The Antiviral Effect of the Chemical Compounds Targeting DED/EDh Motifs of the Viral Proteins on Lymphocytic Choriomeningitis Virus and SARS-CoV-2
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cells and Compounds
2.2. Viruses
2.3. Cell Viability Assay
2.4. Compounds’ Treatments
2.5. Titration of 3rLCMV/GFP
2.6. Plaque Assay for SARS-CoV-2
2.7. Measurement of SARS-CoV-2 RNA in the Cell
2.8. Docking Simulation
2.9. Statistical Analysis
3. Results
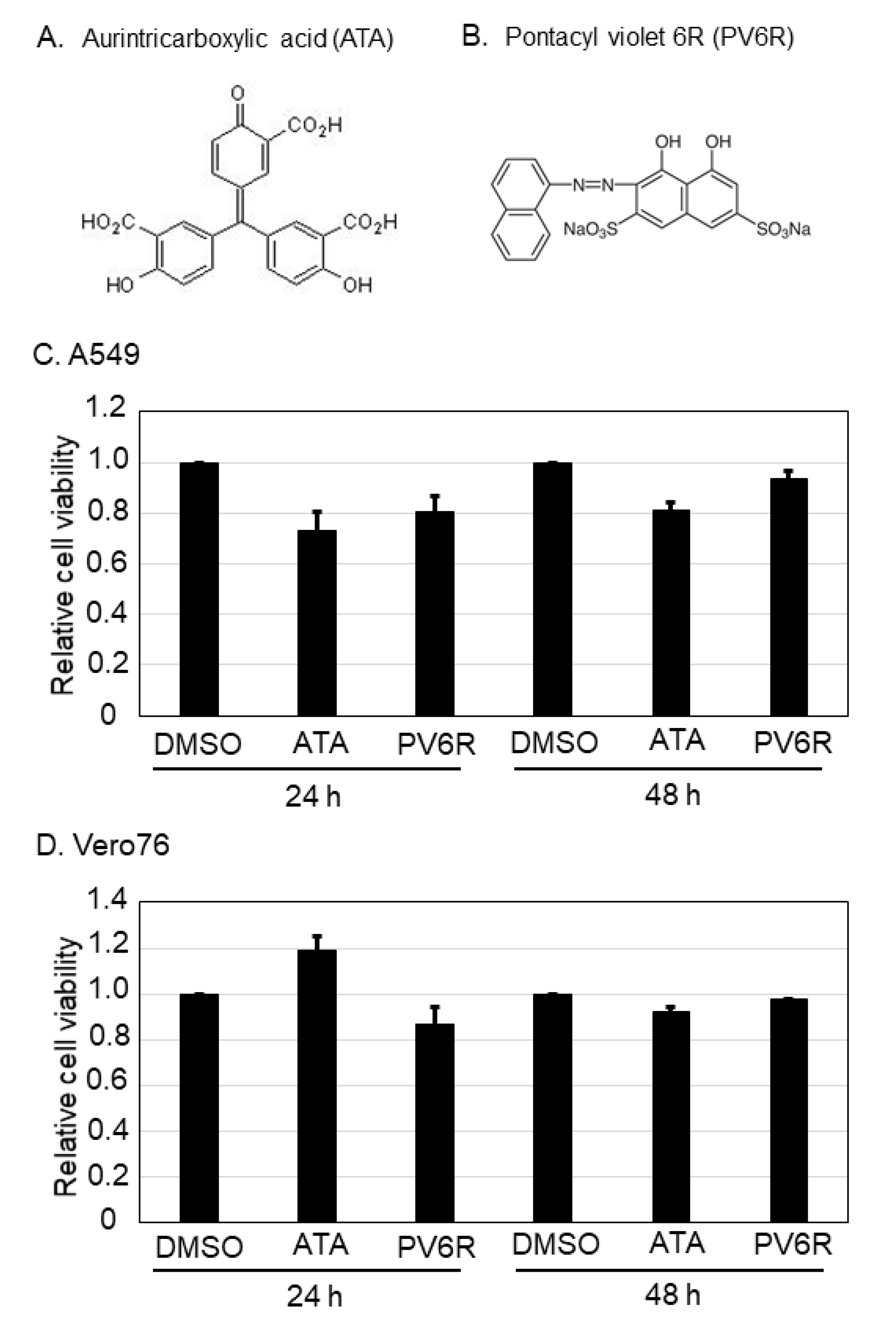
3.1. Cell Viability against ATA and PV6R Treatment
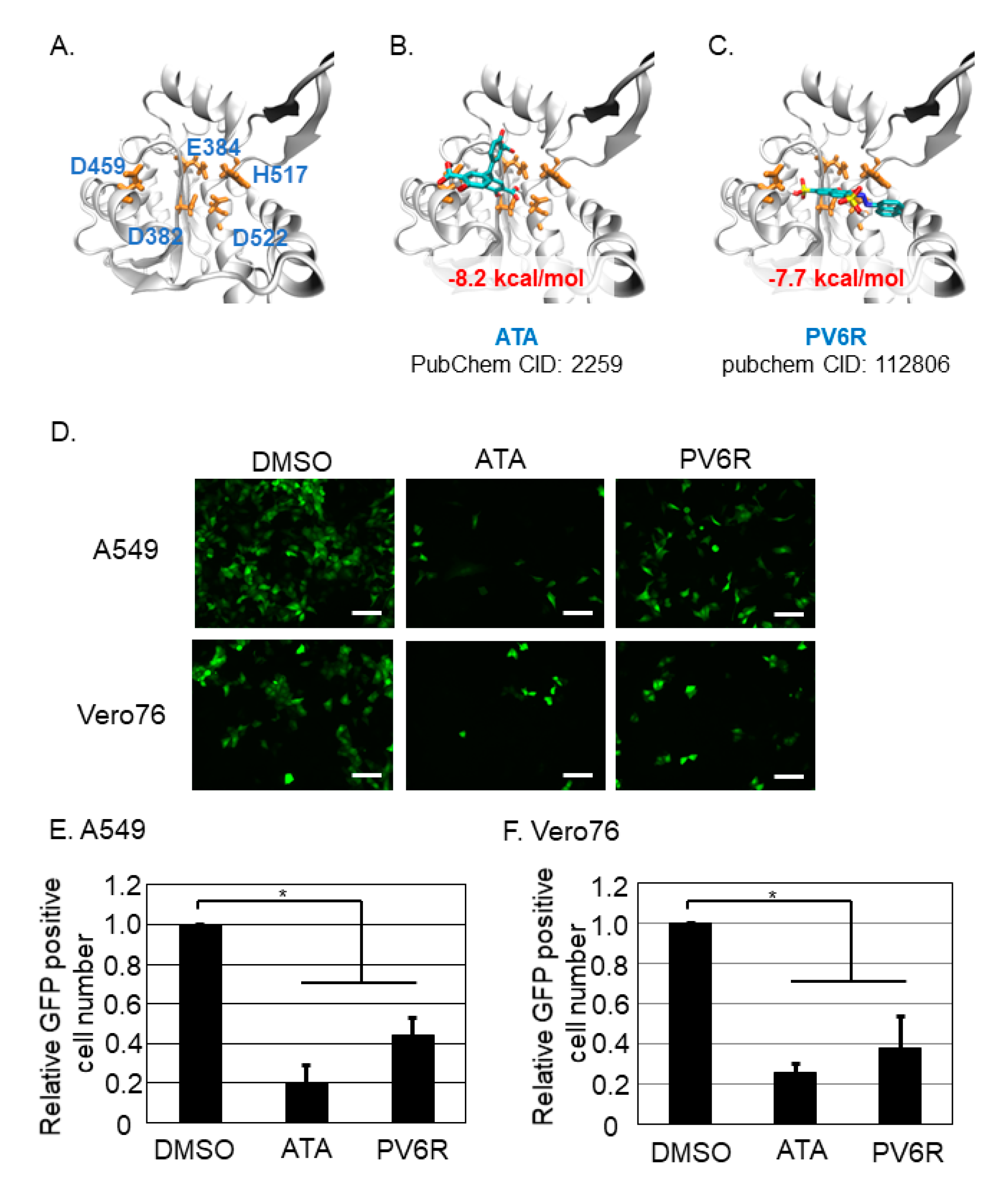
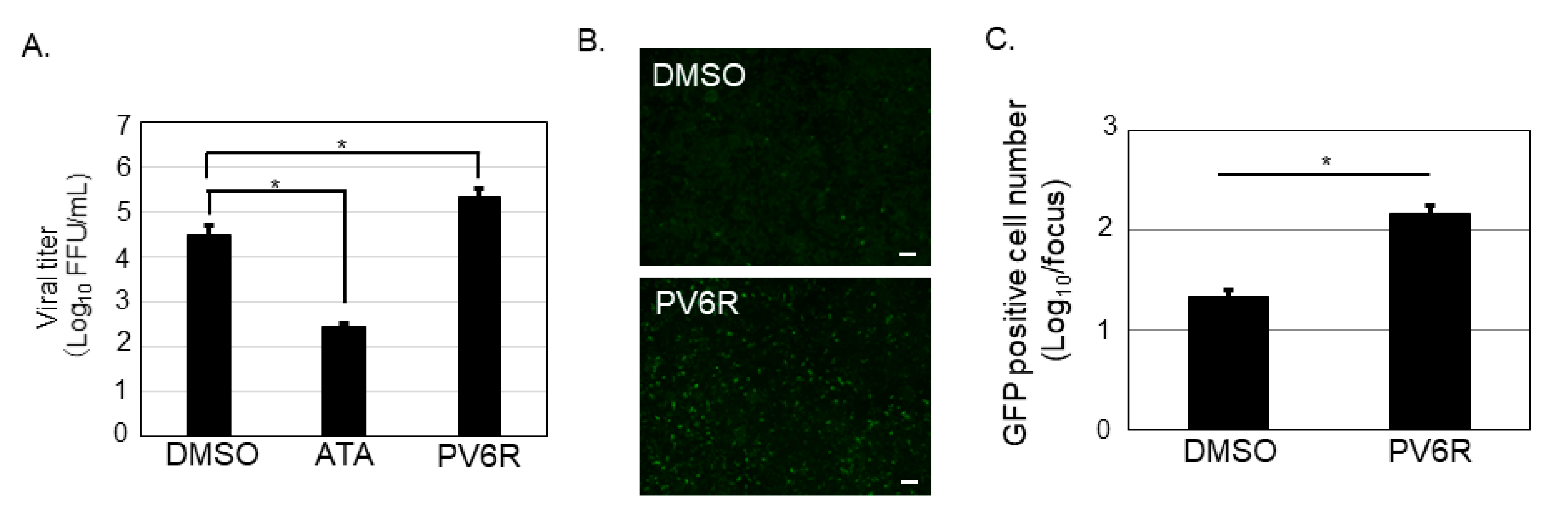
3.2. Both ATA and PV6R Inhibited the Post-Entry Step of the LCMV Infection
3.3. ATA, but Not PV6R, Inhibited the Propagation of the LCMV
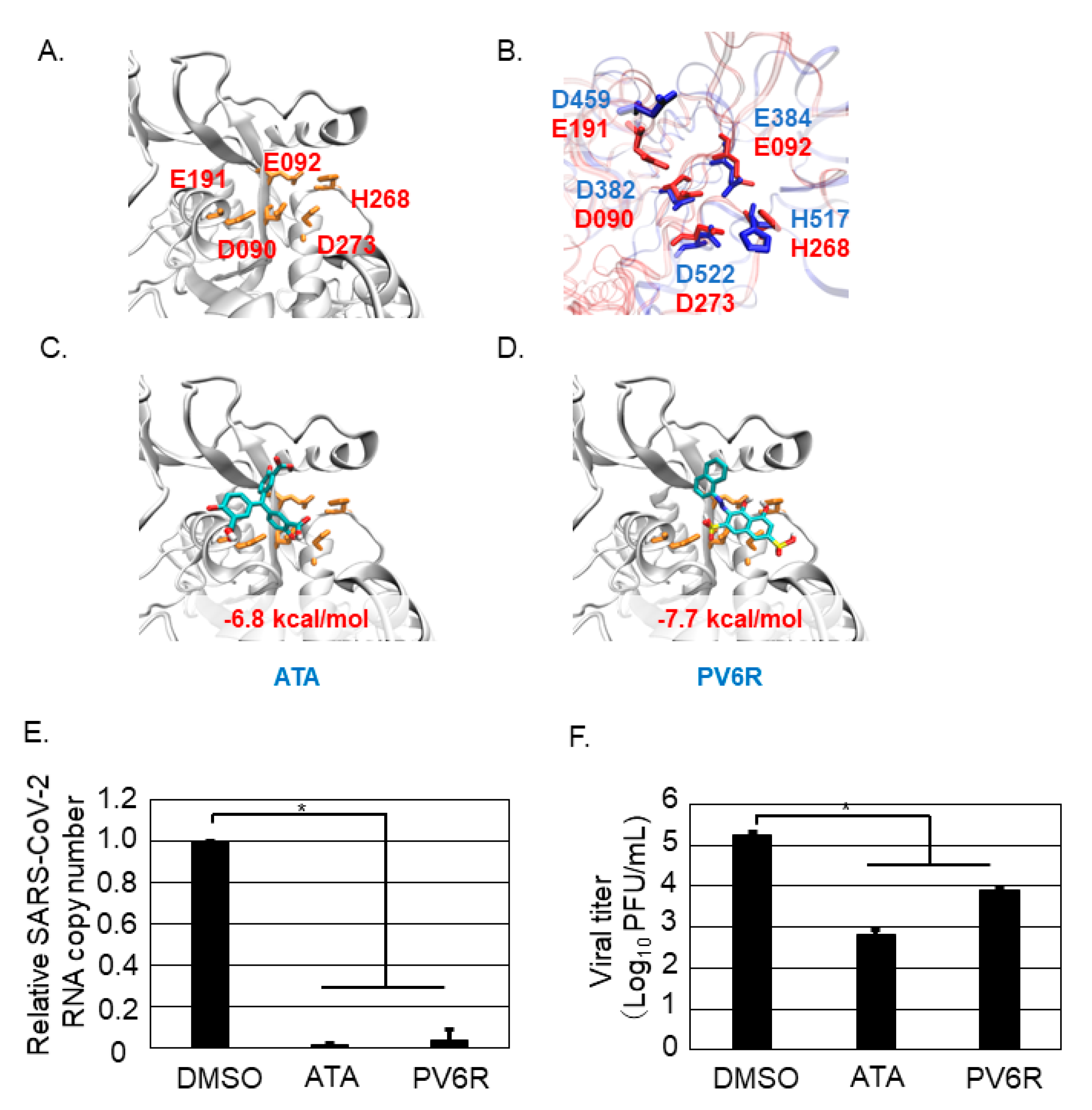
3.4. Both ATA and PV6R Inhibited SARS-CoV-2 Replication and Propagation
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Radoshitzky, S.R.; Buchmeier, M.J.; de la Torre, J.C. Arenaviridae: The Viruses and Their Replication. In Fields Virology, 7th ed.; Springer: Alphen aan den Rijn, The Netherlands, 2020; Volume 1, pp. 784–809. [Google Scholar]
- Perlman, S.; Masters, P.S. Coronaviridae: The Viruses and Their Replication. In Fields Virology, 7th ed.; Springer: Alphen aan den Rijn, The Netherlands, 2020; Volume 1, pp. 410–448. [Google Scholar]
- Watanabe, K.; Ishikawa, T.; Otaki, H.; Mizuta, S.; Hamada, T.; Nakagaki, T.; Ishibashi, D.; Urata, S.; Yasuda, J.; Tanaka, Y.; et al. Structure-based drug discovery for combating influenza virus by targeting the PA-PB1 interaction. Sci. Rep. 2017, 7, 9500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emonet, S.F.; Garidou, L.; McGavern, D.B.; de