Evidence of Transcriptional Shutoff by Pathogenic Viral Haemorrhagic Septicaemia Virus in Rainbow Trout
Abstract
1. Introduction
2. Materials and Methods
2.1. VHSV Cell Culture and Sampling Regime
2.2. Library Prep and RNA Sequencing
2.3. Sequence Preprocessing
2.4. De Novo Genome Assembly of the VHSV J167 Isolate
2.5. Reference Transcriptome
2.6. Differential Gene Expression Analysis
2.7. Functional Annotation Analysis
2.8. Real-Time RT-PCR Analysis
3. Results
3.1. Transcriptome Sequencing Mapping and Annotation
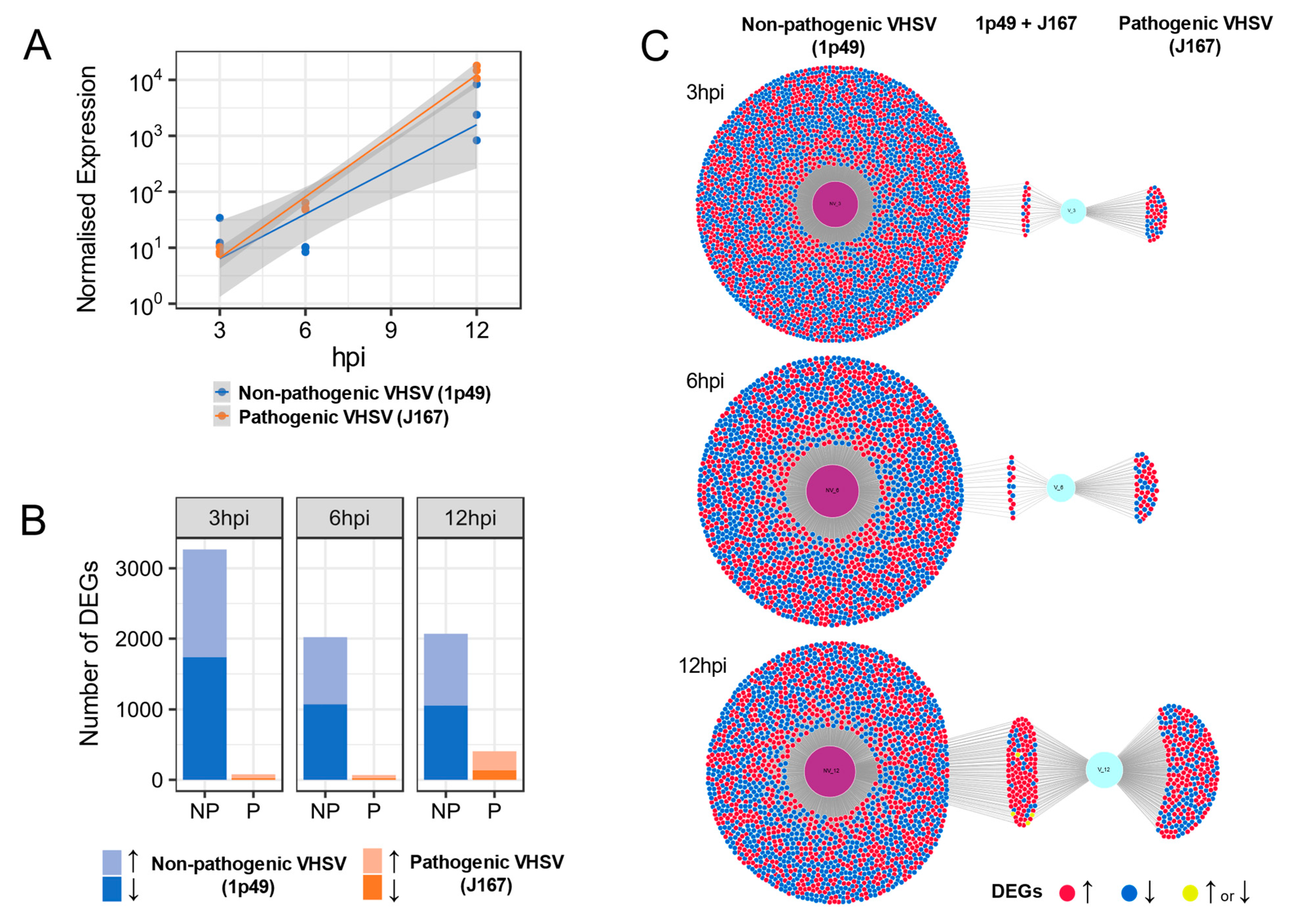
3.2. At Early Times Post Inoculation, Pathogenic and Non-Pathogenic VHSV Isolates Replicate in a Similar Manner in RTG-2 Trout Cells Despite Large Differences in the Host Responses
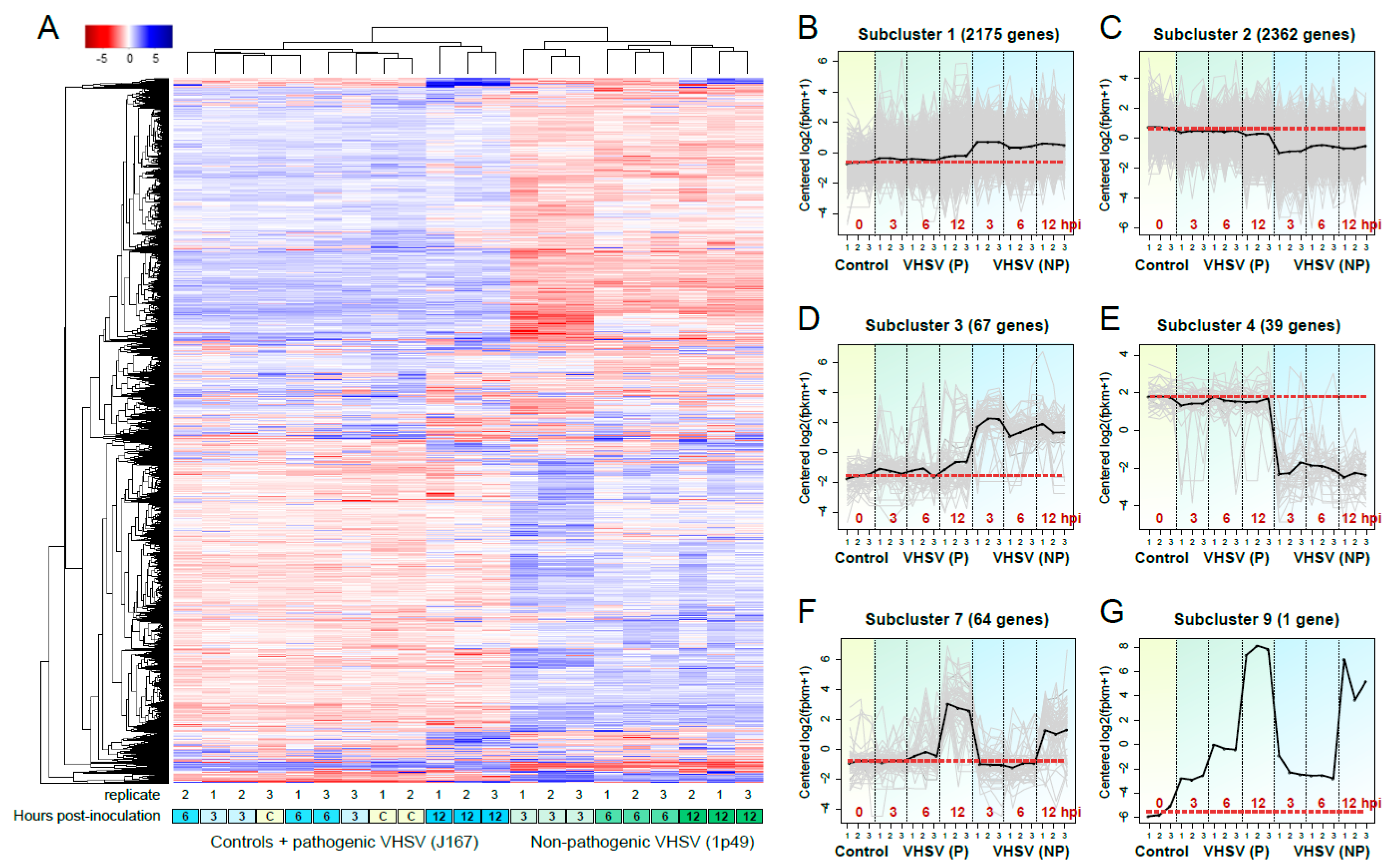
3.3. Hierarchical Clustering Reveals That Pathogen Virulence Is the Main Determinant of Differential Transcription Profiles
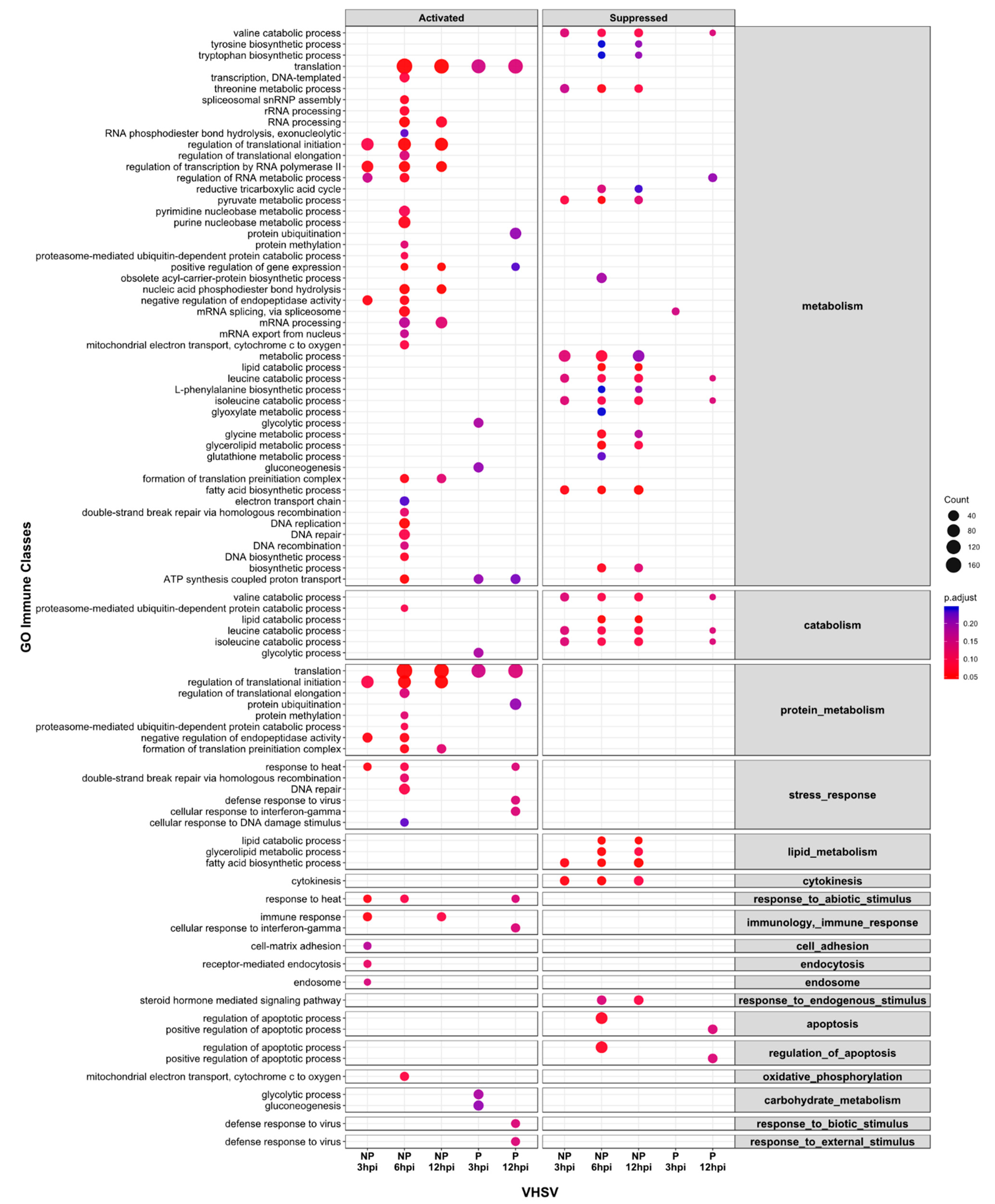
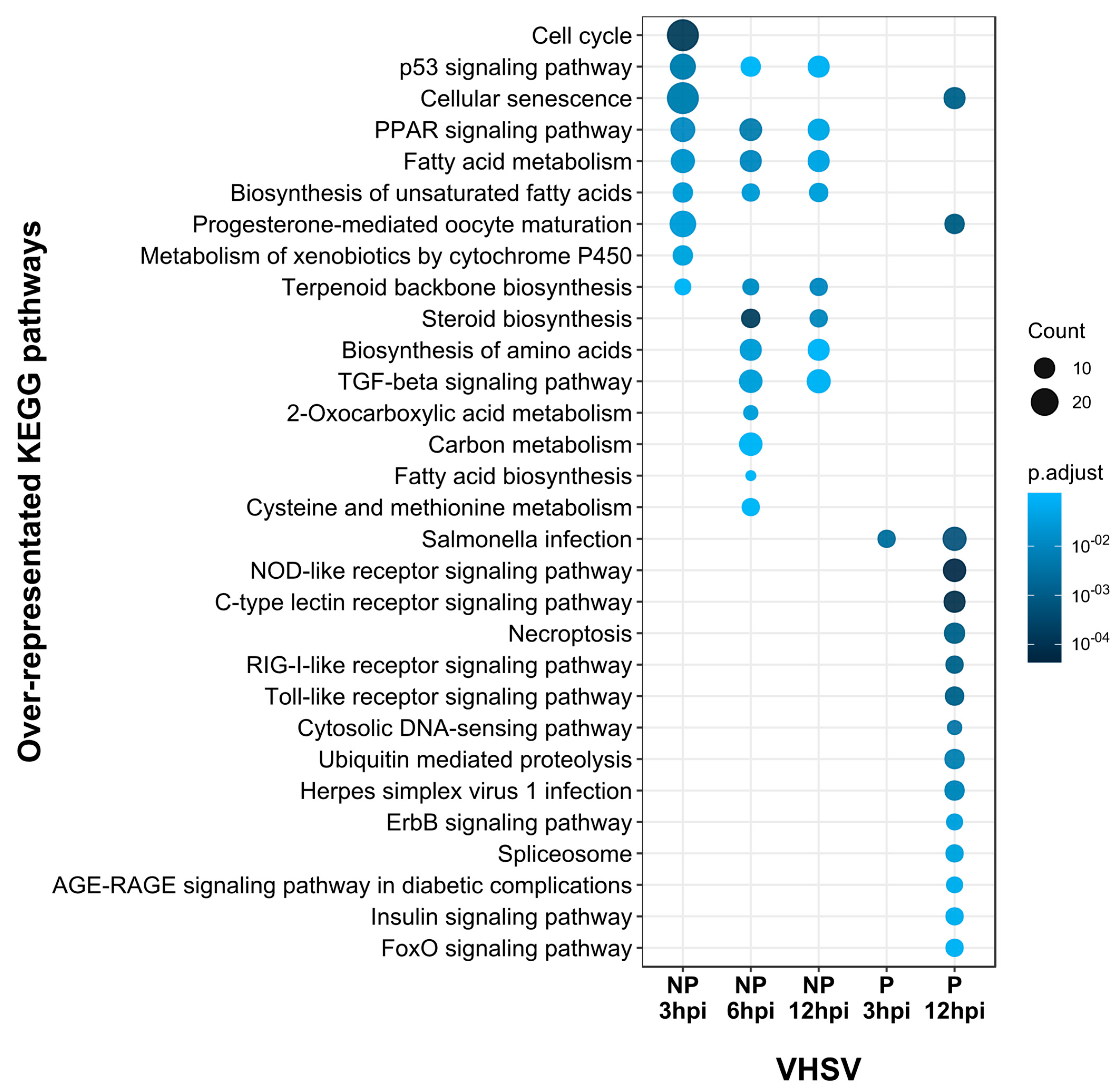
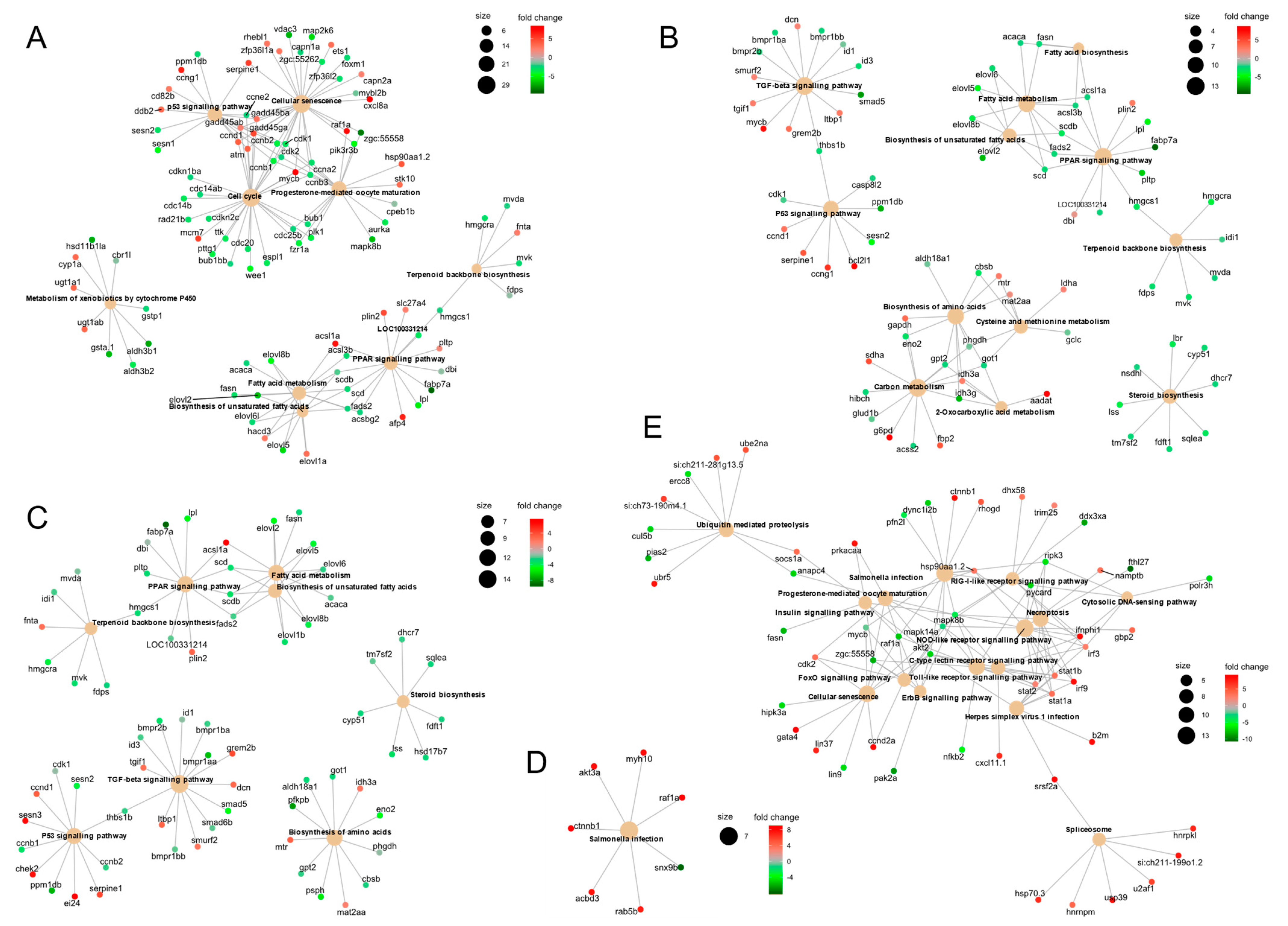
3.4. Gene Set Enrichment Analyses (GSEA) and KEGG Pathway Enrichment Reveal Delayed Activation of Immune Response Pathways in Cells Inoculated with the Pathogenic VHSV Isolate
3.5. Quantitative PCR Validation of Differential Expression Data
4. Discussion
4.1. Transcriptomics Showed Evidence of Cellular Transcriptional Shutoff by the Pathogenic VHSV
4.2. Pathogenicity May Be Associated with Delayed Viral Sensing by Pathogenic VHSV
4.3. VHSV Pathogenicity Is Associated with Endoplasmic Reticulum Stress and Attenuation of Unfolded Protein Response (UPR)
