Scalable, Micro-Neutralization Assay for Assessment of SARS-CoV-2 (COVID-19) Virus-Neutralizing Antibodies in Human Clinical Samples
Abstract
1. Introduction
2. Materials and Methods
2.1. Virus and Cells
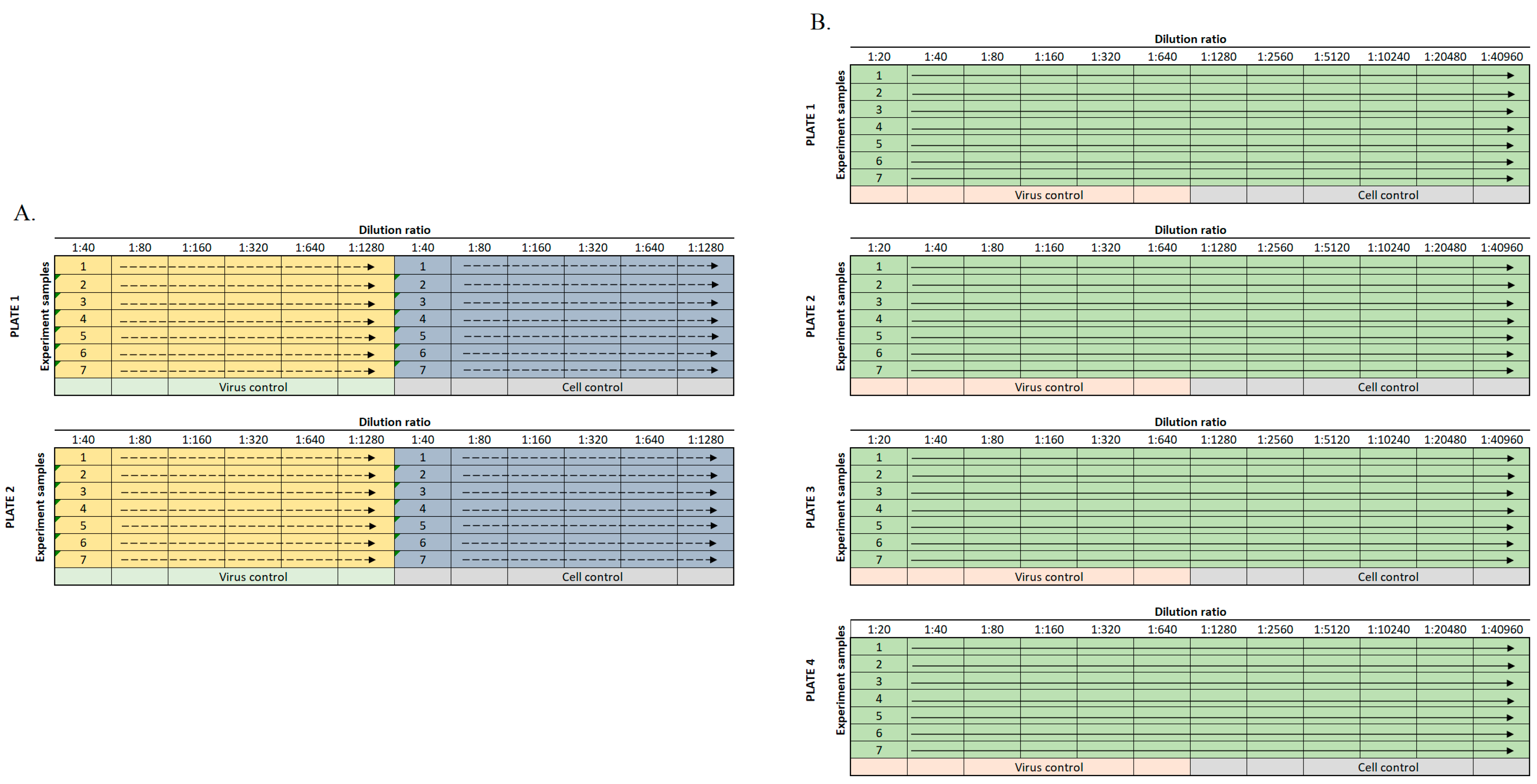
2.2. Sample Dilution
2.3. Cell Staining
2.4. Calculation of Standard NT50 Values
2.5. Calculation of NT50 Values by Regression Analysis
2.6. ELISAs
3. Results
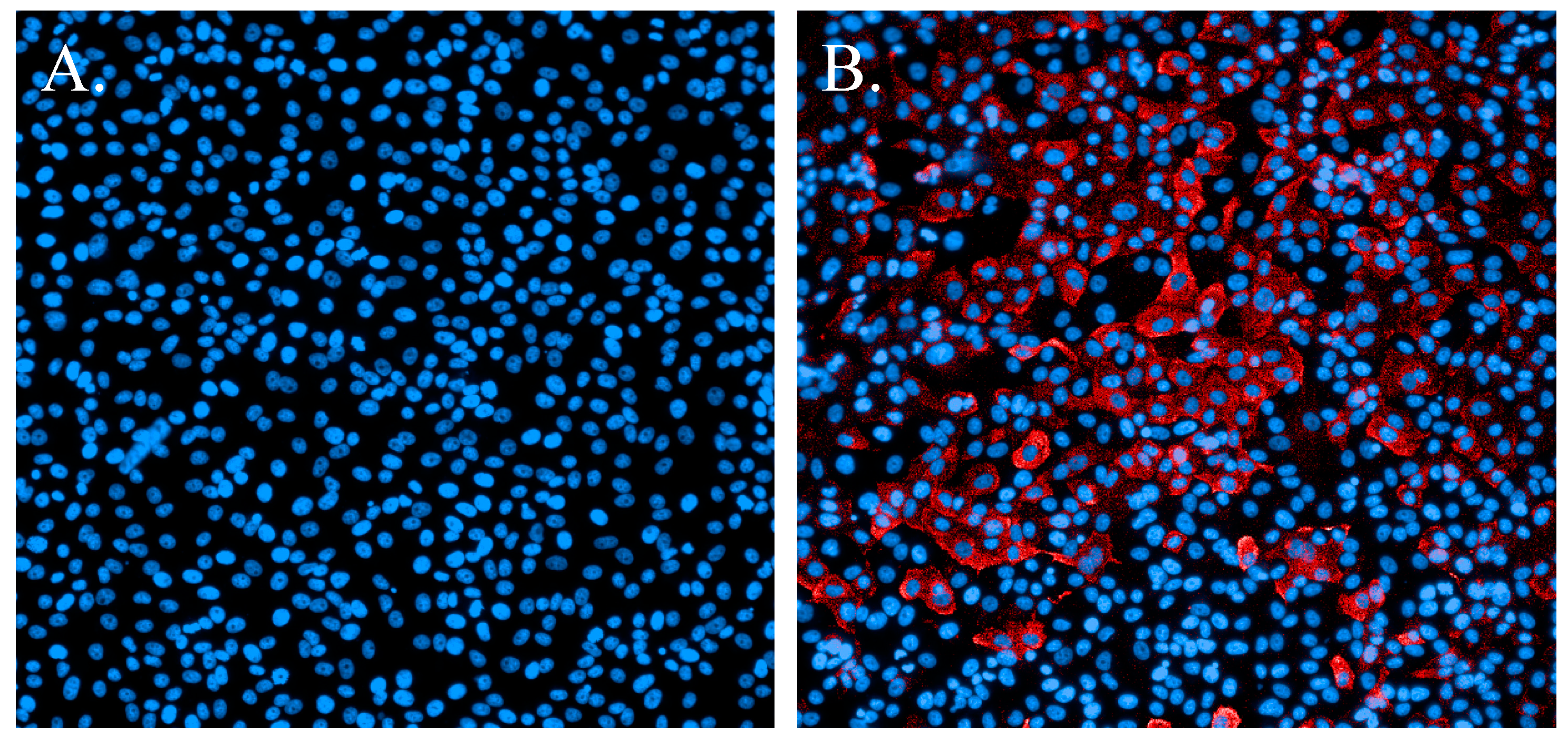
3.1. Immunofluorescence Staining
3.2. Initial Testing of Human Plasma
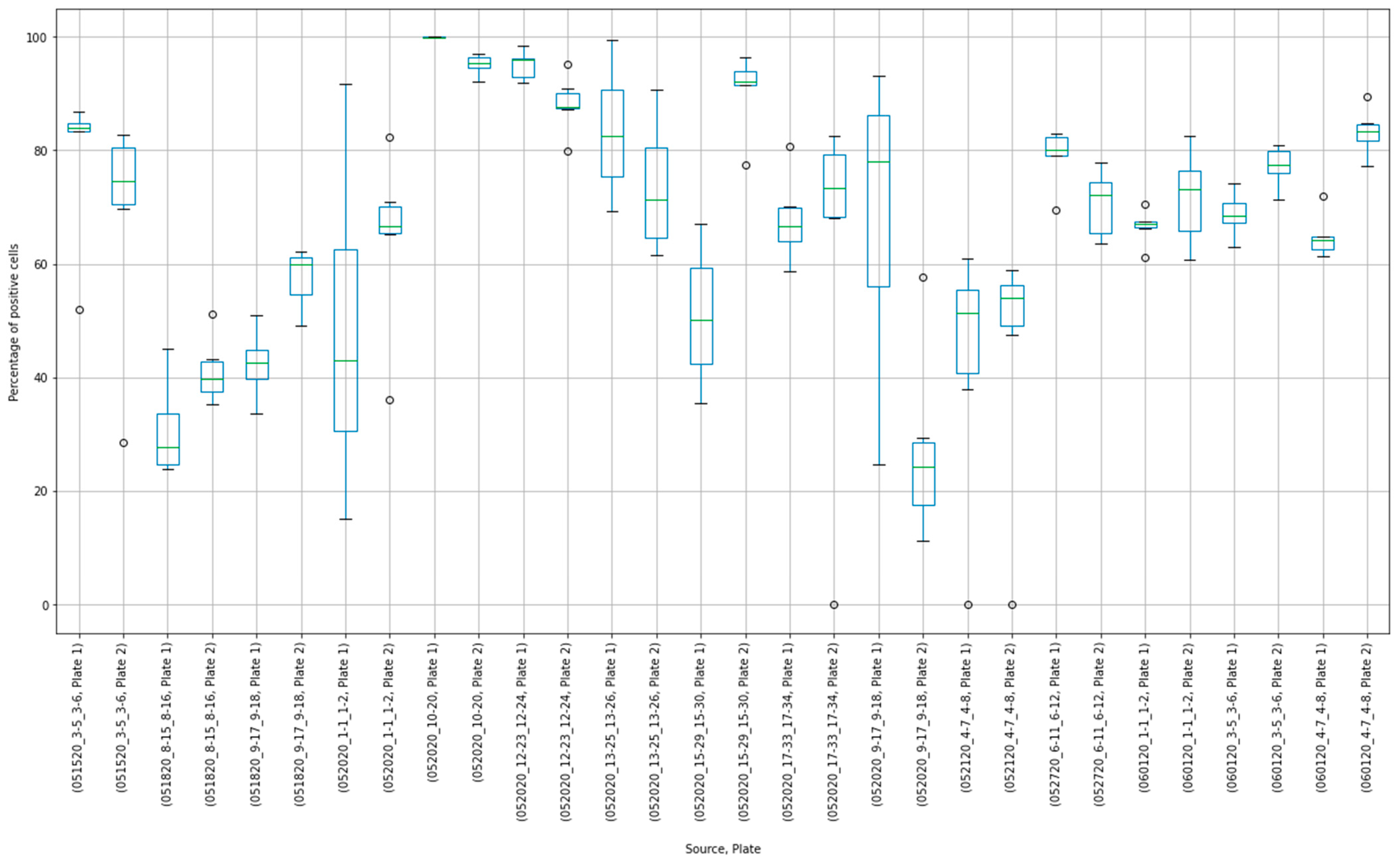
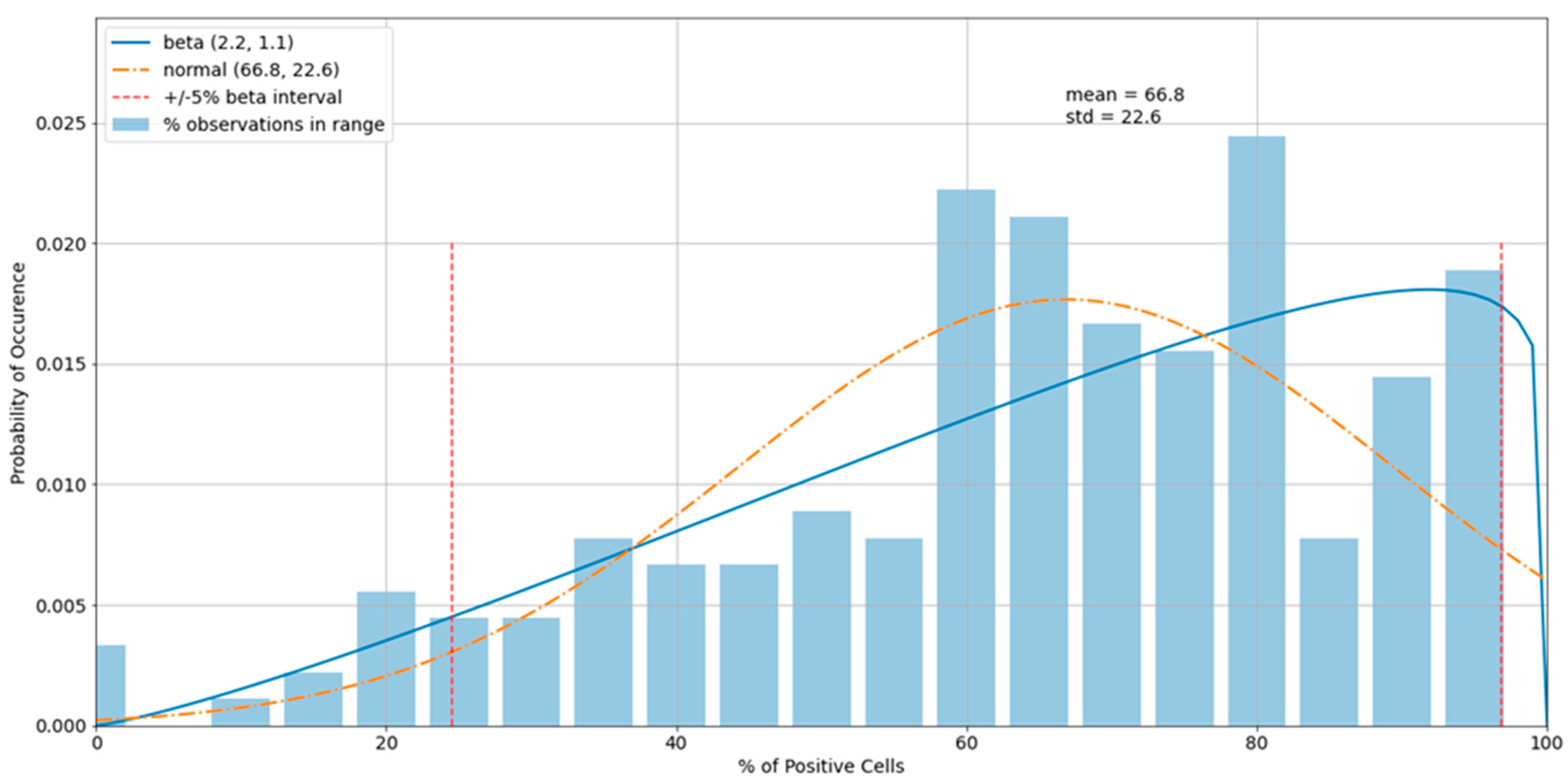
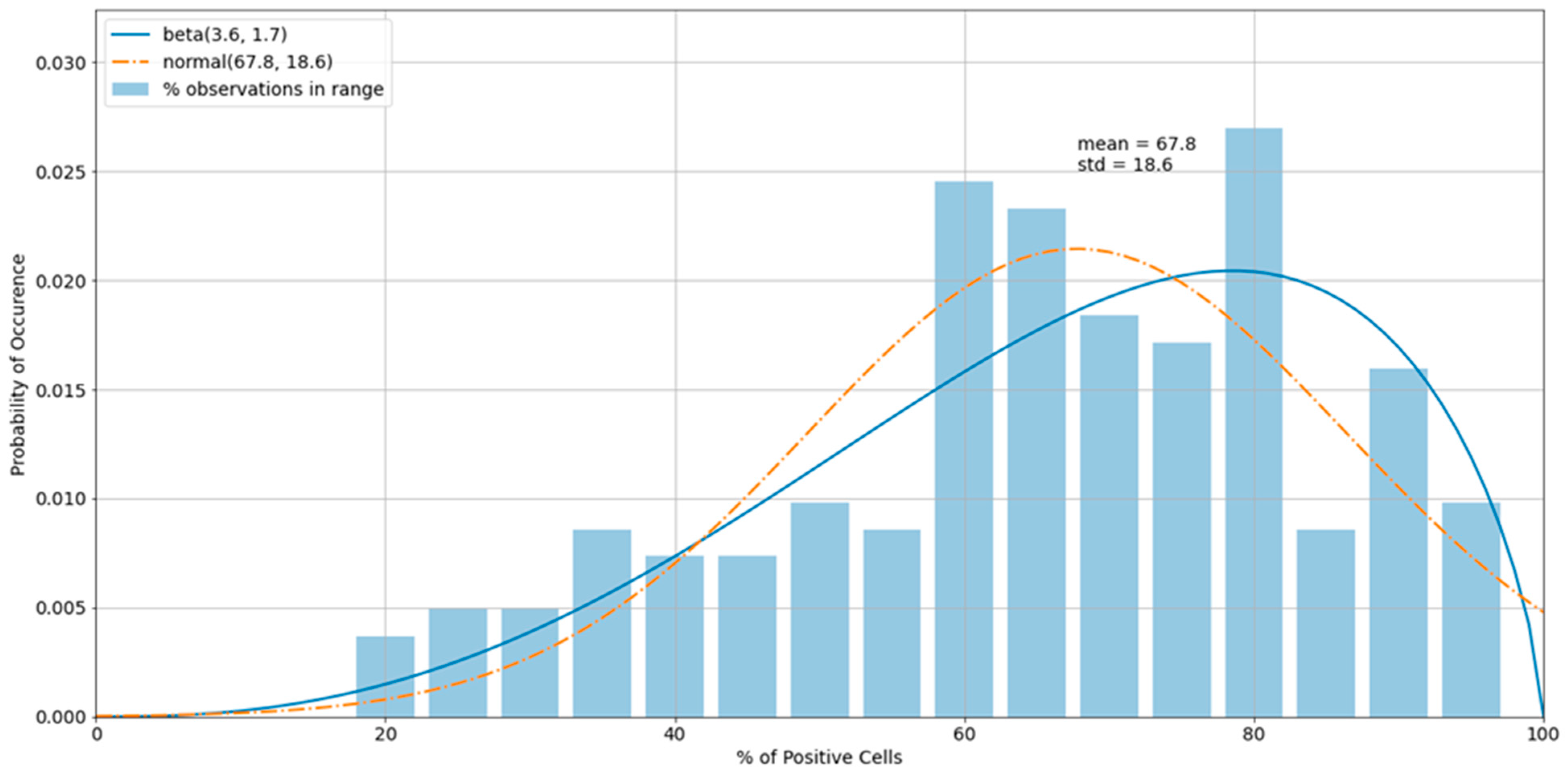
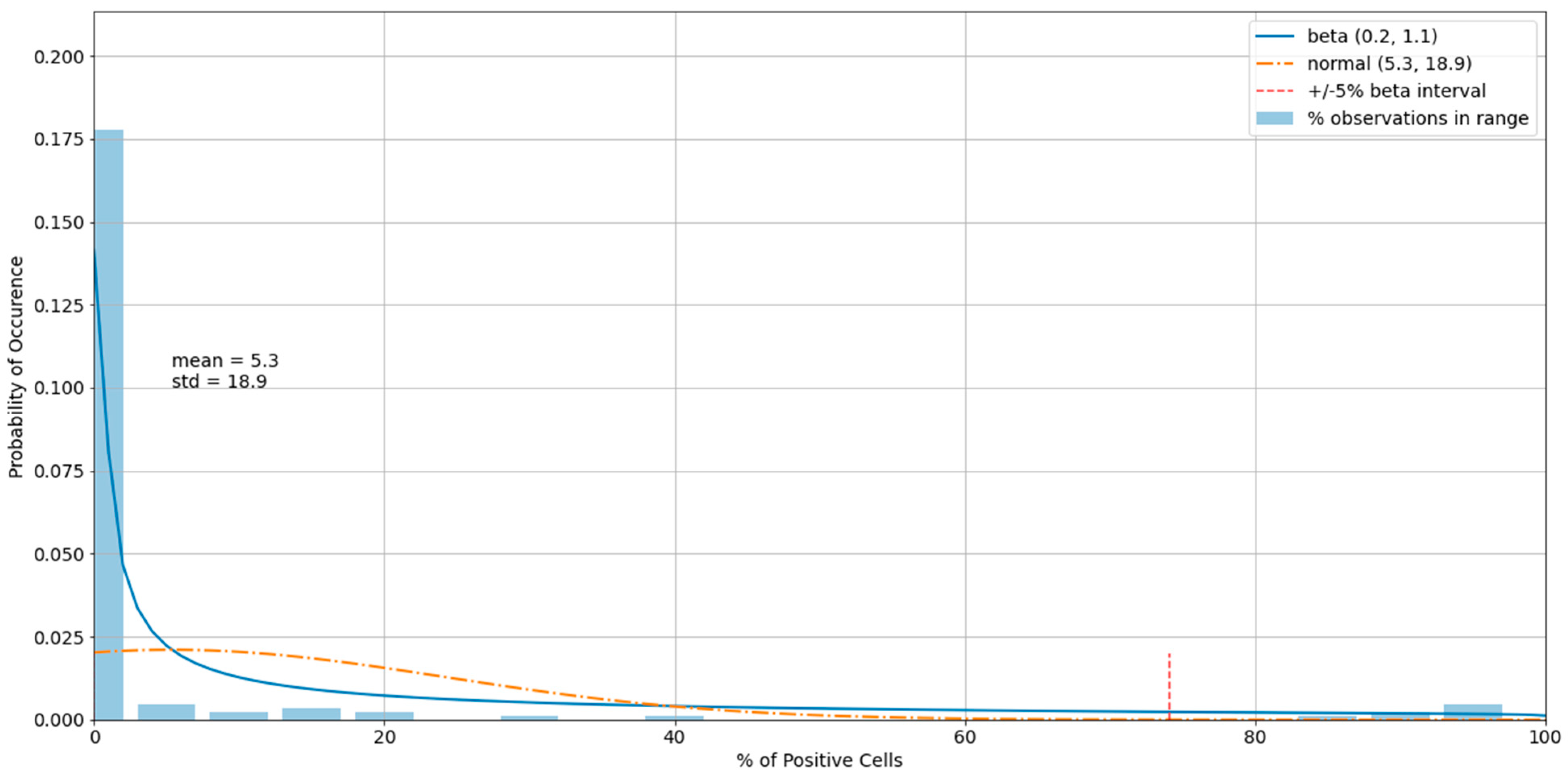
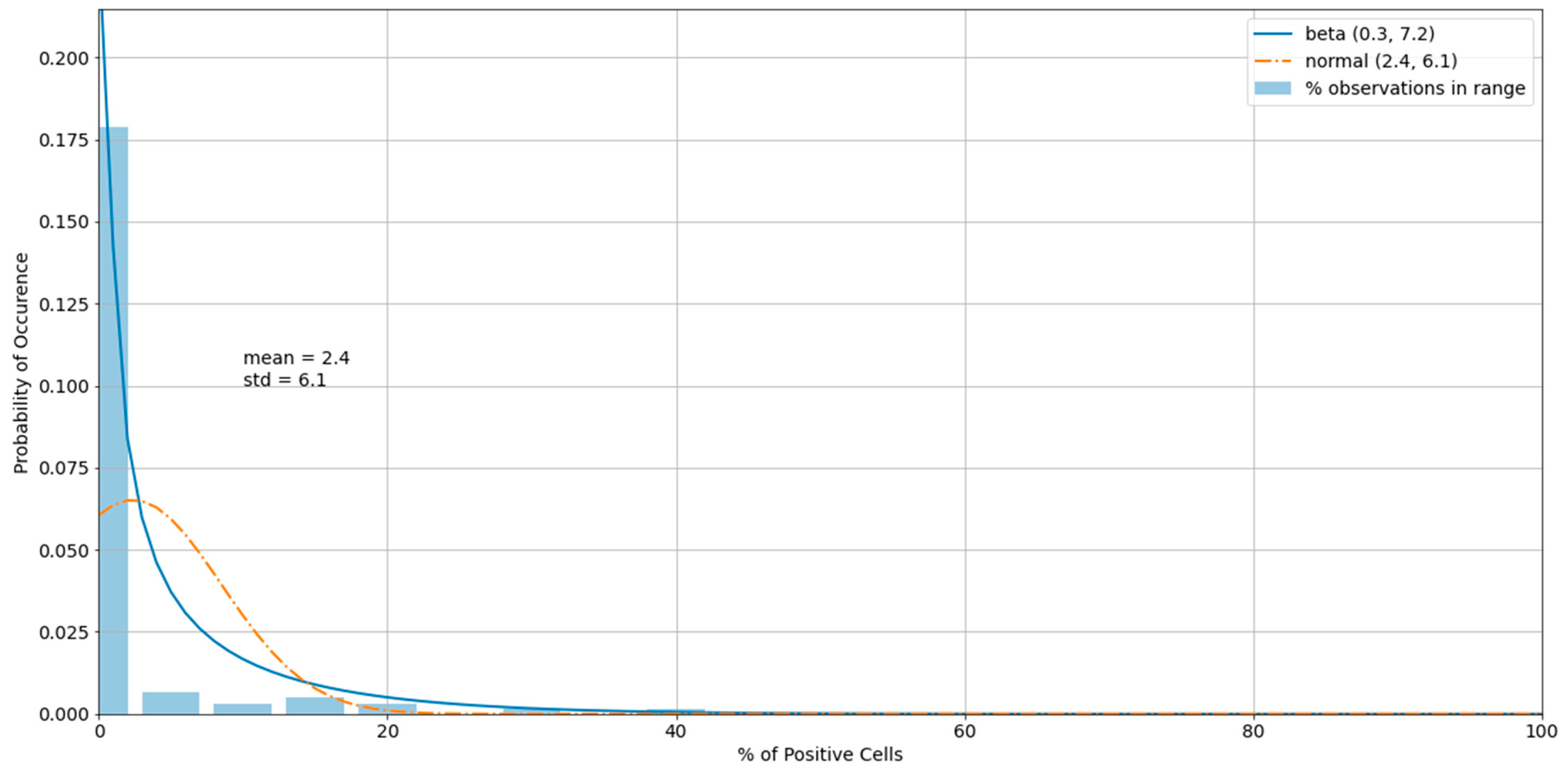