la Torre, J.C. Generation of recombinant lymphocytic choriomeningitis viruses with trisegmented genomes stably expressing two additional genes of interest. Proc. Natl. Acad. Sci. USA 2009, 106, 3473–3478. [Google Scholar] [CrossRef] [Green Version]
- Shirato, K.; Nao, N.; Katano, H.; Takayama, I.; Saito, S.; Kato, F.; Katoh, H.; Sakata, M.; Nakatsu, Y.; Mori, Y.; et al. Development of Genetic Diagnostic Methods for Detection for Novel Coronavirus 2019(nCoV-2019) in Japan. Jpn. J. Infect. Dis. 2020, 73, 304–307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- West, B.R.; Hastie, K.M.; Saphire, E.O. Structure of the LCMV nucleoprotein provides a template for understanding arenavirus replication and immunosuppression. Acta Cryst. D Biol. Cryst. 2014, 70, 1764–1769. [Google Scholar] [CrossRef] [PubMed]
- Sali, A.; Blundell, T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993, 234, 779–815. [Google Scholar] [CrossRef]
- Ma, Y.; Wu, L.; Shaw, N.; Gao, Y.; Wang, J.; Sun, Y.; Lou, Z.; Yan, L.; Zhang, R.; Rao, Z. Structural basis and functional analysis of the SARS coronavirus nsp14-nsp10 complex. Proc. Natl. Acad. Sci. USA 2015, 112, 9436–9441. [Google Scholar] [CrossRef] [Green Version]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [Green Version]
- Huang, K.W.; Hsu, K.C.; Chu, L.Y.; Yang, J.M.; Yuan, H.S.; Hsiao, Y.Y. Identification of Inhibitors for the DEDDh Family of Exonucleases and a Unique Inhibition Mechanism by Crystal Structure Analysis of CRN-4 Bound with 2-Morpholin-4-ylethanesulfonate (MES). J. Med. Chem. 2016, 59, 8019–8029. [Google Scholar] [CrossRef] [Green Version]