4.4. Non-Pathogenic VHSV Replication Triggered Host Stress Response Via the p53 and TGF-β Pathways
4.5. Differential Modulation of Transcription of VHSV Receptors in Pathogenic and Non Pathogenic Isolates
4.6. Suppression of Cholesterol Biosynthesis and Trafficking as a Potential Mechanism of Attenuation of VHSV Virulence
4.7. Limitations of this Study
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wolf, K. Viral hemorrhagic septicemia. In Fish Viruses and Fish Viral Diseases; Wolf, K., Ed.; Cornell University Press: New York, NY, USA, 1988; pp. 217–249. [Google Scholar]
- Olesen, N.J. Sanitation of viral haemorrhagic septicaemia (VHS). J. Appl. Ichthyol. 1998, 14, 173–177. [Google Scholar] [CrossRef]
- Skall, H.F.; Olesen, N.J.; Mellergaard, S. Viral haemorrhagic septicaemia virus in marine fish and its implications for fish farming—A review. J. Fish Dis. 2005, 28, 509–529. [Google Scholar] [CrossRef]
- López-Vázquez, C.; Raynard, R.S.; Bain, N.; Snow, M.; Bandín, I.; Dopazo, C.P. Genotyping of marine viral haemorrhagic septicaemia virus isolated from the Flemish Cap by nucleotide sequence analysis and restriction fragment length polymorphism patterns. Dis. Aquat. Organ. 2006, 73, 23–31. [Google Scholar] [CrossRef]
- Schutze, H.; Mundt, E.; Mettenleiter, T.C. Complete genomic sequence of viral hemorrhagic septicemia virus, a fish rhabdovirus. Virus Genes 1999, 19, 59–65. [Google Scholar] [CrossRef]
