Progress in Anti-Mammarenavirus Drug Development
Abstract
:1. Introduction
2. Mammarenavirus Molecular and Cell Biology
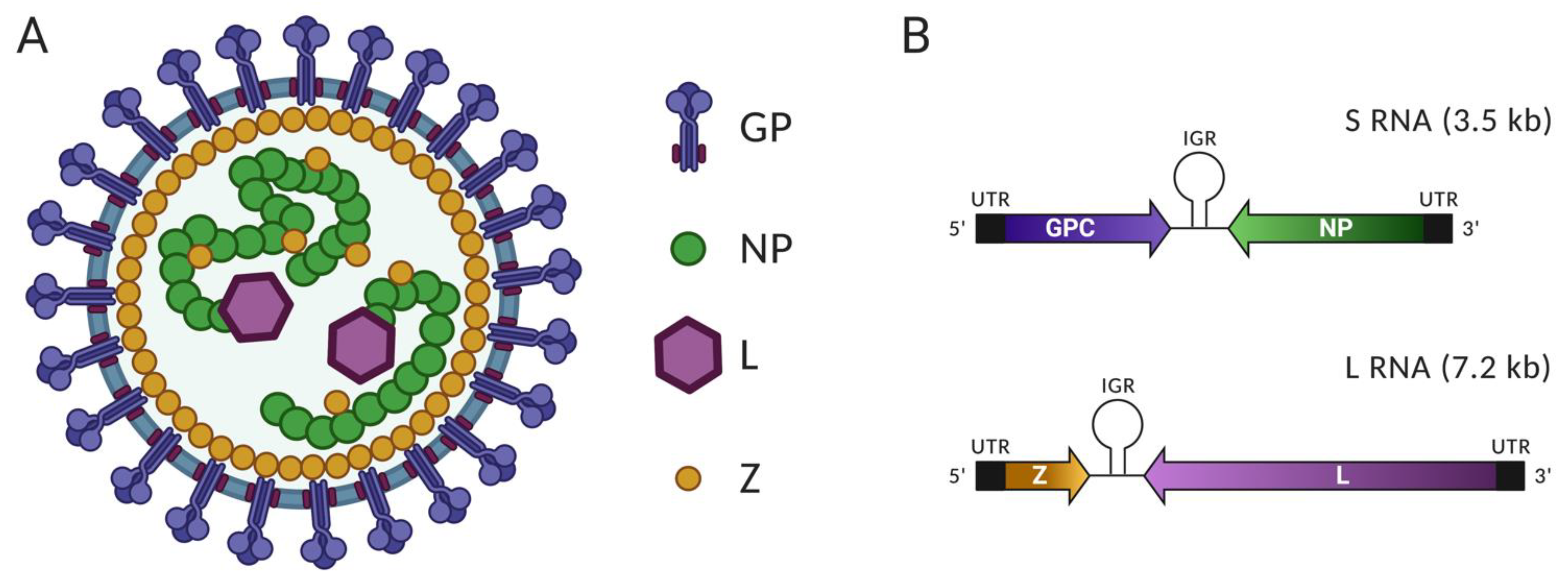
2.1. Virus Particle, Viral Proteins and Genomic Organization
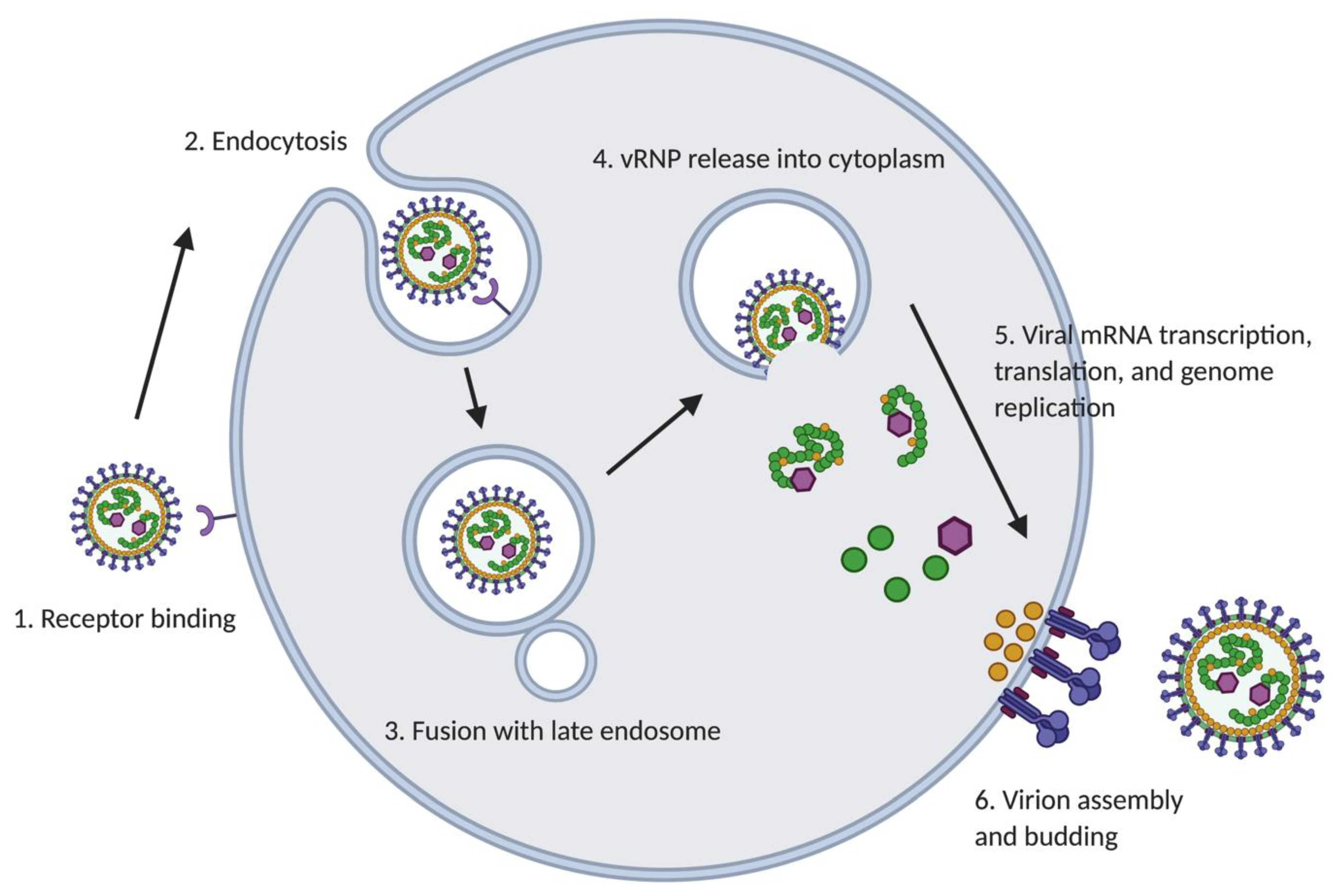
2.2. Mammarenavirus Life Cycle
2.2.1. Cell Entry
2.2.2. Genome Replication and Transcription
2.2.3. Assembly and Budding
3. Drugs Targeting Different Steps of Mammarenavirus Life Cycle
3.1. Cell Entry
3.2. Viral Genome Replication
3.3. Processing of GPC
3.4. Virion Assembly and Cell Egress
3.5. Monoclonal Antibodies
3.6. Targeting Host Factors
4. High-Throughput Screening for Discovery of Anti-Mammarenaviral Drugs
5. Drug Repurposing Strategy
6. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Radoshitzky, S.R.; Buchmeier, M.J.; Charrel, R.N.; Clegg, J.C.S.; Gonzalez, J.J.; Gunther, S.; Hepojoki, J.; Kuhn, J.H.; Lukashevich, I.S.; Romanowski, V.; et al. ICTV Virus Taxonomy Profile: Arenaviridae. J. Gen. Virol. 2019, 100, 1200–1201. [Google Scholar] [CrossRef]
- Buchmeier, M.J.; Peters, C.J.; de la Torre, J.C. Arenaviridae: The viruses and their replication. In Field’s Virology, 5th ed.; Knipe, D.M., Holey, P.M., Eds.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2007; Volume 2, pp. 1791–1851. [Google Scholar]
- Brisse, M.E.; Ly, H. Hemorrhagic Fever-Causing Arenaviruses: Lethal Pathogens and Potent Immune Suppressors. Front. Immunol. 2019, 10, 372. [Google Scholar] [CrossRef] [Green Version]
- Shao, J.; Liang, Y.; Ly, H. Human hemorrhagic Fever causing arenaviruses: Molecular mechanisms contributing to virus virulence and disease pathogenesis. Pathogens 2015, 4, 283–306. [Google Scholar] [CrossRef]
- Schieffelin, J. Treatment of Arenavirus Infections. Curr. Treat. Options Infect. Dis. 2015, 7, 261–270. [Google Scholar] [CrossRef]
- McKee, K.T., Jr.; Oro, J.G.; Kuehne, A.I.; Spisso, J.A.; Mahlandt, B.G. Candid No. 1 Argentine hemorrhagic fever vaccine protects against lethal Junin virus challenge in rhesus macaques. Intervirology 1992, 34, 154–163. [Google Scholar] [CrossRef]
- Maiztegui, J.I.; McKee, K.T., Jr.; Barrera Oro, J.G.; Harrison, L.H.; Gibbs, P.H.; Feuillade, M.R.; Enria, D.A.; Briggiler, A.M.; Levis, S.C.; Ambrosio, A.M.; et al. Protective efficacy of a live attenuated vaccine against Argentine hemorrhagic fever. AHF Study Group. J. Infect. Dis. 1998, 177, 277–283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grant, A.; Seregin, A.; Huang, C.; Kolokoltsova, O.; Brasier, A.; Peters, C.; Paessler, S. Junin virus pathogenesis and virus replication. Viruses 2012, 4, 2317–2339. [Google Scholar] [CrossRef] [Green Version]
- Neuman, B.W.; Adair, B.D.; Burns, J.W.; Milligan, R.A.; Buchmeier, M.J.; Yeager, M. Complementarity in the supramolecular design of arenaviruses and retroviruses revealed by electron cryomicroscopy and image analysis. J. Virol. 2005, 79, 3822–3830. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, S.; Cerny, A.M.; Zacharia, A.; Fitzgerald, K.A.; Kurt-Jones, E.A.; Finberg, R.W. Induction and inhibition of type I interferon responses by distinct components of lymphocytic choriomeningitis virus. J. Virol. 2010, 84, 9452–9462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferron, F.; Weber, F.; de la Torre, J.C.; Reguera, J. Transcription and replication mechanisms of Bunyaviridae and Arenaviridae L proteins. Virus Res. 2017, 234, 118–134. [Google Scholar] [CrossRef] [PubMed]
- Shao, J.; Liang, Y.; Ly, H. Roles of Arenavirus Z Protein in Mediating Virion Budding, Viral Transcription-Inhibition and Interferon-Beta Suppression. Methods Mol. Biol. 2018, 1604, 217–227. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Sun, Z.; Pryce, R.; Parsy, M.L.; Fehling, S.K.; Schlie, K.; Siebert, C.A.; Garten, W.; Bowden, T.A.; Strecker, T.; et al. Acidic pH-Induced Conformations and LAMP1 Binding of the Lassa Virus Glycoprotein Spike. PLoS Pathog. 2016, 12, e1005418. [Google Scholar] [CrossRef] [Green Version]
- Fehling, S.K.; Lennartz, F.; Strecker, T. Multifunctional nature of the arenavirus RING finger protein Z. Viruses 2012, 4, 2973–3011. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hastie, K.M.; Kimberlin, C.R.; Zandonatti, M.A.; MacRae, I.J.; Saphire, E.O. Structure of the Lassa virus nucleoprotein reveals a dsRNA-specific 3’ to 5’ exonuclease activity essential for immune suppression. Proc. Natl. Acad. Sci. USA 2011, 108, 2396–2401. [Google Scholar] [CrossRef] [Green Version]
- Martinez-Sobrido, L.; Zuniga, E.I.; Rosario, D.; Garcia-Sastre, A.; de la Torre, J.C. Inhibition of the type I interferon response by the nucleoprotein of the prototypic arenavirus lymphocytic choriomeningitis virus. J. Virol. 2006, 80, 9192–9199. [Google Scholar] [CrossRef] [Green Version]
