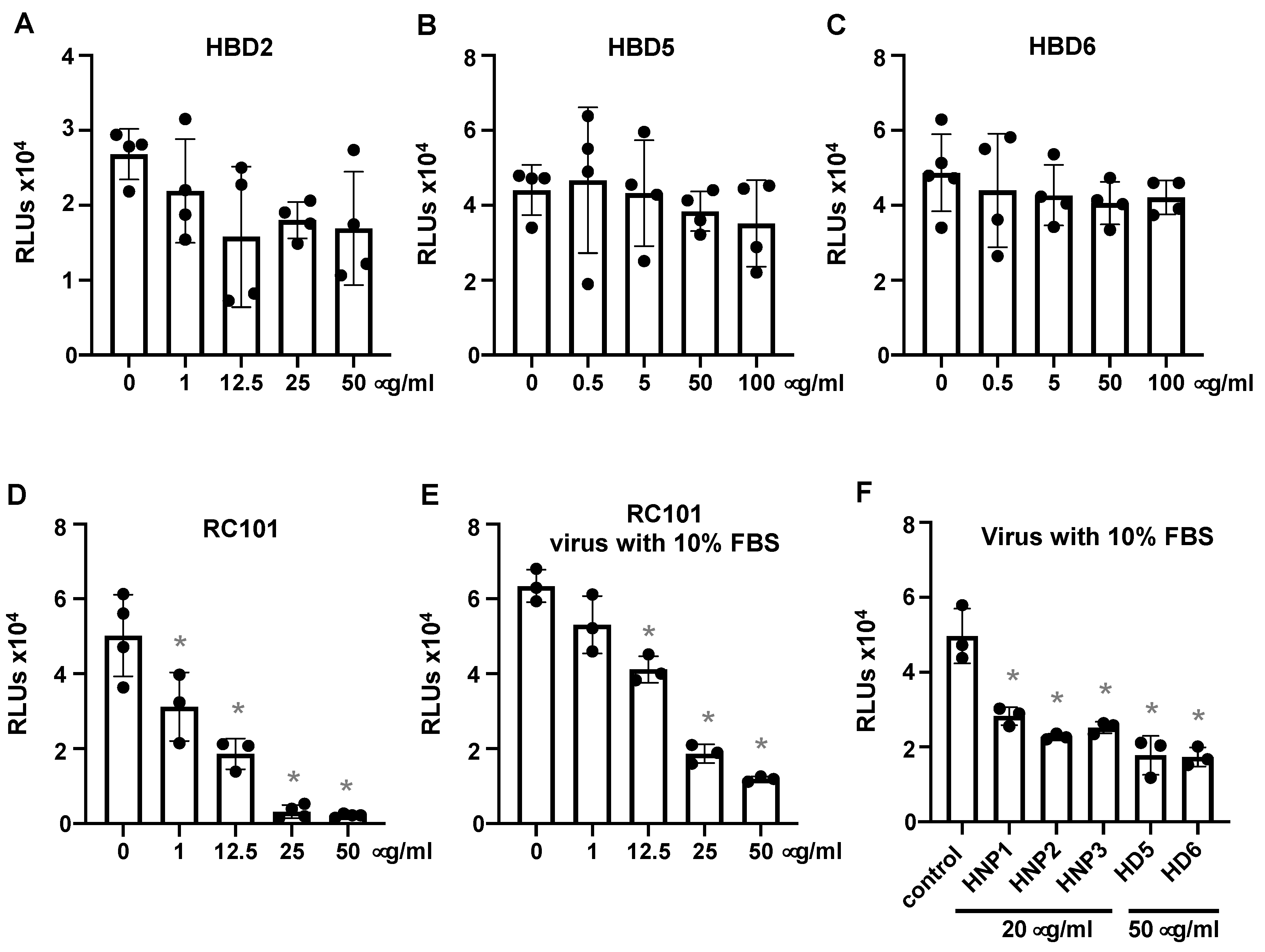
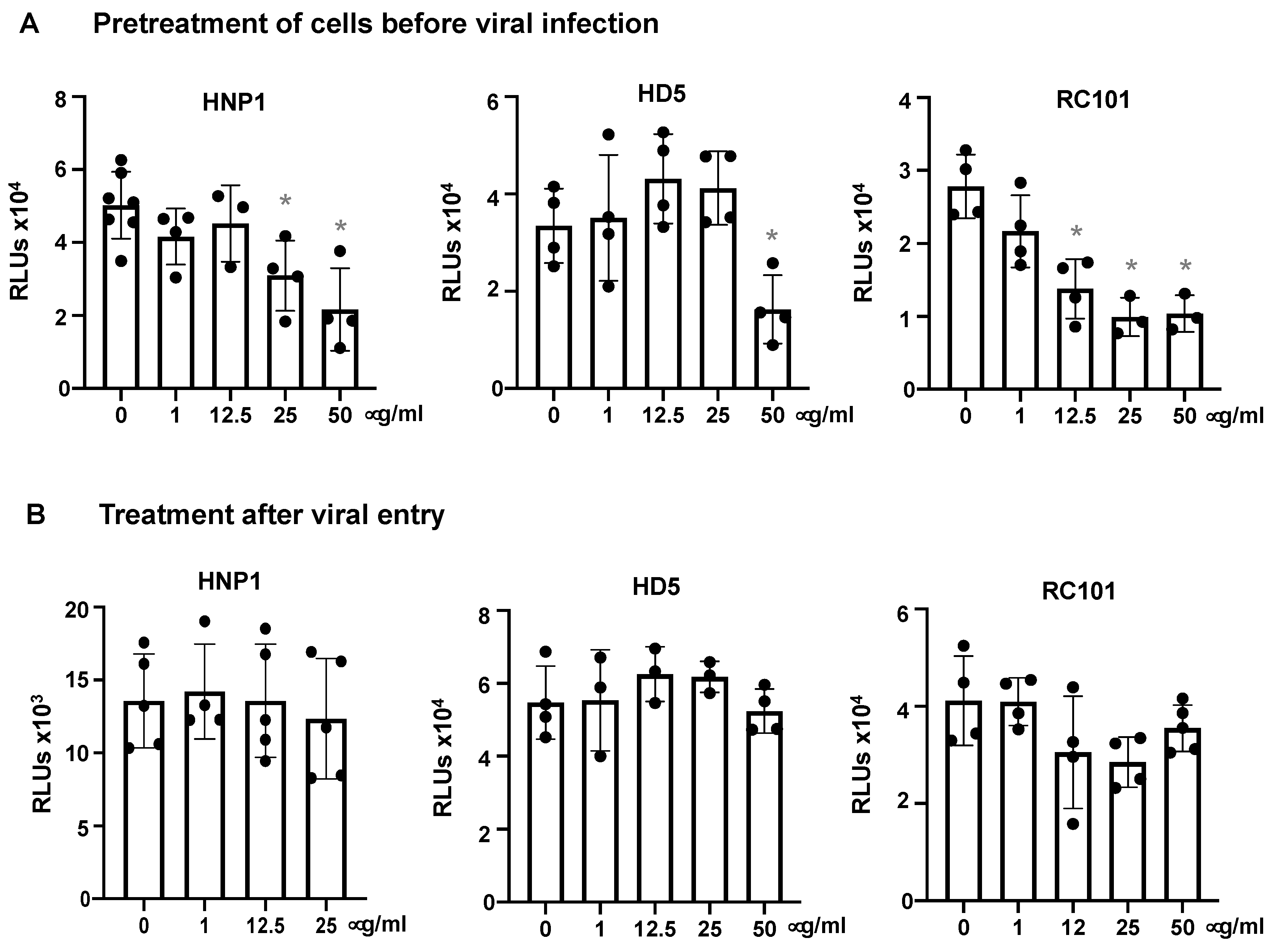
3.1. Human α-Defensins Inhibit SARS-CoV-2 Infection
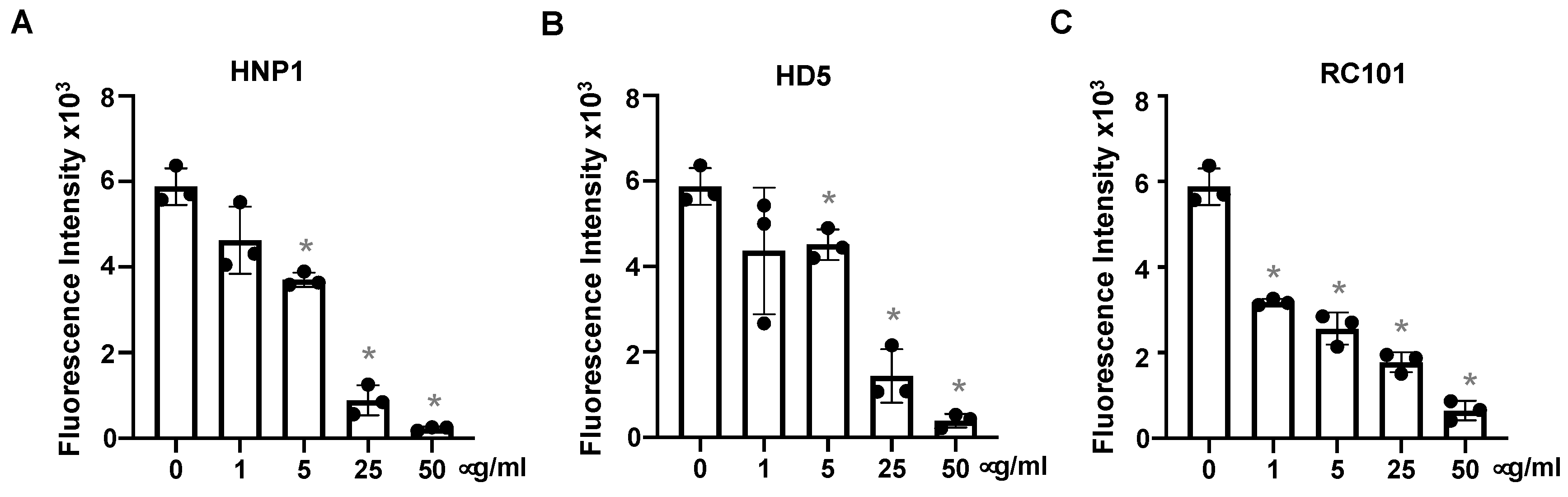