- Raja-Halli, M.; Vehmas, T.K.; Rimaila-Pärnänen, E.; Sainmaa, S.; Skall, H.F.; Olesen, N.J.; Tapiovaara, H. Viral haemorrhagic septicaemia (VHS) outbreaks in Finnish rainbow trout farms. Dis. Aquat. Organ. 2006, 72, 201–211. [Google Scholar] [CrossRef]
- Nishizawa, T.; Savaş, H.; Işidan, H.; Üstündaǧ, C.; Iwamoto, H.; Yoshimizu, M. Genotyping and pathogenicity of viral hemorrhagic septicemia virus from free-living turbot (Psetta maxima) in a Turkish coastal area of the Black Sea. Appl. Environ. Microbiol. 2006, 72, 2373–2378. [Google Scholar] [CrossRef]
- Dale, O.B.; Ørpetveit, I.; Lyngstad, T.M.; Kahns, S.; Skall, H.F.; Olesen, N.J.; Dannevig, B.H. Outbreak of viral haemorrhagic septicaemia (VHS) in seawater-farmed rainbow trout in Norway caused by VHS virus Genotype III. Dis. Aquat. Organ. 2009, 85, 93–103. [Google Scholar] [CrossRef] [PubMed]
- Stone, D.M.; Ferguson, H.W.; Tyson, P.A.; Savage, J.; Wood, G.; Dodge, M.J.; Woolford, G.; Dixon, P.F.; Feist, S.W.; Way, K. The first report of viral haemorrhagic septicaemia in farmed rainbow trout, Oncorhynchus mykiss (Walbaum), in the United Kingdom. J. Fish Dis. 2008, 31, 775–784. [Google Scholar] [CrossRef] [PubMed]
- Einer-Jensen, K.; Ahrens, P.; Forsberg, R.; Lorenzen, N. Evolution of the fish rhabdovirus viral haemorrhagic septicaemia virus. J. Gen. Virol. 2004, 85, 1167–1179. [Google Scholar] [CrossRef] [PubMed]
- Stone, D.M.; Way, K.; Dixon, P.F. Nucleotide sequence of the glycoprotein gene of viral haemorrhagic septicaemia (VHS) viruses from different geographical areas: A link between VHS in farmed fish species and viruses isolated from North Sea cod (Gadus morhua L.). J. Gen. Virol. 1997, 78, 1319–1326. [Google Scholar] [CrossRef]