- Ogando, N.S.; Zevenhoven-Dobbe, J.C.; van der Meer, Y.; Bredenbeek, P.J.; Posthuma, C.C.; Snijder, E.J. The Enzymatic Activity of the nsp14 Exoribonuclease Is Critical for Replication of MERS-CoV and SARS-CoV-2. J. Virol. 2020, 94. [Google Scholar] [CrossRef] [PubMed]
- Minskaia, E.; Hertzig, T.; Gorbalenya, A.E.; Campanacci, V.; Cambillau, C.; Canard, B.; Ziebuhr, J. Discovery of an RNA virus 3′->5′ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc. Natl. Acad. Sci. USA 2006, 103, 5108–5113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Case, J.B.; Li, Y.; Elliott, R.; Lu, X.; Graepel, K.W.; Sexton, N.R.; Smith, E.C.; Weiss, S.R.; Denison, M.R. Murine Hepatitis Virus nsp14 Exoribonuclease Activity Is Required for Resistance to Innate Immunity. J. Virol. 2018, 92. [Google Scholar] [CrossRef] [Green Version]
- Pruijssers, A.J.; Denison, M.R. Nucleoside analogues for the treatment of coronavirus infections. Curr. Opin. Virol. 2019, 35, 57–62. [Google Scholar] [CrossRef]
- Moeller, N.H.; Shi, K.; Demir, O.; Banerjee, S.; Yin, L.; Belica, C.; Durfee, C.; Amaro, R.E.; Aihara, H. Structure and dynamics of SARS-CoV-2 proofreading exoribonuclease ExoN. bioRxiv 2021. [Google Scholar] [CrossRef]
- Munaweera, R.; Hu, Y.S. Computational Characterizations of the Interactions Between the Pontacyl Violet 6R and Exoribonuclease as a Potential Drug Target Against SARS-CoV-2. Front. Chem. 2020, 8, 627340. [Google Scholar] [CrossRef]