3.2. Defensins Inhibit Viral Entry
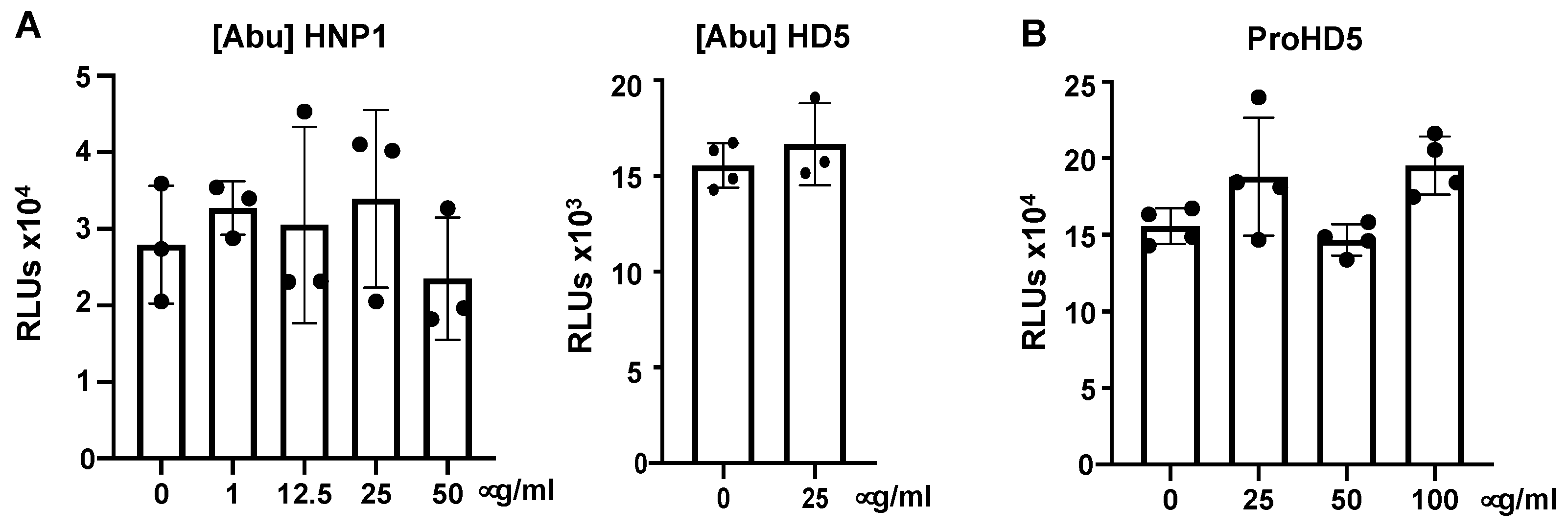
3.3. Native Structure (Disulfide Bonding) of Defensins Is Required to Inhibit SARS-CoV-2 Infection