- Longdon, B.; Brockhurst, M.A.; Russell, C.A.; Welch, J.J.; Jiggins, F.M. The Evolution and Genetics of Virus Host Shifts. PLoS Pathog. 2014, 10, e1004395. [Google Scholar] [CrossRef]
- Kim, M.S.; Kim, K.H. Effects of NV gene knock-out recombinant viral hemorrhagic septicemia virus (VHSV) on Mx gene expression in Epithelioma papulosum cyprini (EPC) cells and olive flounder (Paralichthys olivaceus). Fish Shellfish. Immunol. 2012, 32, 459–463. [Google Scholar] [CrossRef]
- Cano, I.; Collet, B.; Pereira, C.; Paley, R.; Aerle, R.V.; Stone, D.; Taylor, N.G.H. In vivo virulence of viral haemorrhagic septicaemia virus (VHSV) in rainbow trout Oncorhynchus mykiss correlates inversely with in vitro Mx gene expression. Vet. Microbiol. 2016, 187. [Google Scholar] [CrossRef] [PubMed]
- Wargo, A.R.; Garver, K.A.; Kurath, G. Virulence correlates with fitness in vivo for two M group genotypes of Infectious hematopoietic necrosis virus (IHNV). Virology 2010, 404, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Peñaranda, M.M.D.; Wargo, A.R.; Kurath, G. In vivo fitness correlates with host-specific virulence of Infectious hematopoietic necrosis virus (IHNV) in sockeye salmon and rainbow trout. Virology 2011, 417, 312–319. [Google Scholar] [CrossRef] [PubMed]
- Alcami, A.; Koszinowski, U.H. Viral mechanisms of immune evasion. Trends Microbiol. 2000, 8, 410–418. [Google Scholar] [CrossRef]
- Oksayan, S.; Ito, N.; Moseley, G.; Blondel, D. Subcellular Trafficking in Rhabdovirus Infection and Immune Evasion: A Novel Target for Therapeutics. Infect. Disord.-Drug Targets 2012, 12, 38–58. [Google Scholar] [CrossRef] [PubMed]