- Lenz, O.; ter Meulen, J.; Klenk, H.D.; Seidah, N.G.; Garten, W. The Lassa virus glycoprotein precursor GP-C is proteolytically processed by subtilase SKI-1/S1P. Proc. Natl. Acad. Sci. USA 2001, 98, 12701–12705. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eschli, B.; Quirin, K.; Wepf, A.; Weber, J.; Zinkernagel, R.; Hengartner, H. Identification of an N-terminal trimeric coiled-coil core within arenavirus glycoprotein 2 permits assignment to class I viral fusion proteins. J. Virol. 2006, 80, 5897–5907. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kunz, S.; Edelmann, K.H.; de la Torre, J.C.; Gorney, R.; Oldstone, M.B. Mechanisms for lymphocytic choriomeningitis virus glycoprotein cleavage, transport, and incorporation into virions. Virology 2003, 314, 168–178. [Google Scholar] [CrossRef] [Green Version]
- Di Simone, C.; Zandonatti, M.A.; Buchmeier, M.J. Acidic pH triggers LCMV membrane fusion activity and conformational change in the glycoprotein spike. Virology 1994, 198, 455–465. [Google Scholar] [CrossRef]
- Igonet, S.; Vaney, M.C.; Vonrhein, C.; Bricogne, G.; Stura, E.A.; Hengartner, H.; Eschli, B.; Rey, F.A. X-ray structure of the arenavirus glycoprotein GP2 in its postfusion hairpin conformation. Proc. Natl. Acad. Sci. USA 2011, 108, 19967–19972. [Google Scholar] [CrossRef] [Green Version]
- Eichler, R.; Lenz, O.; Garten, W.; Strecker, T. The role of single N-glycans in proteolytic processing and cell surface transport of the Lassa virus glycoprotein GP-C. Virol. J. 2006, 3, 41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- York, J.; Romanowski, V.; Lu, M.; Nunberg, J.H. The signal peptide of the Junin arenavirus envelope glycoprotein is myristoylated and forms an essential subunit of the mature G1-G2 complex. J. Virol. 2004, 78, 10783–10792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ortin, J.; Martin-Benito, J. The RNA synthesis machinery of negative-stranded RNA viruses. Virology 2015, 479–480, 532–544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perez, M.; Craven, R.C.; de la Torre, J.C. The small RING finger protein Z drives arenavirus budding: Implications for antiviral strategies. Proc. Natl. Acad. Sci. USA 2003, 100, 12978–12983. [Google Scholar] [CrossRef] [Green Version]
- Campbell Dwyer, E.J.; Lai, H.; MacDonald, R.C.; Salvato, M.S.; Borden, K.L. The lymphocytic choriomeningitis virus RING protein Z associates with eukaryotic initiation factor 4E and selectively represses translation in a RING-dependent manner. J. Virol. 2000, 74, 3293–3300. [Google Scholar] [CrossRef] [Green Version]
- Volpon, L.; Osborne, M.J.; Capul, A.A.; de la Torre, J.C.; Borden, K.L. Structural characterization of the Z RING-eIF4E complex reveals a distinct mode of control for eIF4E. Proc. Natl. Acad. Sci. USA 2010, 107, 5441–5446. [Google Scholar] [CrossRef] [Green Version]
- Borden, K.L.; Campbell Dwyer, E.J.; Salvato, M.S. An arenavirus RING (zinc-binding) protein binds the oncoprotein promyelocyte leukemia protein (PML) and relocates PML nuclear bodies to the cytoplasm. J. Virol. 1998, 72, 758–766. [Google Scholar] [CrossRef] [Green Version]
- Djavani, M.; Rodas, J.; Lukashevich, I.S.; Horejsh, D.; Pandolfi, P.P.; Borden, K.L.; Salvato, M.S. Role of the promyelocytic leukemia protein PML in the interferon sensitivity of lymphocytic choriomeningitis virus. J. Virol. 2001, 75, 6204–6208. [Google Scholar] [CrossRef] [Green Version]
- Urata, S.; Yasuda, J. Molecular mechanism of arenavirus assembly and budding. Viruses 2012, 4, 2049–2079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kranzusch, P.J.; Whelan, S.P. Arenavirus Z protein controls viral RNA synthesis by locking a polymerase-promoter complex. Proc. Natl. Acad. Sci. USA 2011, 108, 19743–19748. [Google Scholar] [CrossRef] [Green Version]
- Cornu, T.I.; de la Torre, J.C. RING finger Z protein of lymphocytic choriomeningitis virus (LCMV) inhibits transcription and RNA replication of an LCMV S-segment minigenome. J. Virol. 2001, 75, 9415–9426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Capul, A.A.; Perez, M.; Burke, E.; Kunz, S.; Buchmeier, M.J.; de la Torre, J.C. Arenavirus Z-glycoprotein association requires Z myristoylation but not functional RING or late domains. J. Virol. 2007, 81, 9451–9460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ortiz-Riano, E.; Cheng, B.Y.; de la Torre, J.C.; Martinez-Sobrido, L. The C-terminal region of lymphocytic choriomeningitis virus nucleoprotein contains distinct and segregable functional domains involved in NP-Z interaction and counteraction of the type I interferon response. J. Virol. 2011, 85, 13038–13048. [Google Scholar] [CrossRef] [Green Version]