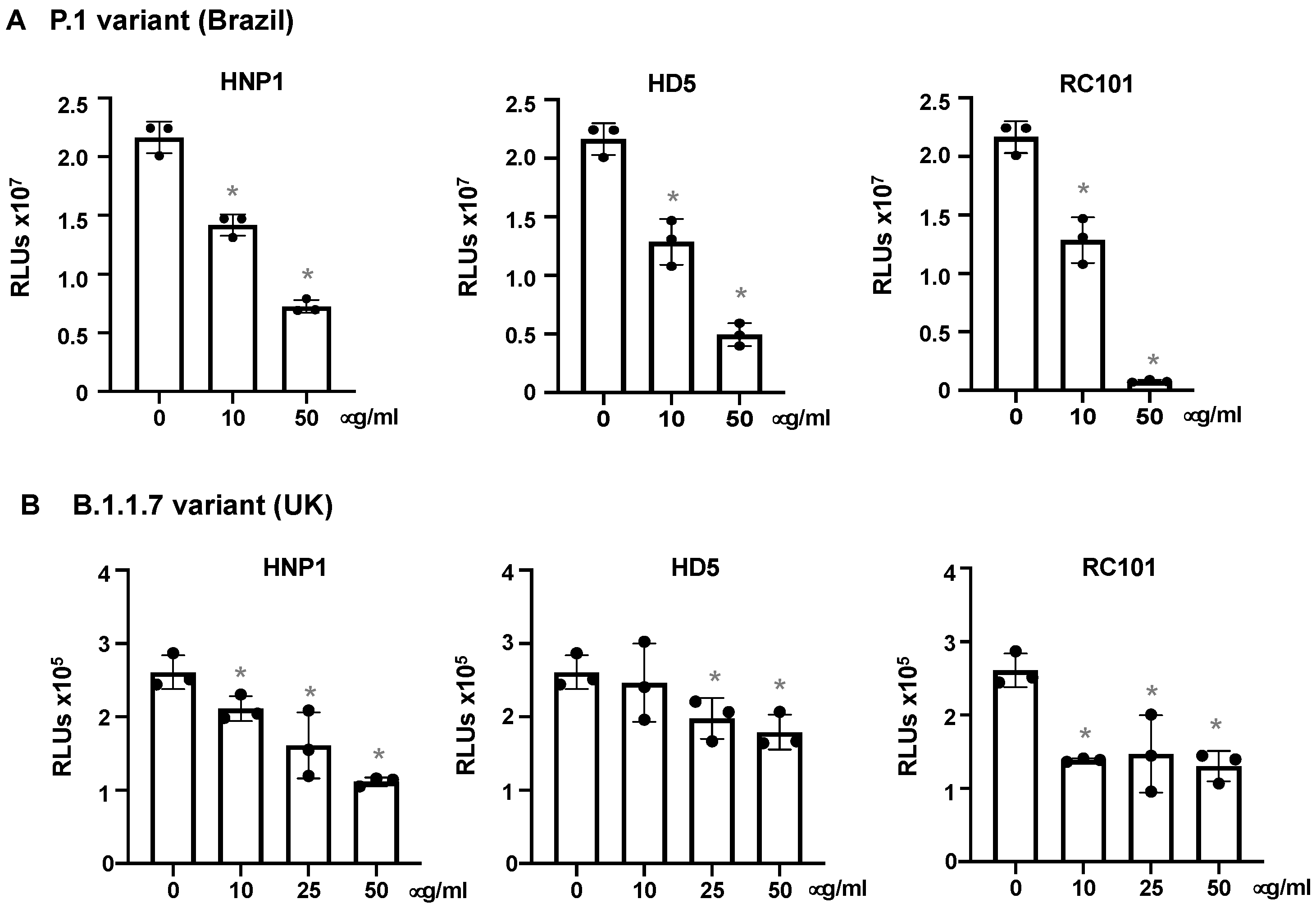
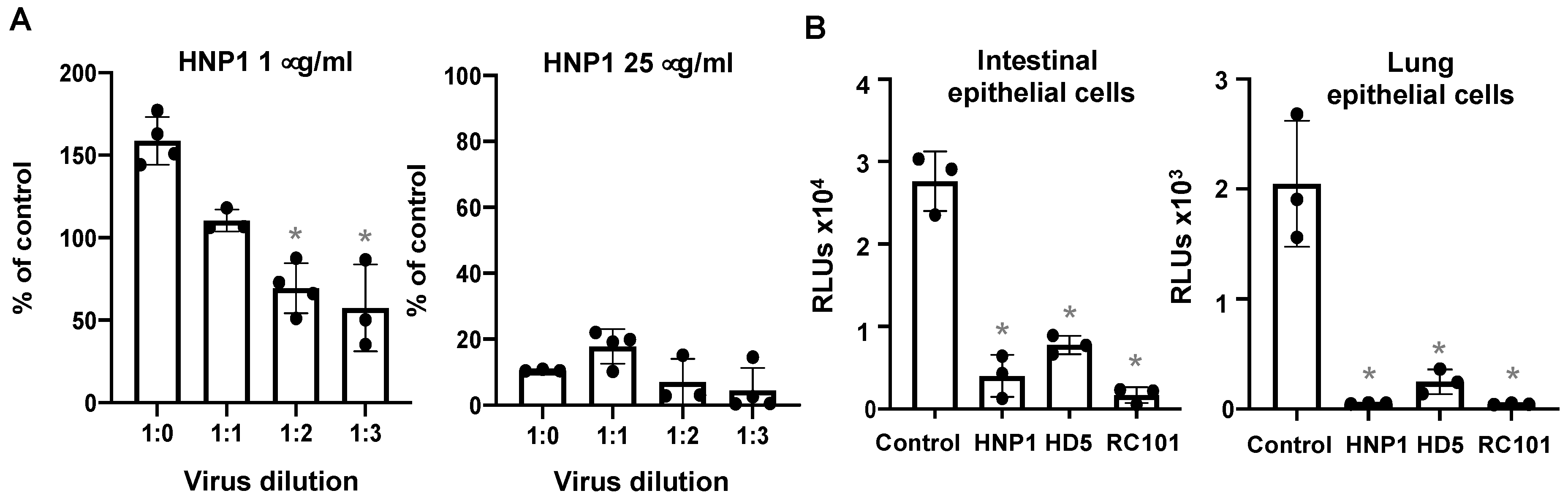
3.4. The effect of Virus Titers and Cell Types on Defensin-Mediated Viral Infection
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Available online: https://coronavirus.jhu.edu/map.html (accessed on 23 June 2021).