- Faul, E.J.; Lyles, D.S.; Schnell, M.J. Interferon response and viral evasion by members of the family rhabdoviridae. Viruses 2009, 10, 832–851. [Google Scholar] [CrossRef]
- Biacchesi, S.; Mérour, E.; Chevret, D.; Lamoureux, A.; Bernard, J.; Brémont, M. NV proteins of fish novirhabdovirus recruit cellular PPM1Bb protein phosphatase and antagonize RIG-I-mediated IFN induction. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Mortensen, H.F.; Heuer, O.E.; Lorenzen, N.; Otte, L.; Olesen, N.J. Isolation of viral haemorrhagic septicaemia virus (VHSV) from wild marine fish species in the Baltic Sea, Kattegat, Skagerrak and the North Sea. Proc. Virus Res. 1999, 63, 95–106. [Google Scholar] [CrossRef]
- Skall, H.F.; Slierendrecht, W.J.; King, J.A.; Olesen, N.J. Experimental infection of rainbow trout Oncorhynchus mykiss with viral haemorrhagic septicaemia virus isolates from European marine and farmed fishes. Dis. Aquat. Organ. 2004, 58, 99–110. [Google Scholar] [CrossRef]
- Wolf, K.; Quimby, M.C. Established eurythermic line of fish cells in vitro. Science 1962, 135, 1065–1066. [Google Scholar] [CrossRef]
- Burleson, F.G.; Chabers, T.M.; Wiederbrauk, D.L. TCID50 (Chapter 12). In Virology. A Laboratory Manua; Academic Press: San Diego, CA, USA, 1992; pp. 58–61. ISBN 9780121447304. [Google Scholar]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online: http://wwwbioinformaticsbabrahamacuk/projects/fastqc/ (accessed on 1 September 2020).