- Martinez-Sobrido, L.; Emonet, S.; Giannakas, P.; Cubitt, B.; Garcia-Sastre, A.; de la Torre, J.C. Identification of amino acid residues critical for the anti-interferon activity of the nucleoprotein of the prototypic arenavirus lymphocytic choriomeningitis virus. J. Virol. 2009, 83, 11330–11340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carnec, X.; Baize, S.; Reynard, S.; Diancourt, L.; Caro, V.; Tordo, N.; Bouloy, M. Lassa virus nucleoprotein mutants generated by reverse genetics induce a robust type I interferon response in human dendritic cells and macrophages. J. Virol. 2011, 85, 12093–12097. [Google Scholar] [CrossRef] [Green Version]
- Qi, X.; Lan, S.; Wang, W.; Schelde, L.M.; Dong, H.; Wallat, G.D.; Ly, H.; Liang, Y.; Dong, C. Cap binding and immune evasion revealed by Lassa nucleoprotein structure. Nature 2010, 468, 779–783. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hastie, K.M.; Kimberlin, C.R.; Zandonatti, M.A.; MacRae, I.J.; Saphire, E.O. Structure of the Lassa virus nucleoprotein reveals a dsRNA-specific 3′ to 5′ exonuclease activity essential for immune suppression. Proc. Natl. Acad. Sci. USA 2011, 108, 2396–2401. [Google Scholar] [CrossRef] [Green Version]
- Jiang, X.; Huang, Q.; Wang, W.; Dong, H.; Ly, H.; Liang, Y.; Dong, C. Structures of arenaviral nucleoproteins with triphosphate dsRNA reveal a unique mechanism of immune suppression. J. Biol. Chem. 2013, 288, 16949–16959. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, J.G.; Avila-Perez, G.; Madere, F.; Hilimire, T.A.; Nogales, A.; Almazan, F.; Martinez-Sobrido, L. Potent Inhibition of Zika Virus Replication by Aurintricarboxylic Acid. Front. Microbiol. 2019, 10, 718. [Google Scholar] [CrossRef]