- Park, A.; Iwasaki, A. Type I and Type III Interferons—Induction, Signaling, Evasion, and Application to Combat COVID-19. Cell Host Microbe 2020, 27, 870–878. [Google Scholar] [CrossRef] [PubMed]
- Channappanavar, R.; Fehr, A.R.; Vijay, R.; Mack, M.; Zhao, J.; Meyerholz, D.K.; Perlman, S. Dysregulated Type I Interferon and Inflammatory Monocyte-Macrophage Responses Cause Lethal Pneumonia in SARS-CoV-Infected Mice. Cell Host Microbe 2016, 19, 181–193. [Google Scholar] [CrossRef] [Green Version]
- Channappanavar, R.; Fehr, A.R.; Zheng, J.; Wohlford-Lenane, C.; Abrahante, J.E.; Mack, M.; Sompallae, R.; McCray, P.B., Jr.; Meyerholz, D.K.; Perlman, S. IFN-I response timing relative to virus replication determines MERS coronavirus infection outcomes. J. Clin. Investig. 2019, 129, 3625–3639. [Google Scholar] [CrossRef]
- Ganz, T. Defensins: Antimicrobial peptides of innate immunity. Nat. Rev. Immunol. 2003, 3, 710–720. [Google Scholar] [CrossRef]
- Selsted, M.E.; Ouellette, A.J. Mammalian defensins in the antimicrobial immune response. Nat. Immunol. 2005, 6, 551–557. [Google Scholar] [CrossRef]
- Lehrer, R.I.; Lu, W. alpha-Defensins in human innate immunity. Immunol. Rev. 2012, 245, 84–112. [Google Scholar] [CrossRef]
- Bao, L.; Deng, W.; Huang, B.; Gao, H.; Liu, J.; Ren, L.; Wei, Q.; Yu, P.; Xu, Y.; Qi, F.; et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature 2020, 583, 830–833. [Google Scholar] [CrossRef]
- Sun, S.-H.; Chen, Q.; Gu, H.-J.; Yang, G.; Wang, Y.-X.; Huang, X.-Y.; Liu, S.-S.; Zhang, N.-N.; Li, X.-F.; Xiong, R.; et al. A Mouse Model of SARS-CoV-2 Infection and Pathogenesis. Cell Host Microbe 2020, 28, 124–133. [Google Scholar] [CrossRef]
- Rockx, B.; Kuiken, T.; Herfst, S.; Bestebroer, T.; Lamers, M.M.; Oude Munnink, B.B.; de Meulder, D.; van Amerongen, G.; van den Brand, J.; Okba, N.M.A.; et al. Comparative pathogenesis of COVID-19, MERS, and SARS in a nonhuman primate model. Science 2020, 368, 1012. [Google Scholar] [CrossRef] [Green Version]
- Barnes, B.J.; Adrover, J.M.; Baxter-Stoltzfus, A.; Borczuk, A.; Cools-Lartigue, J.; Crawford, J.M.; Dassler-Plenker, J.; Guerci, P.; Huynh, C.; Knight, J.S.; et al. Targeting potential drivers of COVID-19: Neutrophil extracellular traps. J. Exp. Med. 2020, 217, e20200652. [Google Scholar] [CrossRef]
- Chang, T.L.; Klotman, M.E. Defensins: Natural anti-HIV peptides. Aids Rev. 2004, 6, 161–168. [Google Scholar]
- Ding, J.; Chou, Y.Y.; Chang, T.L. Defensins in viral infections. J. Innate Immun. 2009, 1, 413–420. [Google Scholar] [CrossRef] [Green Version]
- Klotman, M.E.; Chang, T.L. Defensins in innate antiviral immunity. Nat. Rev. Immunol. 2006, 6, 447–456. [Google Scholar] [CrossRef]
- Wilson, S.S.; Bromme, B.A.; Holly, M.K.; Wiens, M.E.; Gounder, A.P.; Sul, Y.; Smith, J.G. Alpha-defensin-dependent enhancement of enteric viral infection. PLoS Pathog. 2017, 13, e1006446. [Google Scholar] [CrossRef] [PubMed]
- Klotman, M.E.; Rapista, A.; Teleshova, N.; Micsenyi, A.; Jarvis, G.A.; Lu, W.; Porter, E.; Chang, T.L. Neisseria gonorrhoeae-Induced Human Defensins 5 and 6 Increase HIV Infectivity: Role in Enhanced Transmission. J. Immunol. 2008, 180, 6176–6185. [Google Scholar] [CrossRef] [Green Version]
- Lehrer, R.I.; Cole, A.M.; Selsted, M.E. theta-Defensins: Cyclic peptides with endless potential. J. Biol. Chem. 2012, 287, 27014–27019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cole, A.M.; Hong, T.; Boo, L.M.; Nguyen, T.; Zhao, C.; Bristol, G.; Zack, J.A.; Waring, A.J.; Yang, O.O.; Lehrer, R.I. Retrocyclin: A primate peptide that protects cells from infection by T- and M-tropic strains of HIV-1. Proc. Natl. Acad. Sci. USA 2002, 99, 1813–1818. [Google Scholar] [CrossRef] [Green Version]
- Fruitwala, S.; El-Naccache, D.W.; Chang, T.L. Multifaceted immune functions of human defensins and underlying mechanisms. Semin Cell Dev. Biol. 2019, 88, 163–172. [Google Scholar] [CrossRef]
- Yang, D.; Liu, Z.H.; Tewary, P.; Chen, Q.; de la Rosa, G.; Oppenheim, J.J. Defensin participation in innate and adaptive immunity. Curr. Pharm. Des. 2007, 13, 3131–3139. [Google Scholar] [CrossRef]