- Bushmanova, E.; Antipov, D.; Lapidus, A.; Prjibelski, A.D. RnaSPAdes: A de novo transcriptome assembler and its application to RNA-Seq data. Gigascience 2019. [Google Scholar] [CrossRef] [PubMed]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B. Aligning short sequencing reads with Bowtie. Curr. Protoc. Bioinform. 2010. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, M.; Morgan, M. Scalable Genomics with R and Bioconductor. Stat. Sci. 2014, 29, 214–226. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y.; Benjamini, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Sun, L.; Dong, S.; Ge, Y.; Fonseca, J.P.; Robinson, Z.T.; Mysore, K.S.; Mehta, P. Divenn: An interactive and integrated web-based visualization tool for comparing gene lists. Front. Genet. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Powell, D. Software Package: Vennt 2014. Available online: https://drpowell.github.io/vennt/ (accessed on 1 September 2020).
- Robinson, M.D.; Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010, 11. [Google Scholar] [CrossRef]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer International Publishing: Berlin, Germany, 2016; Volume 174, ISBN 978-3-319-24275-0. [Google Scholar]
- Elso, C.M.; Roberts, L.J.; Smyth, G.K.; Thomson, R.J.; Baldwin, T.M.; Foote, S.J.; Handman, E. Leishmaniasis host response loci (lmr1-3) modify disease severity through a Th1/Th2-independent pathway. Genes Immun. 2004, 5, 93–100. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. ClusterProfiler: An R package for comparing biological themes among gene clusters. Omics A J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Booth, D.S.; Szmidt-Middleton, H.; King, N.; Westbrook, M.J.; Young, S.L.; Kuo, A.; Abedin, M.; Chapman, J.; Fairclough, S.R.; Hellsten, U.; et al. RStudio: Integrated Development for R. Nature 2018, 20, 1403–1414. [Google Scholar] [CrossRef]
- Hu, Z.-L.; Bao, J.; Reecy, J. CateGOrizer: A Web-Based Program to Batch Analyze Gene Ontology Classification Categories. Online J. Bioinform. 2008, 9, 108–112. [Google Scholar]
- Marc Carlson Genome Wide Annotation for Zebrafish. R Package Version 3.8.2. 2019. Available online: http://bioconductor.org/packages/release/data/annotation/html/org.Dr.eg.db.html (accessed on 1 September 2020).
- Hellemans, J.; Mortier, G.; De Paepe, A.; Speleman, F.; Vandesompele, J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2008, 8. [Google Scholar] [CrossRef]
- Wertz, G.W.; Youngner, J.S. Interferon Production and Inhibition of Host Synthesis in Cells Infected with Vesicular Stomatitis Virus. J. Virol. 1970. [Google Scholar] [CrossRef]
- Connor, J.H.; Lyles, D.S. Vesicular Stomatitis Virus Infection Alters the eIF4F Translation Initiation Complex and Causes Dephosphorylation of the eIF4E Binding Protein 4E-BP1. J. Virol. 2002. [Google Scholar] [CrossRef] [PubMed]
- Ammayappan, A.; Kurath, G.; Thompson, T.M.; Vakharia, V.N. A Reverse Genetics System for the Great Lakes Strain of Viral Hemorrhagic Septicemia Virus: The NV Gene is Required for Pathogenicity. Mar. Biotechnol. 2011. [Google Scholar] [CrossRef]
- Chiou, P.P.; Kim, C.H.; Ormonde, P.; Leong, J.-A.C. Infectious Hematopoietic Necrosis Virus Matrix Protein Inhibits Host-Directed Gene Expression and Induces Morphological Changes of Apoptosis in Cell Cultures. J. Virol. 2000. [Google Scholar] [CrossRef]
- Gorgoglione, B.; Ringiesn, J.L.; Pham, L.H.; Shepherd, B.S.; Leaman, D.W. Comparative effects of Novirhabdovirus genes on modulating constitutive transcription and innate antiviral responses, in different teleost host cell types. Virol. J. 2020. [Google Scholar] [CrossRef] [PubMed]
- Kesterson, S.P.; Ringiesn, J.; Vakharia, V.N.; Shepherd, B.S.; Leaman, D.W.; Malathi, K. Effect of the viral hemorrhagic septicemia virus nonvirion protein on translation via PERK-eIF2α pathway. Viruses 2020, 12, 499. [Google Scholar] [CrossRef]
- Lamers, M.M.; van den Hoogen, B.G.; Haagmans, B.L. ADAR1: “Editor-in-Chief” of Cytoplasmic Innate Immunity. Front. Immunol. 2019, 10, 1763. [Google Scholar] [CrossRef] [PubMed]
- Samuel, C.E. Adenosine deaminases acting on RNA (ADARs) are both antiviral and proviral. Virology 2011, 411, 180–193. [Google Scholar] [CrossRef]