- Strecker, T.; Eichler, R.; Meulen, J.; Weissenhorn, W.; Dieter Klenk, H.; Garten, W.; Lenz, O. Lassa virus Z protein is a matrix protein and sufficient for the release of virus-like particles [corrected]. J. Virol. 2003, 77, 10700–10705. [Google Scholar] [CrossRef] [Green Version]
- Martinez-Sobrido, L.; de la Torre, J.C. Reporter-Expressing, Replicating-Competent Recombinant Arenaviruses. Viruses 2016, 8. [Google Scholar] [CrossRef]
- Perez, M.; de la Torre, J.C. Characterization of the genomic promoter of the prototypic arenavirus lymphocytic choriomeningitis virus. J. Virol. 2003, 77, 1184–1194. [Google Scholar] [CrossRef] [Green Version]
- Pinschewer, D.D.; Perez, M.; de la Torre, J.C. Dual role of the lymphocytic choriomeningitis virus intergenic region in transcription termination and virus propagation. J. Virol. 2005, 79, 4519–4526. [Google Scholar] [CrossRef] [Green Version]
- Iwasaki, M.; Cubitt, B.; Sullivan, B.M.; de la Torre, J.C. The High Degree of Sequence Plasticity of the Arenavirus Noncoding Intergenic Region (IGR) Enables the Use of a Nonviral Universal Synthetic IGR To Attenuate Arenaviruses. J. Virol. 2016, 90, 3187–3197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spiropoulou, C.F.; Kunz, S.; Rollin, P.E.; Campbell, K.P.; Oldstone, M.B. New World arenavirus clade C, but not clade A and B viruses, utilizes alpha-dystroglycan as its major receptor. J. Virol. 2002, 76, 5140–5146. [Google Scholar] [CrossRef] [Green Version]
- Cao, W.; Henry, M.D.; Borrow, P.; Yamada, H.; Elder, J.H.; Ravkov, E.V.; Nichol, S.T.; Compans, R.W.; Campbell, K.P.; Oldstone, M.B. Identification of alpha-dystroglycan as a receptor for lymphocytic choriomeningitis virus and Lassa fever virus. Science 1998, 282, 2079–2081. [Google Scholar] [CrossRef] [Green Version]
- Radoshitzky, S.R.; Abraham, J.; Spiropoulou, C.F.; Kuhn, J.H.; Nguyen, D.; Li, W.; Nagel, J.; Schmidt, P.J.; Nunberg, J.H.; Andrews, N.C.; et al. Transferrin receptor 1 is a cellular receptor for New World haemorrhagic fever arenaviruses. Nature 2007, 446, 92–96. [Google Scholar] [CrossRef]
- Hallam, S.J.; Koma, T.; Maruyama, J.; Paessler, S. Review of Mammarenavirus Biology and Replication. Front. Microbiol. 2018, 9, 1751. [Google Scholar] [CrossRef]
- Raaben, M.; Jae, L.T.; Herbert, A.S.; Kuehne, A.I.; Stubbs, S.H.; Chou, Y.Y.; Blomen, V.A.; Kirchhausen, T.; Dye, J.M.; Brummelkamp, T.R.; et al. NRP2 and CD63 Are Host Factors for Lujo Virus Cell Entry. Cell Host Microbe 2017, 22, 688–696 e685. [Google Scholar] [CrossRef] [Green Version]
- Bielenberg, D.R.; Pettaway, C.A.; Takashima, S.; Klagsbrun, M. Neuropilins in neoplasms: Expression, regulation, and function. Exp. Cell Res. 2006, 312, 584–593. [Google Scholar] [CrossRef] [PubMed]
- Aung, N.Y.; Ohe, R.; Meng, H.; Kabasawa, T.; Yang, S.; Kato, T.; Yamakawa, M. Specific Neuropilins Expression in Alveolar Macrophages among Tissue-Specific Macrophages. PLoS ONE 2016, 11, e0147358. [Google Scholar] [CrossRef] [PubMed]
- Immormino, R.M.; Lauzier, D.C.; Nakano, H.; Hernandez, M.L.; Alexis, N.E.; Ghio, A.J.; Tilley, S.L.; Doerschuk, C.M.; Peden, D.B.; Cook, D.N.; et al. Neuropilin-2 regulates airway inflammatory responses to inhaled lipopolysaccharide. Am. J. Physiol. Lung Cell Mol. Physiol. 2018, 315, L202–L211. [Google Scholar] [CrossRef]
- Rojek, J.M.; Sanchez, A.B.; Nguyen, N.T.; de la Torre, J.C.; Kunz, S. Different mechanisms of cell entry by human-pathogenic Old World and New World arenaviruses. J. Virol. 2008, 82, 7677–7687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rojek, J.M.; Perez, M.; Kunz, S. Cellular entry of lymphocytic choriomeningitis virus. J. Virol. 2008, 82, 1505–1517. [Google Scholar] [CrossRef] [Green Version]
- Pasqual, G.; Rojek, J.M.; Masin, M.; Chatton, J.Y.; Kunz, S. Old world arenaviruses enter the host cell via the multivesicular body and depend on the endosomal sorting complex required for transport. PLoS Pathog. 2011, 7, e1002232. [Google Scholar] [CrossRef]
- Jae, L.T.; Raaben, M.; Herbert, A.S.; Kuehne, A.I.; Wirchnianski, A.S.; Soh, T.K.; Stubbs, S.H.; Janssen, H.; Damme, M.; Saftig, P.; et al. Virus entry. Lassa virus entry requires a trigger-induced receptor switch. Science 2014, 344, 1506–1510. [Google Scholar] [CrossRef] [Green Version]
- Di Simone, C.; Buchmeier, M.J. Kinetics and pH dependence of acid-induced structural changes in the lymphocytic choriomeningitis virus glycoprotein complex. Virology 1995, 209, 3–9. [Google Scholar] [CrossRef] [Green Version]
- Lopez, N.; Jacamo, R.; Franze-Fernandez, M.T. Transcription and RNA replication of tacaribe virus genome and antigenome analogs require N and L proteins: Z protein is an inhibitor of these processes. J. Virol. 2001, 75, 12241–12251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, K.J.; Novella, I.S.; Teng, M.N.; Oldstone, M.B.; de La Torre, J.C. NP and L proteins of lymphocytic choriomeningitis virus (LCMV) are sufficient for efficient transcription and replication of LCMV genomic RNA analogs. J. Virol. 2000, 74, 3470–3477. [Google Scholar] [CrossRef] [Green Version]