- Wohlford-Lenane, C.L.; Meyerholz, D.K.; Perlman, S.; Zhou, H.; Tran, D.; Selsted, M.E.; McCray, P.B., Jr. Rhesus theta-defensin prevents death in a mouse model of severe acute respiratory syndrome coronavirus pulmonary disease. J. Virol. 2009, 83, 11385–11390. [Google Scholar] [CrossRef] [Green Version]
- Wu, Z.; Ericksen, B.; Tucker, K.; Lubkowski, J.; Lu, W. Synthesis and characterization of human alpha-defensins 4-6. J. Pept. Res. 2004, 64, 118–125. [Google Scholar] [CrossRef] [PubMed]
- Porter, E.M.; Liu, L.; Oren, A.; Anton, P.A.; Ganz, T. Localization of human intestinal defensin 5 in Paneth cell granules. Infect. Immun. 1997, 65, 2389–2395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.M.; Wang, W.; Song, Z.G.; Hu, Y.; Tao, Z.W.; Tian, J.H.; Pei, Y.Y.; et al. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sood, C.; Marin, M.; Chande, A.; Pizzato, M.; Melikyan, G.B. SERINC5 protein inhibits HIV-1 fusion pore formation by promoting functional inactivation of envelope glycoproteins. J. Biol. Chem. 2017, 292, 6014–6026. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moore, M.J.; Dorfman, T.; Li, W.; Wong, S.K.; Li, Y.; Kuhn, J.H.; Coderre, J.; Vasilieva, N.; Han, Z.; Greenough, T.C.; et al. Retroviruses pseudotyped with the severe acute respiratory syndrome coronavirus spike protein efficiently infect cells expressing angiotensin-converting enzyme 2. J. Virol. 2004, 78, 10628–10635. [Google Scholar] [CrossRef] [Green Version]
- Rogers, T.F.; Zhao, F.; Huang, D.; Beutler, N.; Burns, A.; He, W.-t.; Limbo, O.; Smith, C.; Song, G.; Woehl, J.; et al. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science 2020, 369, 956–963. [Google Scholar] [CrossRef]
- Xu, C.; Wang, A.; Geng, K.; Honnen, W.; Wang, X.; Bruiners, N.; Singh, S.; Ferrara, F.; D’Angelo, S.; Bradbury, A.R.M.; et al. Human Immunodeficiency Viruses Pseudotyped with SARS-CoV-2 Spike Proteins Infect a Broad Spectrum of Human Cell Lines through Multiple Entry Mechanisms. Viruses 2021, 13, 953. [Google Scholar] [CrossRef]
- Daher, K.A.; Selsted, M.E.; Lehrer, R.I. Direct inactivation of viruses by human granulocyte defensins. J. Virol. 1986, 60, 1068–1074. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, T.L.; Vargas, J., Jr.; DelPortillo, A.; Klotman, M.E. Dual role of alpha-defensin-1 in anti-HIV-1 innate immunity. J. Clin. Investig. 2005, 115, 765–773. [Google Scholar] [CrossRef] [Green Version]
- Xie, X.; Muruato, A.; Lokugamage, K.G.; Narayanan, K.; Zhang, X.; Zou, J.; Liu, J.; Schindewolf, C.; Bopp, N.E.; Aguilar, P.V.; et al. An Infectious cDNA Clone of SARS-CoV-2. Cell Host Microbe 2020, 27, 841–848.e3. [Google Scholar] [CrossRef]
- Xu, C.; Wang, A.; Hoskin, E.R.; Cugini, C.; Markowitz, K.; Chang, T.L.; Fine, D.H. Differential Effects of Antiseptic Mouth Rinses on SARS-CoV-2 Infectivity In Vitro. Pathogens 2021, 10, 272. [Google Scholar] [CrossRef] [PubMed]
- de la Vega, M.; Marin, M.; Kondo, N.; Miyauchi, K.; Kim, Y.; Epand, R.F.; Epand, R.M.; Melikyan, G.B. Inhibition of HIV-1 endocytosis allows lipid mixing at the plasma membrane, but not complete fusion. Retrovirology 2011, 8, 99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghosh, D.; Porter, E.; Shen, B.; Lee, S.K.; Wilk, D.; Drazba, J.; Yadav, S.P.; Crabb, J.W.; Ganz, T.; Bevins, C.L. Paneth cell trypsin is the processing enzyme for human defensin-5. Nat. Immunol. 2002, 3, 583–590. [Google Scholar] [CrossRef]
- Burki, T. Understanding variants of SARS-CoV-2. Lancet 2021, 397, 462. [Google Scholar] [CrossRef]
- Faria, N.R.; Mellan, T.A.; Whittaker, C.; Claro, I.M.; Candido, D.d.S.; Mishra, S.; Crispim, M.A.E.; Sales, F.C.S.; Hawryluk, I.; McCrone, J.T.; et al. Genomics and epidemiology of the P.1 SARS-CoV-2 lineage in Manaus, Brazil. Science 2021, 372, 815–821. [Google Scholar] [CrossRef]
- Volz, E.; Hill, V.; McCrone, J.T.; Price, A.; Jorgensen, D.; O’Toole, Á.; Southgate, J.; Johnson, R.; Jackson, B.; Nascimento, F.F.; et al. Evaluating the Effects of SARS-CoV-2 Spike Mutation D614G on Transmissibility and Pathogenicity. Cell 2021, 184, 64–75.e11. [Google Scholar] [CrossRef]
- Washington, N.L.; Gangavarapu, K.; Zeller, M.; Bolze, A.; Cirulli, E.T.; Schiabor Barrett, K.M.; Larsen, B.B.; Anderson, C.; White, S.; Cassens, T.; et al. Emergence and rapid transmission of SARS-CoV-2 B.1.1.7 in the United States. Cell 2021, 184, 2587–2594.e7. [Google Scholar] [CrossRef]