3.3. Outliers in the Virus Control and Cell Control Observations
3.4. Detecting Outliers in the Sample Observations
3.5. Assay Variability
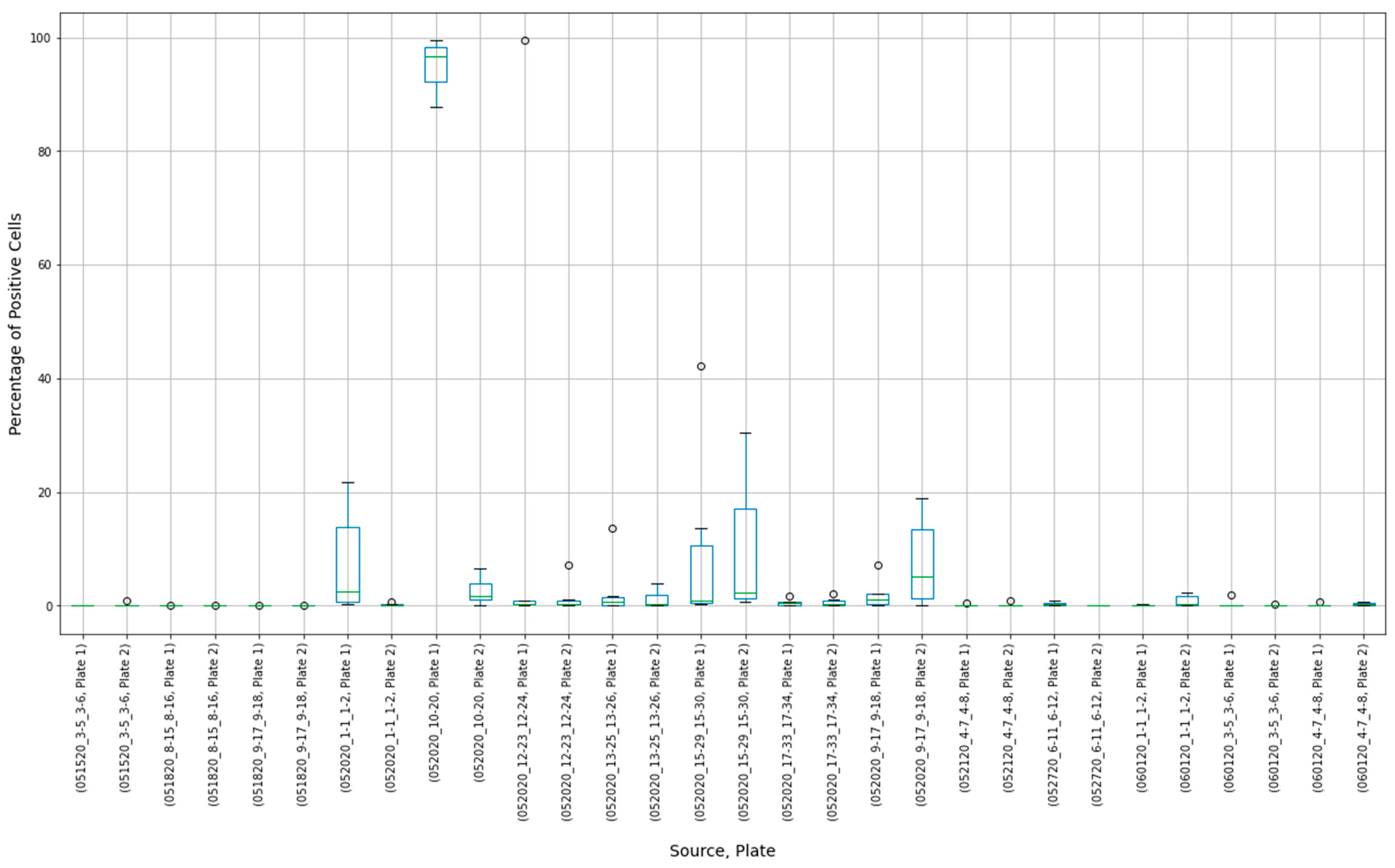
3.6. Specificity
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shi, X.; Chen, M.; Zhang, Y. The cardiovascular disorders and prognostic cardiac biomarkers in COVID-19. Mol. Biol. Rep. 2021, 22, 1–9. [Google Scholar]
- Harapan, B.N.; Yoo, H.J. Neurological symptoms, manifestations, and complications associated with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease 19 (COVID-19). J. Neurol. 2021, 23, 1–3. [Google Scholar]