- King, B.R.; Samacoits, A.; Eisenhauer, P.L.; Ziegler, C.M.; Bruce, E.A.; Zenklusen, D.; Zimmer, C.; Mueller, F.; Botten, J. Visualization of Arenavirus RNA Species in Individual Cells by Single-Molecule Fluorescence In Situ Hybridization Suggests a Model of Cyclical Infection and Clearance during Persistence. J. Virol. 2018, 92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hass, M.; Golnitz, U.; Muller, S.; Becker-Ziaja, B.; Gunther, S. Replicon system for Lassa virus. J. Virol. 2004, 78, 13793–13803. [Google Scholar] [CrossRef] [Green Version]
- Lopez, N.; Franze-Fernandez, M.T. A single stem-loop structure in Tacaribe arenavirus intergenic region is essential for transcription termination but is not required for a correct initiation of transcription and replication. Virus Res. 2007, 124, 237–244. [Google Scholar] [CrossRef]
- Raju, R.; Raju, L.; Hacker, D.; Garcin, D.; Compans, R.; Kolakofsky, D. Nontemplated bases at the 5’ ends of Tacaribe virus mRNAs. Virology 1990, 174, 53–59. [Google Scholar] [CrossRef]
- Lelke, M.; Brunotte, L.; Busch, C.; Gunther, S. An N-terminal region of Lassa virus L protein plays a critical role in transcription but not replication of the virus genome. J. Virol. 2010, 84, 1934–1944. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vogel, D.; Rosenthal, M.; Gogrefe, N.; Reindl, S.; Gunther, S. Biochemical characterization of the Lassa virus L protein. J. Biol. Chem. 2019, 294, 8088–8100. [Google Scholar] [CrossRef] [Green Version]
- Emonet, S.E.; Urata, S.; de la Torre, J.C. Arenavirus reverse genetics: New approaches for the investigation of arenavirus biology and development of antiviral strategies. Virology 2011, 411, 416–425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marq, J.B.; Hausmann, S.; Veillard, N.; Kolakofsky, D.; Garcin, D. Short double-stranded RNAs with an overhanging 5’ ppp-nucleotide, as found in arenavirus genomes, act as RIG-I decoys. J. Biol. Chem. 2011, 286, 6108–6116. [Google Scholar] [CrossRef] [Green Version]
- Linero, F.; Welnowska, E.; Carrasco, L.; Scolaro, L. Participation of eIF4F complex in Junin virus infection: Blockage of eIF4E does not impair virus replication. Cell Microbiol. 2013, 15, 1766–1782. [Google Scholar] [CrossRef]
- Bieniasz, P.D. Late budding domains and host proteins in enveloped virus release. Virology 2006, 344, 55–63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shtanko, O.; Imai, M.; Goto, H.; Lukashevich, I.S.; Neumann, G.; Watanabe, T.; Kawaoka, Y. A role for the C terminus of Mopeia virus nucleoprotein in its incorporation into Z protein-induced virus-like particles. J. Virol. 2010, 84, 5415–5422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Urata, S.; Noda, T.; Kawaoka, Y.; Yokosawa, H.; Yasuda, J. Cellular factors required for Lassa virus budding. J. Virol. 2006, 80, 4191–4195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Votteler, J.; Sundquist, W.I. Virus budding and the ESCRT pathway. Cell Host Microbe. 2013, 14, 232–241. [Google Scholar] [CrossRef] [Green Version]
- Meng, B.; Lever, A.M.L. The Interplay between ESCRT and Viral Factors in the Enveloped Virus Life Cycle. Viruses 2021, 13, 324. [Google Scholar] [CrossRef] [PubMed]
- Bolken, T.C.; Laquerre, S.; Zhang, Y.; Bailey, T.R.; Pevear, D.C.; Kickner, S.S.; Sperzel, L.E.; Jones, K.F.; Warren, T.K.; Amanda Lund, S.; et al. Identification and characterization of potent small molecule inhibitor of hemorrhagic fever New World arenaviruses. Antivir. Res. 2006, 69, 86–97. [Google Scholar] [CrossRef]
- Larson, R.A.; Dai, D.; Hosack, V.T.; Tan, Y.; Bolken, T.C.; Hruby, D.E.; Amberg, S.M. Identification of a broad-spectrum arenavirus entry inhibitor. J. Virol. 2008, 82, 10768–10775. [Google Scholar] [CrossRef] [Green Version]
- Cashman, K.A.; Smith, M.A.; Twenhafel, N.A.; Larson, R.A.; Jones, K.F.; Allen, R.D., III; Dai, D.; Chinsangaram, J.; Bolken, T.C.; Hruby, D.E.; et al. Evaluation of Lassa antiviral compound ST-193 in a guinea pig model. Antivir. Res. 2011, 90, 70–79. [Google Scholar] [CrossRef]
- Madu, I.G.; Files, M.; Gharaibeh, D.N.; Moore, A.L.; Jung, K.H.; Gowen, B.B.; Dai, D.; Jones, K.F.; Tyavanagimatt, S.R.; Burgeson, J.R.; et al. A potent Lassa virus antiviral targets an arenavirus virulence determinant. PLoS Pathog. 2018, 14, e1007439. [Google Scholar] [CrossRef] [Green Version]
- Ngo, N.; Henthorn, K.S.; Cisneros, M.I.; Cubitt, B.; Iwasaki, M.; de la Torre, J.C.; Lama, J. Identification and Mechanism of Action of a Novel Small-Molecule Inhibitor of Arenavirus Multiplication. J. Virol. 2015, 89, 10924–10933. [Google Scholar] [CrossRef] [Green Version]