- He, M.; Zhang, H.; Li, Y.; Wang, G.; Tang, B.; Zhao, J.; Huang, Y.; Zheng, J. Cathelicidin-derived antimicrobial peptides inhibit Zika virus through direct inactivation and interferon pathway. Front. Immunol. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Cai, Z.; Cai, L.; Jiang, J.; Chang, K.-S.; van der Westhuyzen, D.R.; Luo, G. Human Serum Amyloid A Protein Inhibits Hepatitis C Virus Entry into Cells. J. Virol. 2007, 81, 6128–6133. [Google Scholar] [CrossRef]
- Mousavinezhad-Moghaddam, M.; Amin, A.A.; Rafatpanah, H.; Rezaee, S.A.R. A new insight into viral proteins as immunomodulatory therapeutic agents: KSHV vOX2 a homolog of human CD200 as a potent anti-inflammatory protein. Iran. J. Basic Med. Sci. 2016, 19, 2–13. [Google Scholar]
- Koutsakos, M.; McWilliam, H.E.G.; Aktepe, T.E.; Fritzlar, S.; Illing, P.T.; Mifsud, N.A.; Purcell, A.W.; Rockman, S.; Reading, P.C.; Vivian, J.P.; et al. Downregulation of MHC class I expression by influenza A and B viruses. Front. Immunol. 2019. [Google Scholar] [CrossRef]
- Zügel, U.; Kaufmann, S.H.E. Role of heat shock proteins in protection from and pathogenesis of infectious diseases. Clin. Microbiol. Rev. 1999, 12, 19–39. [Google Scholar] [CrossRef]
- Kim, M.Y.; Oglesbee, M. Virus-Heat Shock Protein Interaction and a Novel Axis for Innate Antiviral Immunity. Cells 2012, 1, 646–666. [Google Scholar] [CrossRef] [PubMed]
- Basu, N.; Todgham, A.E.; Ackerman, P.A.; Bibeau, M.R.; Nakano, K.; Schulte, P.M.; Iwama, G.K. Heat shock protein genes and their functional significance in fish. Gene 2002, 295, 173–183. [Google Scholar] [CrossRef]
- Mayer, M.P. Recruitment of Hsp70 chaperones: A crucial part of viral survival strategies. Rev. Physiol. Biochem. Pharmacol. 2005, 153, 1–46. [Google Scholar] [CrossRef] [PubMed]
- Lahaye, X.; Vidy, A.; Fouquet, B.; Blondel, D. Hsp70 Protein Positively Regulates Rabies Virus Infection. J. Virol. 2012. [Google Scholar] [CrossRef] [PubMed]
- Chan, S.W.; Egan, P.A. Effects of hepatitis C virus envelope glycoprotein unfolded protein response activation on translation and transcription. Arch. Virol. 2009, 154, 1631–1640. [Google Scholar] [CrossRef]
- Chan, S.W. Unfolded protein response in hepatitis C virus infection. Front. Microbiol. 2014, 5, 233. [Google Scholar] [CrossRef]
- Jheng, J.-R.; Wang, S.-C.; Jheng, C.-R.; Horng, J.-T. Enterovirus 71 induces dsRNA/PKR-dependent cytoplasmic redistribution of GRP78/BiP to promote viral replication. Emerg. Microbes Infect. 2016, 5, e23. [Google Scholar] [CrossRef]
- Tan, Z.; Zhang, W.; Sun, J.; Fu, Z.; Ke, X.; Zheng, C.; Zhang, Y.; Li, P.; Liu, Y.; Hu, Q.; et al. ZIKV infection activates the IRE1-XBP1 and ATF6 pathways of unfolded protein response in neural cells. J. Neuroinflamm. 2018. [Google Scholar] [CrossRef] [PubMed]
- Elston, R.; Inman, G.J. Crosstalk between p53 and TGF- β Signalling. J. Signal Transduct. 2012. [Google Scholar] [CrossRef]
- Lee, H.-R.; Toth, Z.; Shin, Y.C.; Lee, J.-S.; Chang, H.; Gu, W.; Oh, T.-K.; Kim, M.H.; Jung, J.U. Kaposi’s Sarcoma-Associated Herpesvirus Viral Interferon Regulatory Factor 4 Targets MDM2 To Deregulate the p53 Tumor Suppressor Pathway. J. Virol. 2009. [Google Scholar] [CrossRef] [PubMed]
- Thomson, B.J. Viruses and apoptosis. Int. J. Exp. Pathol. 2001, 82, 65–76. [Google Scholar] [CrossRef]
- Aranda, M.; Maule, A. Virus-induced host gene shutoff in animals and plants. Virology 1998, 261–267. [Google Scholar] [CrossRef]
- Lazo, P.A.; Santos, C.R. Interference with p53 functions in human viral infections, a target for novel antiviral strategies? Rev. Med. Virol. 2011, 21, 285–300. [Google Scholar] [CrossRef] [PubMed]
- Ma-Lauer, Y.; Carbajo-Lozoya, J.; Hein, M.Y.; Müller, M.A.; Deng, W.; Lei, J.; Meyer, B.; Kusov, Y.; Von Brunn, B.; Bairad, D.R.; et al. P53 down-regulates SARS coronavirus replication and is targeted by the SARS-unique domain and PLpro via E3 ubiquitin ligase RCHY1. Proc. Natl. Acad. Sci. USA 2016, 113, E5192–E5201. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.H.; Li, H.C.; Ku, T.S.; Wu, P.C.; Yeh, Y.J.; Cheng, J.C.; Lin, T.Y.; Lo, S.Y. Hepatitis C virus down-regulates SERPINE1/PAI-1 expression to facilitate its replication. J. Gen. Virol. 2017, 98, 2274–2286. [Google Scholar] [CrossRef]