- Karuppan, M.K.M.; Devadoss, D.; Nair, M.; Chand, H.S.; Lakshmana, M.K. SARS-CoV-2 Infection in the Central and Peripheral Nervous System-Associated Morbidities and Their Potential Mechanism. Mol. Neurobiol. 2021, 13, 1–6. [Google Scholar]
- Brandao, S.C.S.; Ramos, J.O.X.; de Arruda, G.F.A.; Godoi, E.; Carreira, L.; Lopes, R.W.; Grossman, G.B.; de Souza Leao Lima, R. Mapping COVID-19 functional sequelae: The perspective of nuclear medicine. Am. J. Nucl. Med. Mol. Imaging 2020, 10, 319–333. [Google Scholar]
- Sun, P.; Lu, X.; Xu, C.; Sun, W.; Pan, B. Understanding of COVID-19 based on current evidence. J. Med. Virol. 2020, 92, 548–551. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Soleimani, J.; Herasevich, S.; Pinevich, Y.; Pennington, K.M.; Dong, Y.; Pickering, B.W.; Barwise, A.K. Clinical Characteristics, Treatment, and Outcomes of Critically Ill Patients With COVID-19: A Scoping Review. Mayo Clin. Proc. 2021, 96, 183–202. [Google Scholar] [CrossRef] [PubMed]
- Bastard, P.; Rosen, L.B.; Zhang, Q.; Michailidis, E.; Hoffmann, H.-H.; Zhang, Y.; Dorgham, K.; Philippot, Q.; Rosain, J.; Béziat, V.; et al. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science 2020, 370, eabd4585. [Google Scholar] [CrossRef]
- Zhang, Q.; Bastard, P.; Liu, Z.; Le Pen, J.; Moncada-Velez, M.; Chen, J.; Ogishi, M.; Sabli, I.K.D.; Hodeib, S.; Korol, C.; et al. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science 2020, 370, eabd4570. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, S.C.A.; Yang, F.; Jackson, K.J.L.; Hoh, R.A.; Roltgen, K.; Jean, G.H.; Stevens, B.A.; Lee, J.Y.; Rustagi, A.; Rogers, A.J.; et al. Human B Cell Clonal Expansion and Convergent Antibody Responses to SARS-CoV-2. Cell Host Microbe 2020, 28, 516–525.e5. [Google Scholar] [CrossRef] [PubMed]
- Galipeau, Y.; Greig, M.; Liu, G.; Driedger, M.; Langlois, M.A. Humoral Responses and Serological Assays in SARS-CoV-2 Infections. Front. Immunol. 2020, 11, 610688. [Google Scholar] [CrossRef]
- Sun, B.; Feng, Y.; Mo, X.; Zheng, P.; Wang, Q.; Li, P.; Peng, P.; Liu, X.; Chen, Z.; Huang, H.; et al. Kinetics of SARS-CoV-2 specific IgM and IgG responses in COVID-19 patients. Emerg. Microbes Infect. 2020, 9, 940–948. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://www.uscovidplasma.org/ (accessed on 15 February 2021).
- Joyner, M.J.; Senefeld, J.W.; Klassen, S.A.; Mills, J.R.; Johnson, P.W.; Theel, E.S.; Wiggins, C.C.; Bruno, K.A.; Klompas, A.M.; Lesser, E.R.; et al. Effect of Convalescent Plasma on Mortality among Hospitalized Patients with COVID-19: Initial Three-Month Experience. medRxiv 2020. [Google Scholar] [CrossRef]
- Available online: https://www.fda.gov/media/141477/download (accessed on 10 February 2021).
- Available online: https://www.fda.gov/media/143891/download (accessed on 15 February 2021).
- Available online: https://www.fda.gov/media/143602/download (accessed on 15 February 2021).