- Spence, J.S.; Melnik, L.I.; Badani, H.; Wimley, W.C.; Garry, R.F. Inhibition of arenavirus infection by a glycoprotein-derived peptide with a novel mechanism. J. Virol. 2014, 88, 8556–8564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blaising, J.; Polyak, S.J.; Pecheur, E.I. Arbidol as a broad-spectrum antiviral: An update. Antivir. Res. 2014, 107, 84–94. [Google Scholar] [CrossRef] [PubMed]
- Pecheur, E.I.; Borisevich, V.; Halfmann, P.; Morrey, J.D.; Smee, D.F.; Prichard, M.; Mire, C.E.; Kawaoka, Y.; Geisbert, T.W.; Polyak, S.J. The Synthetic Antiviral Drug Arbidol Inhibits Globally Prevalent Pathogenic Viruses. J. Virol. 2016, 90, 3086–3092. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hulseberg, C.E.; Feneant, L.; Szymanska-de Wijs, K.M.; Kessler, N.P.; Nelson, E.A.; Shoemaker, C.J.; Schmaljohn, C.S.; Polyak, S.J.; White, J.M. Arbidol and Other Low-Molecular-Weight Drugs That Inhibit Lassa and Ebola Viruses. J. Virol. 2019, 93. [Google Scholar] [CrossRef] [Green Version]
- Tang, K.; He, S.; Zhang, X.; Guo, J.; Chen, Q.; Yan, F.; Banadyga, L.; Zhu, W.; Qiu, X.; Guo, Y. Tangeretin, an extract from Citrus peels, blocks cellular entry of arenaviruses that cause viral hemorrhagic fever. Antivir. Res. 2018, 160, 87–93. [Google Scholar] [CrossRef] [PubMed]
- Torriani, G.; Trofimenko, E.; Mayor, J.; Fedeli, C.; Moreno, H.; Michel, S.; Heulot, M.; Chevalier, N.; Zimmer, G.; Shrestha, N.; et al. Identification of Clotrimazole Derivatives as Specific Inhibitors of Arenavirus Fusion. J. Virol. 2019, 93. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Yan, F.; Tang, K.; Chen, Q.; Guo, J.; Zhu, W.; He, S.; Banadyga, L.; Qiu, X.; Guo, Y. Identification of a clinical compound losmapimod that blocks Lassa virus entry. Antivir. Res. 2019, 167, 68–77. [Google Scholar] [CrossRef] [PubMed]
- Moeschler, S.; Locher, S.; Zimmer, G. 1-Benzyl-3-cetyl-2-methylimidazolium Iodide (NH125) Is a Broad-Spectrum Inhibitor of Virus Entry with Lysosomotropic Features. Viruses 2018, 10. [Google Scholar] [CrossRef] [Green Version]
- Friesland, A.; Zhao, Y.; Chen, Y.H.; Wang, L.; Zhou, H.; Lu, Q. Small molecule targeting Cdc42-intersectin interaction disrupts Golgi organization and suppresses cell motility. Proc. Natl. Acad. Sci. USA 2013, 110, 1261–1266. [Google Scholar] [CrossRef] [Green Version]
- Chou, Y.Y.; Cuevas, C.; Carocci, M.; Stubbs, S.H.; Ma, M.; Cureton, D.K.; Chao, L.; Evesson, F.; He, K.; Yang, P.L.; et al. Identification and Characterization of a Novel Broad-Spectrum Virus Entry Inhibitor. J. Virol. 2016, 90, 4494–4510. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Tang, K.; Guo, Y. The antifungal isavuconazole inhibits the entry of lassa virus by targeting the stable signal peptide-GP2 subunit interface of lassa virus glycoprotein. Antivir. Res. 2020, 174, 104701. [Google Scholar] [CrossRef]
- Wang, M.K.; Ren, T.; Liu, H.; Lim, S.Y.; Lee, K.; Honko, A.; Zhou, H.; Dyall, J.; Hensley, L.; Gartin, A.K.; et al. Critical role for cholesterol in Lassa fever virus entry identified by a novel small molecule inhibitor targeting the viral receptor LAMP1. PLoS Pathog. 2018, 14, e1007322. [Google Scholar] [CrossRef] [PubMed]
- McCormick, J.B.; King, I.J.; Webb, P.A.; Scribner, C.L.; Craven, R.B.; Johnson, K.M.; Elliott, L.H.; Belmont-Williams, R. Lassa fever. Effective therapy with ribavirin. N. Engl. J. Med. 1986, 314, 20–26. [Google Scholar] [CrossRef] [PubMed]
- Oestereich, L.; Rieger, T.; Ludtke, A.; Ruibal, P.; Wurr, S.; Pallasch, E.; Bockholt, S.; Krasemann, S.; Munoz-Fontela, C.; Gunther, S. Efficacy of Favipiravir Alone and in Combination With Ribavirin in a Lethal, Immunocompetent Mouse Model of Lassa Fever. J. Infect. Dis. 2016, 213, 934–938. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feld, J.J.; Hoofnagle, J.H. Mechanism of action of interferon and ribavirin in treatment of hepatitis C. Nature 2005, 436, 967–972. [Google Scholar] [CrossRef]
- Lau, J.Y.; Tam, R.C.; Liang, T.J.; Hong, Z. Mechanism of action of ribavirin in the combination treatment of chronic HCV infection. Hepatology 2002, 35, 1002–1009. [Google Scholar] [CrossRef]
- Carrillo-Bustamante, P.; Nguyen, T.H.T.; Oestereich, L.; Gunther, S.; Guedj, J.; Graw, F. Determining Ribavirin’s mechanism of action against Lassa virus infection. Sci. Rep. 2017, 7, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Furuta, Y.; Komeno, T.; Nakamura, T. Favipiravir (T-705), a broad spectrum inhibitor of viral RNA polymerase. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2017, 93, 449–463. [Google Scholar] [CrossRef] [Green Version]