- Ray, S.; Broor, S.L.; Vaishnav, Y.; Sarkar, C.; Girish, R.; Dar, L.; Seth, P.; Broor, S. Transforming growth factor beta in hepatitis C virus infection: In vivo and in vitro findings. J. Gastroenterol. Hepatol. 2003, 18, 393–403. [Google Scholar] [CrossRef]
- Bearzotti, M.; Delmas, B.; Lamoureux, A.; Loustau, A.-M.; Chilmonczyk, S.; Bremont, M. Fish Rhabdovirus Cell Entry Is Mediated by Fibronectin. J. Virol. 1999, 73, 7703–7709. [Google Scholar] [CrossRef]
- Albertini, A.A.V.; Baquero, E.; Ferlin, A.; Gaudin, Y. Molecular and cellular aspects of rhabdovirus entry. Viruses 2012, 4, 117–139. [Google Scholar] [CrossRef]
- Löffler, S.; Lottspeich, F.; Lanza, F.; Azorsa, D.O.; ter Meulen, V.; Schneider-Schaulies, J. CD9, a tetraspan transmembrane protein, renders cells susceptible to canine distemper virus. J. Virol. 1997, 71, 42–49. [Google Scholar] [CrossRef]
- Earnest, J.T.; Hantak, M.P.; Li, K.; McCray, P.B.; Perlman, S.; Gallagher, T. The tetraspanin CD9 facilitates MERS-coronavirus entry by scaffolding host cell receptors and proteases. PLoS Pathog. 2017, 71, e1006546. [Google Scholar] [CrossRef]
- Bearer, E.L.; Satpute-Krishnan, P. The role of the cytoskeleton in the life cycle of viruses and intracellular bacteria: Tracks, motors, and polymerization machines. Curr. Drug Targets. Infect. Disord. 2002, 2, 247–264. [Google Scholar] [CrossRef]
- Guiney, D.G.; Lesnick, M. Targeting of the actin cytoskeleton during infection by Salmonella strains. Clin. Immunol. 2005, 114, 248–255. [Google Scholar] [CrossRef]
- Stella, A.O.; Turville, S. All-round manipulation of the actin cytoskeleton by HIV. Viruses 2018, 10, 63. [Google Scholar] [CrossRef]
- Taylor, H.E.; Linde, M.E.; Khatua, A.K.; Popik, W.; Hildreth, J.E.K. Sterol Regulatory Element-Binding Protein 2 Couples HIV-1 Transcription to Cholesterol Homeostasis and T Cell Activation. J. Virol. 2011, 85, 7699–7709. [Google Scholar] [CrossRef] [PubMed]
- Park, C.Y.; Jun, H.J.; Wakita, T.; Cheong, J.H.; Hwang, S.B. Hepatitis C virus nonstructural 4B protein modulates sterol regulatory element-binding protein signaling via the AKT pathway. J. Biol. Chem. 2009, 284, 9237–9246. [Google Scholar] [CrossRef]
- Syed, G.H.; Tang, H.; Khan, M.; Hassanein, T.; Liu, J.; Siddiqui, A.; Diamond, M.S. Hepatitis C Virus Stimulates Low-Density Lipoprotein Receptor Expression To Facilitate Viral Propagation. J. Virol. 2014, 88, 2519–2529. [Google Scholar] [CrossRef]
- Aquilino, C.; Castro, R.; Fischer, U.; Tafalla, C. Transcriptomic responses in rainbow trout gills upon infection with viral hemorrhagic septicemia virus (VHSV). Dev. Comp. Immunol. 2014. [Google Scholar] [CrossRef] [PubMed]
- Verrier, E.R.; Genet, C.; Laloë, D.; Jaffrezic, F.; Rau, A.; Esquerre, D.; Dechamp, N.; Ciobotaru, C.; Hervet, C.; Krieg, F.; et al. Genetic and transcriptomic analyses provide new insights on the early antiviral response to VHSV in resistant and susceptible rainbow trout. BMC Genom. 2018. [Google Scholar] [CrossRef]
- Tang, X.; Huang, Y.; Lei, J.; Luo, H.; Zhu, X. The single-cell sequencing: New developments and medical applications. Cell Biosci. 2019, 9, 53. [Google Scholar] [CrossRef] [PubMed]
- Vogel, C.; Marcotte, E.M. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet. 2012. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cano, I.; Santos, E.M.; Moore, K.; Farbos, A.; van Aerle, R. Evidence of Transcriptional Shutoff by Pathogenic Viral Haemorrhagic Septicaemia Virus in Rainbow Trout. Viruses 2021, 13, 1129. https://doi.org/10.3390/v13061129
Cano I, Santos EM, Moore K, Farbos A, van Aerle R. Evidence of Transcriptional Shutoff by Pathogenic Viral Haemorrhagic Septicaemia Virus in Rainbow Trout. Viruses. 2021; 13(6):1129. https://doi.org/10.3390/v13061129
Chicago/Turabian StyleCano, Irene, Eduarda M. Santos, Karen Moore, Audrey Farbos, and Ronny van Aerle. 2021. "Evidence of Transcriptional Shutoff by Pathogenic Viral Haemorrhagic Septicaemia Virus in Rainbow Trout" Viruses 13, no. 6: 1129. https://doi.org/10.3390/v13061129
APA StyleCano, I., Santos, E. M., Moore, K., Farbos, A., & van Aerle, R. (2021). Evidence of Transcriptional Shutoff by Pathogenic Viral Haemorrhagic Septicaemia Virus in Rainbow Trout. Viruses, 13(6), 1129. https://doi.org/10.3390/v13061129