- Myskiw, C.; Deschambault, Y.; Jefferies, K.; He, R.; Cao, J. Aurintricarboxylic acid inhibits the early stage of vaccinia virus replication by targeting both cellular and viral factors. J. Virol. 2007, 81, 3027–3032. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Bopda-Waffo, A.; Basu, A.; Krishnan, R.; Silberstein, E.; Taylor, D.R.; Talele, T.T.; Arora, P.; Kaushik-Basu, N. Characterization of aurintricarboxylic acid as a potent hepatitis C virus replicase inhibitor. Antivir. Chem. Chemother. 2009, 20, 19–36. [Google Scholar] [CrossRef] [Green Version]
- Hashem, A.M.; Flaman, A.S.; Farnsworth, A.; Brown, E.G.; Van Domselaar, G.; He, R.; Li, X. Aurintricarboxylic acid is a potent inhibitor of influenza A and B virus neuraminidases. PLoS ONE 2009, 4, e8350. [Google Scholar] [CrossRef] [PubMed]
- He, R.; Adonov, A.; Traykova-Adonova, M.; Cao, J.; Cutts, T.; Grudesky, E.; Deschambaul, Y.; Berry, J.; Drebot, M.; Li, X. Potent and selective inhibition of SARS coronavirus replication by aurintricarboxylic acid. Biochem. Biophys. Res. Commun. 2004, 320, 1199–1203. [Google Scholar] [CrossRef] [PubMed]
- Yap, Y.; Zhang, X.; Andonov, A.; He, R. Structural analysis of inhibition mechanisms of aurintricarboxylic acid on SARS-CoV polymerase and other proteins. Comput. Biol. Chem. 2005, 29, 212–219. [Google Scholar] [CrossRef]
- Ibrahim, H.; Wilusz, J.; Wilusz, C.J. RNA recognition by 3′-to-5′ exonucleases: The substrate perspective. Biochim. Biophys. Acta 2008, 1779, 256–265. [Google Scholar] [CrossRef] [Green Version]
- Walther, W.; Stein, U.; Siegel, R.; Fichtner, I.; Schlag, P.M. Use of the nuclease inhibitor aurintricarboxylic acid (ATA) for improved non-viral intratumoral in vivo gene transfer by jet-injection. J. Gene. Med. 2005, 7, 477–485. [Google Scholar] [CrossRef]