- Furuta, Y.; Gowen, B.B.; Takahashi, K.; Shiraki, K.; Smee, D.F.; Barnard, D.L. Favipiravir (T-705), a novel viral RNA polymerase inhibitor. Antivir. Res. 2013, 100, 446–454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Warren, T.K.; Wells, J.; Panchal, R.G.; Stuthman, K.S.; Garza, N.L.; Van Tongeren, S.A.; Dong, L.; Retterer, C.J.; Eaton, B.P.; Pegoraro, G.; et al. Protection against filovirus diseases by a novel broad-spectrum nucleoside analogue BCX4430. Nature 2014, 508, 402–405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neuman, B.W.; Bederka, L.H.; Stein, D.A.; Ting, J.P.; Moulton, H.M.; Buchmeier, M.J. Development of peptide-conjugated morpholino oligomers as pan-arenavirus inhibitors. Antimicrob. Agents Chemother. 2011, 55, 4631–4638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, K.W.; Hsu, K.C.; Chu, L.Y.; Yang, J.M.; Yuan, H.S.; Hsiao, Y.Y. Identification of Inhibitors for the DEDDh Family of Exonucleases and a Unique Inhibition Mechanism by Crystal Structure Analysis of CRN-4 Bound with 2-Morpholin-4-ylethanesulfonate (MES). J. Med. Chem. 2016, 59, 8019–8029. [Google Scholar] [CrossRef] [Green Version]
- Saez-Ayala, M.; Laban Yekwa, E.; Mondielli, C.; Roux, L.; Hernandez, S.; Bailly, F.; Cotelle, P.; Rogolino, D.; Canard, B.; Ferron, F.; et al. Metal chelators for the inhibition of the lymphocytic choriomeningitis virus endonuclease domain. Antivir. Res. 2019, 162, 79–89. [Google Scholar] [CrossRef] [PubMed]
- Sepulveda, C.S.; Garcia, C.C.; Damonte, E.B. Inhibition of arenavirus infection by thiuram and aromatic disulfides. Antivir. Res. 2010, 87, 329–337. [Google Scholar] [CrossRef]
- Sepulveda, C.S.; Garcia, C.C.; Levingston Macleod, J.M.; Lopez, N.; Damonte, E.B. Targeting of arenavirus RNA synthesis by a carboxamide-derivatized aromatic disulfide with virucidal activity. PLoS ONE 2013, 8, e81251. [Google Scholar] [CrossRef] [PubMed]
- Pasquato, A.; Cendron, L.; Kunz, S. Cleavage of the Glycoprotein of Arenaviruses. In Activation of Viruses by Host Proteases; Böttcher-Friebertshäuser, E., Garten, W., Klenk, H.D., Eds.; Springer International Publishing: Cham, Germany, 2018; pp. 47–70. [Google Scholar]
- Pasquato, A.; Pullikotil, P.; Asselin, M.C.; Vacatello, M.; Paolillo, L.; Ghezzo, F.; Basso, F.; Di Bello, C.; Dettin, M.; Seidah, N.G. The proprotein convertase SKI-1/S1P. In vitro analysis of Lassa virus glycoprotein-derived substrates and ex vivo validation of irreversible peptide inhibitors. J. Biol. Chem. 2006, 281, 23471–23481. [Google Scholar] [CrossRef] [Green Version]
- Rojek, J.M.; Pasqual, G.; Sanchez, A.B.; Nguyen, N.T.; de la Torre, J.C.; Kunz, S. Targeting the proteolytic processing of the viral glycoprotein precursor is a promising novel antiviral strategy against arenaviruses. J. Virol. 2010, 84, 573–584. [Google Scholar] [CrossRef] [Green Version]
- Urata, S.; Yun, N.; Pasquato, A.; Paessler, S.; Kunz, S.; de la Torre, J.C. Antiviral activity of a small-molecule inhibitor of arenavirus glycoprotein processing by the cellular site 1 protease. J. Virol. 2011, 85, 795–803. [Google Scholar] [CrossRef] [Green Version]
- Pasquato, A.; Rochat, C.; Burri, D.J.; Pasqual, G.; de la Torre, J.C.; Kunz, S. Evaluation of the anti-arenaviral activity of the subtilisin kexin isozyme-1/site-1 protease inhibitor PF-429242. Virology 2012, 423, 14–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Urata, S.; de la Torre, J.C. Arenavirus budding. Adv. Virol. 2011, 2011, 180326. [Google Scholar] [CrossRef] [Green Version]
- Perez, M.; Greenwald, D.L.; de la Torre, J.C. Myristoylation of the RING finger Z protein is essential for arenavirus budding. J. Virol. 2004, 78, 11443–11448. [Google Scholar] [CrossRef] [Green Version]
- Russo, R.; Kemp, M.; Bhatti, U.F.; Pai, M.; Wakam, G.; Biesterveld, B.; Alam, H.B. Life on the battlefield: Valproic acid for combat applications. J. Trauma Acute Care Surg. 2020, 89, S69–S76. [Google Scholar] [CrossRef]
- Vazquez-Calvo, A.; Martin-Acebes, M.A.; Saiz, J.C.; Ngo, N.; Sobrino, F.; de la Torre, J.C. Inhibition of multiplication of the prototypic arenavirus LCMV by valproic acid. Antivir. Res. 2013, 99, 172–179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, J.; Han, Z.; Liu, Y.; Liu, W.; Lee, M.S.; Olson, M.A.; Ruthel, G.; Freedman, B.D.; Harty, R.N. A host-oriented inhibitor of Junin Argentine hemorrhagic fever virus egress. J. Virol. 2014, 88, 4736–4743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yasuda, J.; Hunter, E.; Nakao, M.; Shida, H. Functional involvement of a novel Nedd4-like ubiquitin ligase on retrovirus budding. EMBO Rep. 2002, 3, 636–640. [Google Scholar] [CrossRef] [Green Version]
- Han, Z.; Lu, J.; Liu, Y.; Davis, B.; Lee, M.S.; Olson, M.A.; Ruthel, G.; Freedman, B.D.; Schnell, M.J.; Wrobel, J.E.; et al. Small-molecule probes targeting the viral PPxY-host Nedd4 interface block egress of a broad range of RNA viruses. J. Virol. 2014, 88, 7294–7306. [Google Scholar] [CrossRef] [Green Version]