- Smith, J.G.; Silvestry, M.; Lindert, S.; Lu, W.; Nemerow, G.R.; Stewart, P.L. Insight into the mechanisms of adenovirus capsid disassembly from studies of defensin neutralization. PLoS Pathog. 2010, 6, e1000959. [Google Scholar] [CrossRef] [Green Version]
- Porter, E.; Yang, H.; Yavagal, S.; Preza, G.C.; Murillo, O.; Lima, H.; Greene, S.; Mahoozi, L.; Klein-Patel, M.; Diamond, G.; et al. Distinct defensin profiles in Neisseria gonorrhoeae and Chlamydia trachomatis urethritis reveal novel epithelial cell-neutrophil interactions. Infect. Immun. 2005, 73, 4823–4833. [Google Scholar] [CrossRef] [Green Version]
- Rapista, A.; Ding, J.; Benito, B.; Lo, Y.T.; Neiditch, M.B.; Lu, W.; Chang, T.L. Human defensins 5 and 6 enhance HIV-1 infectivity through promoting HIV attachment. Retrovirology 2011, 8, 45. [Google Scholar] [CrossRef] [Green Version]
- Xu, D.; Lu, W. Defensins: A Double-Edged Sword in Host Immunity. Front. Immunol. 2020, 11, 764. [Google Scholar] [CrossRef]
- Huang, Y.; Yang, C.; Xu, X.-f.; Xu, W.; Liu, S.-w. Structural and functional properties of SARS-CoV-2 spike protein: Potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020, 41, 1141–1149. [Google Scholar] [CrossRef]
- Dugan, A.S.; Maginnis, M.S.; Jordan, J.A.; Gasparovic, M.L.; Manley, K.; Page, R.; Williams, G.; Porter, E.; O’Hara, B.A.; Atwood, W.J. Human alpha-defensins inhibit BK virus infection by aggregating virions and blocking binding to host cells. J. Biol. Chem. 2008, 283, 31125–31132. [Google Scholar] [CrossRef] [Green Version]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Kruger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef]
- Cantuti-Castelvetri, L.; Ojha, R.; Pedro, L.D.; Djannatian, M.; Franz, J.; Kuivanen, S.; van der Meer, F.; Kallio, K.; Kaya, T.; Anastasina, M.; et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science 2020, 370, 856. [Google Scholar] [CrossRef]
- Zang, R.; Gomez Castro, M.F.; McCune, B.T.; Zeng, Q.; Rothlauf, P.W.; Sonnek, N.M.; Liu, Z.; Brulois, K.F.; Wang, X.; Greenberg, H.B.; et al. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci. Immunol. 2020, 5, eabc3582. [Google Scholar] [CrossRef]
- Daly, J.L.; Simonetti, B.; Klein, K.; Chen, K.-E.; Williamson, M.K.; Antón-Plágaro, C.; Shoemark, D.K.; Simón-Gracia, L.; Bauer, M.; Hollandi, R.; et al. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science 2020, 370, 861. [Google Scholar] [CrossRef]
- Clausen, T.M.; Sandoval, D.R.; Spliid, C.B.; Pihl, J.; Perrett, H.R.; Painter, C.D.; Narayanan, A.; Majowicz, S.A.; Kwong, E.M.; McVicar, R.N.; et al. SARS-CoV-2 Infection Depends on Cellular Heparan Sulfate and ACE2. Cell 2020, 183, 1043–1057.e15. [Google Scholar] [CrossRef]
- Zamorano Cuervo, N.; Grandvaux, N. ACE2: Evidence of role as entry receptor for SARS-CoV-2 and implications in comorbidities. Elife 2020, 9, e61390. [Google Scholar] [CrossRef]
- Rothan, H.A.; Han, H.C.; Ramasamy, T.S.; Othman, S.; Rahman, N.A.; Yusof, R. Inhibition of dengue NS2B-NS3 protease and viral replication in Vero cells by recombinant retrocyclin-1. BMC Infect. Dis. 2012, 12, 314. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.; Chen, W.; Zhang, Z.; Deng, Y.; Lian, J.-Q.; Du, P.; Wei, D.; Zhang, Y.; Sun, X.-X.; Gong, L.; et al. CD147-spike protein is a novel route for SARS-CoV-2 infection to host cells. Signal Transduct. Target. Ther. 2020, 5, 283. [Google Scholar] [CrossRef]
- Wang, S.; Qiu, Z.; Hou, Y.; Deng, X.; Xu, W.; Zheng, T.; Wu, P.; Xie, S.; Bian, W.; Zhang, C.; et al. AXL is a candidate receptor for SARS-CoV-2 that promotes infection of pulmonary and bronchial epithelial cells. Cell Res. 2021, 31, 126–140. [Google Scholar] [CrossRef]
- Guaní-Guerra, E.; Santos-Mendoza, T.; Lugo-Reyes, S.O.; Terán, L.M. Antimicrobial peptides: General overview and clinical implications in human health and disease. Clin. Immunol. 2010, 135, 1–11. [Google Scholar] [CrossRef]
- Lehrer, R.I.; Ganz, T. Antimicrobial peptides in mammalian and insect host defence. Curr. Opin. Immunol. 1999, 11, 23–27. [Google Scholar] [CrossRef]
- Huan, Y.; Kong, Q.; Mou, H.; Yi, H. Antimicrobial Peptides: Classification, Design, Application and Research Progress in Multiple Fields. Front. Microbiol. 2020, 11, 582779. [Google Scholar] [CrossRef] [PubMed]