- Cushman, M.; Wang, P.L.; Chang, S.H.; Wild, C.; De Clercq, E.; Schols, D.; Goldman, M.E.; Bowen, J.A. Preparation and anti-HIV activities of aurintricarboxylic acid fractions and analogues: Direct correlation of antiviral potency with molecular weight. J. Med. Chem. 1991, 34, 329–337. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Gonzalez, A.; Munoz-Velasco, I.; Cottom-Salas, W.; Becerra, A.; Campillo-Balderas, J.A.; Hernandez-Morales, R.; Vazquez-Salazar, A.; Jacome, R.; Lazcano, A. Structural analysis of viral ExoN domains reveals polyphyletic hijacking events. PLoS ONE 2021, 16, e0246981. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ngwe Tun, M.M.; Morita, K.; Ishikawa, T.; Urata, S. The Antiviral Effect of the Chemical Compounds Targeting DED/EDh Motifs of the Viral Proteins on Lymphocytic Choriomeningitis Virus and SARS-CoV-2. Viruses 2021, 13, 1220. https://doi.org/10.3390/v13071220
Ngwe Tun MM, Morita K, Ishikawa T, Urata S. The Antiviral Effect of the Chemical Compounds Targeting DED/EDh Motifs of the Viral Proteins on Lymphocytic Choriomeningitis Virus and SARS-CoV-2. Viruses. 2021; 13(7):1220. https://doi.org/10.3390/v13071220
Chicago/Turabian StyleNgwe Tun, Mya Myat, Kouichi Morita, Takeshi Ishikawa, and Shuzo Urata. 2021. "The Antiviral Effect of the Chemical Compounds Targeting DED/EDh Motifs of the Viral Proteins on Lymphocytic Choriomeningitis Virus and SARS-CoV-2" Viruses 13, no. 7: 1220. https://doi.org/10.3390/v13071220
APA StyleNgwe Tun, M. M., Morita, K., Ishikawa, T., & Urata, S. (2021). The Antiviral Effect of the Chemical Compounds Targeting DED/EDh Motifs of the Viral Proteins on Lymphocytic Choriomeningitis Virus and SARS-CoV-2. Viruses, 13(7), 1220. https://doi.org/10.3390/v13071220