- Urata, S.; Ngo, N.; de la Torre, J.C. The PI3K/Akt pathway contributes to arenavirus budding. J. Virol. 2012, 86, 4578–4585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xing, J.; Ly, H.; Liang, Y. The Z proteins of pathogenic but not nonpathogenic arenaviruses inhibit RIG-I-like receptor-dependent interferon production. J. Virol. 2015, 89, 2944–2955. [Google Scholar] [CrossRef] [Green Version]
- Ziegler, C.M.; Eisenhauer, P.; Kelly, J.A.; Dang, L.N.; Beganovic, V.; Bruce, E.A.; King, B.R.; Shirley, D.J.; Weir, M.E.; Ballif, B.A.; et al. A Proteomics Survey of Junin Virus Interactions with Human Proteins Reveals Host Factors Required for Arenavirus Replication. J. Virol. 2018, 92. [Google Scholar] [CrossRef] [Green Version]
- Helguera, G.; Jemielity, S.; Abraham, J.; Cordo, S.M.; Martinez, M.G.; Rodriguez, J.A.; Bregni, C.; Wang, J.J.; Farzan, M.; Penichet, M.L.; et al. An antibody recognizing the apical domain of human transferrin receptor 1 efficiently inhibits the entry of all new world hemorrhagic Fever arenaviruses. J. Virol. 2012, 86, 4024–4028. [Google Scholar] [CrossRef] [Green Version]
- Cross, R.W.; Mire, C.E.; Branco, L.M.; Geisbert, J.B.; Rowland, M.M.; Heinrich, M.L.; Goba, A.; Momoh, M.; Grant, D.S.; Fullah, M.; et al. Treatment of Lassa virus infection in outbred guinea pigs with first-in-class human monoclonal antibodies. Antivir. Res. 2016, 133, 218–222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amanat, F.; Duehr, J.; Huang, C.; Paessler, S.; Tan, G.S.; Krammer, F. Monoclonal Antibodies with Neutralizing Activity and Fc-Effector Functions against the Machupo Virus Glycoprotein. J. Virol. 2020, 94. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.E.; Hastie, K.M.; Cross, R.W.; Yenni, R.E.; Elliott, D.H.; Rouelle, J.A.; Kannadka, C.B.; Smira, A.A.; Garry, C.E.; Bradley, B.T.; et al. Most neutralizing human monoclonal antibodies target novel epitopes requiring both Lassa virus glycoprotein subunits. Nat. Commun. 2016, 7, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Ortiz-Riano, E.; Ngo, N.; Devito, S.; Eggink, D.; Munger, J.; Shaw, M.L.; de la Torre, J.C.; Martinez-Sobrido, L. Inhibition of arenavirus by A3, a pyrimidine biosynthesis inhibitor. J. Virol. 2014, 88, 878–889. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sepulveda, C.S.; Garcia, C.C.; Damonte, E.B. Antiviral activity of A771726, the active metabolite of leflunomide, against Junin virus. J. Med. Virol. 2018, 90, 819–827. [Google Scholar] [CrossRef] [PubMed]
- Sepulveda, C.S.; Garcia, C.C.; Fascio, M.L.; D’Accorso, N.B.; Docampo Palacios, M.L.; Pellon, R.F.; Damonte, E.B. Inhibition of Junin virus RNA synthesis by an antiviral acridone derivative. Antivir. Res. 2012, 93, 16–22. [Google Scholar] [CrossRef] [PubMed]
- Nair, V.; Chi, G.; Shu, Q.; Julander, J.; Smee, D.F. A heterocyclic molecule with significant activity against dengue virus. Bioorg. Med. Chem. Lett. 2009, 19, 1425–1427. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Smith, A. l-like 3-deazaneplanocin analogues: Synthesis and antiviral properties. Bioorg. Med. Chem. Lett. 2019, 29, 126613. [Google Scholar] [CrossRef]
- Iwasaki, M.; Minder, P.; Cai, Y.; Kuhn, J.H.; Yates, J.R., III; Torbett, B.E.; de la Torre, J.C. Interactome analysis of the lymphocytic choriomeningitis virus nucleoprotein in infected cells reveals ATPase Na+/K+ transporting subunit Alpha 1 and prohibitin as host-cell factors involved in the life cycle of mammarenaviruses. PLoS Pathog. 2018, 14, e1006892. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, A.M.; Rojek, J.M.; Spiropoulou, C.F.; Gundersen, A.T.; Jin, W.; Shaginian, A.; York, J.; Nunberg, J.H.; Boger, D.L.; Oldstone, M.B.; et al. Unique small molecule entry inhibitors of hemorrhagic fever arenaviruses. J. Biol. Chem. 2008, 283, 18734–18742. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.; Liu, Y.; Zhang, G.; Wang, S.; Guo, J.; Cao, J.; Jia, X.; Zhang, L.; Xiao, G.; Wang, W. Screening and Identification of Lassa Virus Entry Inhibitors from an FDA-Approved Drug Library. J. Virol. 2018, 92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lay Mendoza, M.F.; Acciani, M.D.; Levit, C.N.; Santa Maria, C.; Brindley, M.A. Monitoring Viral Entry in Real-Time Using a Luciferase Recombinant Vesicular Stomatitis Virus Producing SARS-CoV-2, EBOV, LASV, CHIKV, and VSV Glycoproteins. Viruses 2020, 12. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Sobrido, L.; Cheng, B.Y.; de la Torre, J.C. Reverse Genetics Approaches to Control Arenavirus. Methods Mol. Biol. 2016, 1403, 313–351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cubitt, B.; Ortiz-Riano, E.; Cheng, B.Y.; Kim, Y.J.; Yeh, C.D.; Chen, C.Z.; Southall, N.O.E.; Zheng, W.; Martinez-Sobrido, L.; de la Torre, J.C. A cell-based, infectious-free, platform to identify inhibitors of lassa virus ribonucleoprotein (vRNP) activity. Antivir. Res. 2020, 173, 104667. [Google Scholar] [CrossRef] [PubMed]