- Machado, L.R.; Ottolini, B. An evolutionary history of defensins: A role for copy number variation in maximizing host innate and adaptive immune responses. Front. Immunol. 2015, 6, 115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hollox, E.J.; Barber, J.C.; Brookes, A.J.; Armour, J.A. Defensins and the dynamic genome: What we can learn from structural variation at human chromosome band 8p23.1. Genome Res. 2008, 18, 1686–1697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lehrer, R.I. Multispecific myeloid defensins. Curr. Opin. Hematol. 2007, 14, 16–21. [Google Scholar] [CrossRef]
- Taudien, S.; Galgoczy, P.; Huse, K.; Reichwald, K.; Schilhabel, M.; Szafranski, K.; Shimizu, A.; Asakawa, S.; Frankish, A.; Loncarevic, I.F.; et al. Polymorphic segmental duplications at 8p23.1 challenge the determination of individual defensin gene repertoires and the assembly of a contiguous human reference sequence. BMC Genom 2004, 5, 92. [Google Scholar] [CrossRef] [Green Version]
- Butcher, S.; Chahel, H.; Lord, J.M. Review article: Ageing and the neutrophil: No appetite for killing? Immunology 2000, 100, 411–416. [Google Scholar] [CrossRef]
- Wenisch, C.; Patruta, S.; Daxbock, F.; Krause, R.; Horl, W. Effect of age on human neutrophil function. J. Leukoc. Biol. 2000, 67, 40–45. [Google Scholar] [CrossRef]
- Obama, T.; Scalia, R.; Eguchi, S. Targeting neutrophil: New approach against hypertensive cardiac remodeling? Hypertension 2014, 63, 1171–1172. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Zhang, Q.; Wu, H.; Du, H.; Liu, L.; Shi, H.; Wang, C.; Xia, Y.; Guo, X.; Li, C.; et al. Blood Neutrophil to Lymphocyte Ratio as a Predictor of Hypertension. Am. J. Hypertens. 2015, 28, 1339–1346. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Yang, J.; Zhao, F.; Zhi, L.; Wang, X.; Liu, L.; Bi, Z.; Zhao, Y. Prevalence and impact of cardiovascular metabolic diseases on COVID-19 in China. Clin. Res. Cardiol. 2020, 109, 531–538. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zheng, Y.; Gou, X.; Pu, K.; Chen, Z.; Guo, Q.; Ji, R.; Wang, H.; Wang, Y.; Zhou, Y. Prevalence of comorbidities in the novel Wuhan coronavirus (COVID-19) infection: A systematic review and meta-analysis. Int. J. Infect. Dis. 2020, 127, 104371. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; McGoogan, J.M. Characteristics of and Important Lessons from the Coronavirus Disease 2019 (COVID-19) Outbreak in China: Summary of a Report of 72314 Cases from the Chinese Center for Disease Control and Prevention. JAMA 2020, 323, 1239–1242. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Tong, Z.; Guan, X.; Du, B.; Qiu, H. Clinical Characteristics of Patients Who Died of Coronavirus Disease 2019 in China. JAMA Netw. Open 2020, 3, e205619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bdeir, K.; Cane, W.; Canziani, G.; Chaiken, I.; Weisel, J.; Koschinsky, M.L.; Lawn, R.M.; Bannerman, P.G.; Sachais, B.S.; Kuo, A.; et al. Defensin promotes the binding of lipoprotein(a) to vascular matrix. Blood 1999, 94, 2007–2019. [Google Scholar] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, C.; Wang, A.; Marin, M.; Honnen, W.; Ramasamy, S.; Porter, E.; Subbian, S.; Pinter, A.; Melikyan, G.B.; Lu, W.; et al. Human Defensins Inhibit SARS-CoV-2 Infection by Blocking Viral Entry. Viruses 2021, 13, 1246. https://doi.org/10.3390/v13071246
Xu C, Wang A, Marin M, Honnen W, Ramasamy S, Porter E, Subbian S, Pinter A, Melikyan GB, Lu W, et al. Human Defensins Inhibit SARS-CoV-2 Infection by Blocking Viral Entry. Viruses. 2021; 13(7):1246. https://doi.org/10.3390/v13071246
Chicago/Turabian StyleXu, Chuan, Annie Wang, Mariana Marin, William Honnen, Santhamani Ramasamy, Edith Porter, Selvakumar Subbian, Abraham Pinter, Gregory B. Melikyan, Wuyuan Lu, and et al. 2021. "Human Defensins Inhibit SARS-CoV-2 Infection by Blocking Viral Entry" Viruses 13, no. 7: 1246. https://doi.org/10.3390/v13071246
APA StyleXu, C., Wang, A., Marin, M., Honnen, W., Ramasamy, S., Porter, E., Subbian, S., Pinter, A., Melikyan, G. B., Lu, W., & Chang, T. L. (2021). Human Defensins Inhibit SARS-CoV-2 Infection by Blocking Viral Entry. Viruses, 13(7), 1246. https://doi.org/10.3390/v13071246