- Luke, T.; Wu, H.; Zhao, J.; Channappanavar, R.; Coleman, C.M.; Jiao, J.A.; Matsushita, H.; Liu, Y.; Postnikova, E.N.; Ork, B.L.; et al. Human polyclonal immunoglobulin G from transchromosomic bovines inhibits MERS-CoV in vivo. Sci. Transl. Med. 2016, 8, 326ra21. [Google Scholar] [CrossRef] [PubMed]
- Postnikova, E.N.; Pettitt, J.; Van Ryn, C.J.; Holbrook, M.R.; Bollinger, L.; Yu, S.; Cai, Y.; Liang, J.; Sneller, M.C.; Jahrling, P.B.; et al. Scalable, semi-automated fluorescence reduction neutralization assay for qualitative assessment of Ebola virus-neutralizing antibodies in human clinical samples. PLoS ONE 2019, 14, e0221407. [Google Scholar] [CrossRef]
- Available online: https://www.fda.gov/media/137609/download (accessed on 15 February 2021).
- Available online: https://resources.rndsystems.com/pdfs/datasheets/dsr200.pdf (accessed on 15 February 2021).
- Virtanen, P.; Gommers, R.; Oliphant, T.E.; Haberland, M.; Reddy, T.; Cournapeau, D.; Burovski, E.; Peterson, P.; Weckesser, W.; Bright, J.; et al. Author Correction: SciPy 1.0: Fundamental algorithms for scientific computing in Python. Nat. Methods 2020, 17, 352. [Google Scholar] [CrossRef] [PubMed]
- Virtanen, P.; Gommers, R.; Oliphant, T.E.; Haberland, M.; Reddy, T.; Cournapeau, D.; Burovski, E.; Peterson, P.; Weckesser, W.; Bright, J.; et al. SciPy 1.0: Fundamental algorithms for scientific computing in Python. Nat. Methods 2020, 17, 261–272. [Google Scholar] [CrossRef]
- Dean, R.B.; Dixon, W.J. Simplified Statistics for Small Numbers of Observations. Anal. Chem. 1951, 23, 636–638. [Google Scholar] [CrossRef]
- Bond, K.; Nicholson, S.; Lim, S.M.; Karapanagiotidis, T.; Williams, E.; Johnson, D.; Hoang, T.; Sia, C.; Purcell, D.; Mordant, F.; et al. Evaluation of Serological Tests for SARS-CoV-2: Implications for Serology Testing in a Low-Prevalence Setting. J. Infect Dis. 2020, 222, 1280–1288. [Google Scholar] [CrossRef] [PubMed]
- Harvala, H.; Robb, M.L.; Watkins, N.; Ijaz, S.; Dicks, S.; Patel, M.; Supasa, P.; Wanwisa, D.; Liu, C.; Mongkolsapaya, J.; et al. Convalescent plasma therapy for the treatment of patients with COVID-19: Assessment of methods available for antibody detection and their correlation with neutralising antibody levels. Transfus. Med. 2020. [Google Scholar] [CrossRef]
- Mendrone-Junior, A.; Dinardo, C.L.; Ferreira, S.C.; Nishya, A.; Salles, N.A.; de Almeida Neto, C.; Hamasaki, D.T.; Facincani, T.; de Oliveira Alves, L.B.; Machado, R.R.G.; et al. Correlation between SARS-COV-2 antibody screening by immunoassay and neutralizing antibody testing. Transfusion 2021, 61, 1181–1190. [Google Scholar] [CrossRef]
- Riepler, L.; Rossler, A.; Falch, A.; Volland, A.; Borena, W.; von Laer, D.; Kimpel, J. Comparison of Four SARS-CoV-2 Neutralization Assays. Vaccines 2020, 9, 13. [Google Scholar] [CrossRef] [PubMed]
- Weidner, L.; Gansdorfer, S.; Unterweger, S.; Weseslindtner, L.; Drexler, C.; Farcet, M.; Witt, V.; Schistal, E.; Schlenke, P.; Kreil, T.R.; et al. Quantification of SARS-CoV-2 antibodies with eight commercially available immunoassays. J. Clin. Virol. 2020, 129, 104540. [Google Scholar] [CrossRef] [PubMed]
- Gundlapalli, A.V.; Salerno, R.M.; Brooks, J.T.; Averhoff, F.; Petersen, L.R.; McDonald, L.C.; Iademarco, M.F.; CDC COVID-19 Response. SARS-CoV-2 Serologic Assay Needs for the Next Phase of the US COVID-19 Pandemic Response. Open Forum Infect. Dis. 2021, 8, ofaa555. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Muruato, A.; Lokugamage, K.G.; Narayanan, K.; Zhang, X.; Zou, J.; Liu, J.; Schindewolf, C.; Bopp, N.E.; Aguilar, P.V.; et al. An Infectious cDNA Clone of SARS-CoV-2. Cell Host Microbe 2020, 27, 841–848.e3. [Google Scholar] [CrossRef]
| Parameter | Value |
|---|---|
| Cell seeding density | 30,000 cells per well |
| Dulbecco’s Modified Eagle Medium without calcium | |
| Virus multiplicity of infection | 0.5 |
| Virus/sample neutralization period in dilution block | 1 h, 37 °C, 5% CO2 |
| Virus/sample incubation with permissive cells | 24 h, 37 °C, 5% CO2 |
| Step | Purpose | Actions |
|---|---|---|
| 1 | Mask virus control outliers. | Exclude values outside of the critical region (<5%, >95%) of the beta distribution estimated for virus control observations. |
| 2 | Quality check the plates. | If the number of non-masked values of per plate is less than 3, then discard the results of entire experiment. Otherwise, go to Step 3. |
| 3 | Calculate the mean of virus control. | Use non-masked values from both plates to calculate the mean of virus control. |
| 4 | Calculate FRNA50. | Divide the mean of virus control by 2. |
| Step | Purpose | Actions |
|---|---|---|
| 1 | Mask cell control outliers. | Exclude values outside of the critical region (>95%) of the beta distribution estimated for cell control observations. |