- Wendt, L.; Bostedt, L.; Hoenen, T.; Groseth, A. High-throughput screening for negative-stranded hemorrhagic fever viruses using reverse genetics. Antivir. Res. 2019, 170, 104569. [Google Scholar] [CrossRef] [PubMed]
- Capul, A.A.; de la Torre, J.C. A cell-based luciferase assay amenable to high-throughput screening of inhibitors of arenavirus budding. Virology 2008, 382, 107–114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ashburn, T.T.; Thor, K.B. Drug repositioning: Identifying and developing new uses for existing drugs. Nat. Rev. Drug Discov. 2004, 3, 673–683. [Google Scholar] [CrossRef]
- Mercorelli, B.; Palu, G.; Loregian, A. Drug Repurposing for Viral Infectious Diseases: How Far Are We? Trends Microbiol. 2018, 26, 865–876. [Google Scholar] [CrossRef]
- Pandey, A.; Nikam, A.N.; Shreya, A.B.; Mutalik, S.P.; Gopalan, D.; Kulkarni, S.; Padya, B.S.; Fernandes, G.; Mutalik, S.; Prassl, R. Potential therapeutic targets for combating SARS-CoV-2: Drug repurposing, clinical trials and recent advancements. Life Sci. 2020, 256, 117883. [Google Scholar] [CrossRef] [PubMed]
- Madrid, P.B.; Chopra, S.; Manger, I.D.; Gilfillan, L.; Keepers, T.R.; Shurtleff, A.C.; Green, C.E.; Iyer, L.V.; Dilks, H.H.; Davey, R.A.; et al. A systematic screen of FDA-approved drugs for inhibitors of biological threat agents. PLoS ONE 2013, 8, e60579. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.J.; Cubitt, B.; Chen, E.; Hull, M.V.; Chatterjee, A.K.; Cai, Y.; Kuhn, J.H.; de la Torre, J.C. The ReFRAME library as a comprehensive drug repurposing library to identify mammarenavirus inhibitors. Antivir. Res. 2019, 169, 104558. [Google Scholar] [CrossRef]
- Riva, L.; Yuan, S.; Yin, X.; Martin-Sancho, L.; Matsunaga, N.; Pache, L.; Burgstaller-Muehlbacher, S.; De Jesus, P.D.; Teriete, P.; Hull, M.V.; et al. Discovery of SARS-CoV-2 antiviral drugs through large-scale compound repurposing. Nature 2020, 586, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Oppliger, J.; Torriani, G.; Herrador, A.; Kunz, S. Lassa Virus Cell Entry via Dystroglycan Involves an Unusual Pathway of Macropinocytosis. J. Virol. 2016, 90, 6412–6429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raabe, V.N.; Kann, G.; Ribner, B.S.; Morales, A.; Varkey, J.B.; Mehta, A.K.; Lyon, G.M.; Vanairsdale, S.; Faber, K.; Becker, S.; et al. Favipiravir and Ribavirin Treatment of Epidemiologically Linked Cases of Lassa Fever. Clin. Infect. Dis. 2017, 65, 855–859. [Google Scholar] [CrossRef] [Green Version]
- Herring, S.; Oda, J.M.; Wagoner, J.; Kirchmeier, D.; O’Connor, A.; Nelson, E.A.; Huang, Q.; Liang, Y.; DeWald, L.E.; Johansen, L.M.; et al. Inhibition of Arenaviruses by Combinations of Orally Available Approved Drugs. Antimicrob. Agents Chemother. 2021, 65. [Google Scholar] [CrossRef]
- Sun, X.; Vilar, S.; Tatonetti, N.P. High-throughput methods for combinatorial drug discovery. Sci. Transl. Med. 2013, 5, 205rv1. [Google Scholar] [CrossRef]
| Target | Drug Name | Mechanism |
|---|---|---|
| Viral entry | ST-193 |
|
| ST-294 | ||
| ST-336 | ||
| LHF-535 | ||
| F3406 | ||
| AVP-p | ||
| arbidol | ||
| tangeretin | ||
| TRAM-34 |
| |
| losmapimod |
| |
| NH125 |
| |
| ZCL278 |
| |
| isavuconazole |
| |
| dec-RRLL-CMK |
| |
| PF-429242 | ||
| adamantyl diphenyl piperazine 3.3 |
| |
| Viral genomereplication | ribavirin |
|
| favipiravir |
| |
| BCX4430 | ||
| PPMO |
| |
| ATA |
| |
| PV6R | ||
| diketo acids |
| |
| polyphenols | ||
| N-hydroxyisoquinoline-1,3-diones | ||
| NSC4492 |
| |
| KP-146 | ||
| A3 |
| |
| A771726 | ||
| acridone |
| |
| bredinin | ||
| 3-deazaneplanocin |
| |
| ouabain |
| |
| bufalin | ||
| rocaglamide |
| |
| Virion assembly and budding | 2-hydroxymyristic acid |
|
| valproic acid |
| |
| compound0013 |
| |
| compound1 |
| |
| BEZ-235 |
| |
| KP-146 |
|
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, Y.-J.; Venturini, V.; de la Torre, J.C. Progress in Anti-Mammarenavirus Drug Development. Viruses 2021, 13, 1187. https://doi.org/10.3390/v13071187
Kim Y-J, Venturini V, de la Torre JC. Progress in Anti-Mammarenavirus Drug Development. Viruses. 2021; 13(7):1187. https://doi.org/10.3390/v13071187
Chicago/Turabian StyleKim, Yu-Jin, Victor Venturini, and Juan C. de la Torre. 2021. "Progress in Anti-Mammarenavirus Drug Development" Viruses 13, no. 7: 1187. https://doi.org/10.3390/v13071187
APA StyleKim, Y.-J., Venturini, V., & de la Torre, J. C. (2021). Progress in Anti-Mammarenavirus Drug Development. Viruses, 13(7), 1187. https://doi.org/10.3390/v13071187