| 2 | Quality check of the plates. | If the number of non-masked values of per plate is less than 3, then discard the results of the entire experiment. Otherwise, go to Step 3. |
| 3 | Calculate the mean of cell control. | Use non-masked values from both plates to calculate the mean of cell control. |
| 4 | Calculate FRNA100. | Use the mean of cell control. |
| Step | Purpose | Actions |
|---|---|---|
| 1 | Obtain the maximum value, Qmax. | Obtain the difference between the maximum of four observations and the second largest value. Divide it by the range between the maximum and the minimum. |
| 2 | Obtain the minimum value, Qmin. | Obtain the difference between the second smallest value and the minimum of four observations. Divide it by the range between the maximum and the minimum. |
| 3 | Compare with Q95 at 95% confidence level. | Q95 is 0.829. If Qmax or Qmin is above Q95, then mask that observation. If both are masked, discard the sample. |
| Step | Purpose | Actions |
|---|---|---|
| 1 | Check controls. | If at least one plate from virus control or cell control fails, discard the results. Otherwise, go to Step 2. |
| 2 | For each dilution ratio, check four observations of the sample. | Use Dixon’s Q test to check whether the minimum and maximum values of the sample are outliers. If both are rejected, then discard the results. If one is rejected, then remove it from calculations and go to Step 3. |
| 3 | Calculate the means. | Use non-masked values to calculate the mean of each dilution ratio. |
| 4 | Compare with FRNA thresholds. | Compare dilution means with FRNA50 and FRNA100. |
| Run | NT50 | Run | NT50 | |
|---|---|---|---|---|
| 1 | 173 | 11 | 335 | |
| 2 | 267 | 12 | 262 | |
| 3 | 230 | 13 | 359 | |
| 4 | 274 | 14 | 219 | |
| 5 | 337 | 15 | 205 | |
| 6 | 300 | 16 | 231 | |
| 7 | 257 | 17 | 147 | |
| 8 | 258 | 18 | 292 | |
| 9 | 403 | 19 | 191 | |
| 10 | 108 | 20 | 185 | |
| Mean: | 252 | |||
| Standard Deviation: | 74 |
| Sample | FRNA NT50 | R&D ELISA SARS-CoV-2 | EURO ELISA SARS-CoV-2 | |||||
|---|---|---|---|---|---|---|---|---|
| Rep 1 | Rep 2 | Rep 1 | Rep 2 | Rep 3 | Rep 1 | Rep 2 | Rep 3 | |
| 1 | <40 | 80 | + | + | + | + | + | + |
| 2 * | 40 | 40 | + | - | - | + | + | + |
| 3 | <40 | <40 | - | + | + | - | - | - |
| 4 | <40 | <40 | + | + | + | + | + | + |
| 5 | <40 | <40 | - | - | - | - | - | - |
| 6 | <40 | <40 | + | + | + | + | + | + |
| 7 | 160 | 80 | + | + | + | + | + | + |
| 8 | 80 | 40 | + | + | + | + | + | + |
| 9 | 640 | 320 | + | + | + | + | + | + |
| 10 | 80 | 80 | + | + | + | + | + | + |
| 11 | 160 | 80 | + | + | + | + | + | + |
| 12 | 320 | 320 | + | + | + | + | + | + |
| 13 | <40 | <40 | - | - | - | - | - | - |
| 14 | <40 | <40 | - | - | - | - | - | - |
| 15 | 80 | <40 | + | + | + | + | + | + |
| 16 | <40 | <40 | - | - | - | - | - | - |
| 17 | <40 | <40 | - | - | - | - | - | - |
| 18 | 40 | <40 | + | + | + | + | + | + |
| 19 | 320 | 40 | + | + | + | + | + | + |
| 20 | <40 | 40 | - | - | - | - | - | - |
| 21 | 80 | <40 | + | + | + | + | + | + |
| 22 | 80 | 80 | + | + | + | + | + | + |
| 23 | <40 | <40 | - | - | - | - | - | - |
| 24 | 40 | <40 | + | + | + | + | + | + |
| 25 | <40 | <40 | - | - | - | - | - | - |
| 26 | <40 | <40 | - | - | - | - | - | - |
| 27 | <40 | <40 | + | + | + | + | + | + |
| 28 | 80 | <40 | + | + | + | + | + | + |
| 29 | 80 | 80 | + | + | + | + | + | + |
| 30 | <40 | <40 | + | + | + | + | + | + |
| Positive Control | 80 | 80 | + | + | + | + | + | + |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bennett, R.S.; Postnikova, E.N.; Liang, J.; Gross, R.; Mazur, S.; Dixit, S.; Kocher, G.; Yu, S.; Georgia-Clark, S.; Gerhardt, D.; et al. Scalable, Micro-Neutralization Assay for Assessment of SARS-CoV-2 (COVID-19) Virus-Neutralizing Antibodies in Human Clinical Samples. Viruses 2021, 13, 893. https://doi.org/10.3390/v13050893
Bennett RS, Postnikova EN, Liang J, Gross R, Mazur S, Dixit S, Kocher G, Yu S, Georgia-Clark S, Gerhardt D, et al. Scalable, Micro-Neutralization Assay for Assessment of SARS-CoV-2 (COVID-19) Virus-Neutralizing Antibodies in Human Clinical Samples. Viruses. 2021; 13(5):893. https://doi.org/10.3390/v13050893
Chicago/Turabian StyleBennett, Richard S., Elena N. Postnikova, Janie Liang, Robin Gross, Steven Mazur, Saurabh Dixit, Gregory Kocher, Shuiqing Yu, Shalamar Georgia-Clark, Dawn Gerhardt, and et al. 2021. "Scalable, Micro-Neutralization Assay for Assessment of SARS-CoV-2 (COVID-19) Virus-Neutralizing Antibodies in Human Clinical Samples" Viruses 13, no. 5: 893. https://doi.org/10.3390/v13050893
APA StyleBennett, R. S., Postnikova, E. N., Liang, J., Gross, R., Mazur, S., Dixit, S., Kocher, G., Yu, S., Georgia-Clark, S., Gerhardt, D., Cai, Y., Marron, L., Lukin, V. V., & Holbrook, M. R. (2021). Scalable, Micro-Neutralization Assay for Assessment of SARS-CoV-2 (COVID-19) Virus-Neutralizing Antibodies in Human Clinical Samples. Viruses, 13(5), 893. https://doi.org/10.3390/v13050893






