Inhibitors of Protein Glycosylation Are Active against the Coronavirus Severe Acute Respiratory Syndrome Coronavirus SARS-CoV-2
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells, Virus and Antiviral Assay
2.2. Drugs and Proteins
2.3. Plaque Reduction Assay
2.4. Immunofluorescence, Immunoblotting, and Flow-Cytometry
2.5. Viral RNA Quantification
2.6. High Content Assay
2.7. Statistics
3. Results
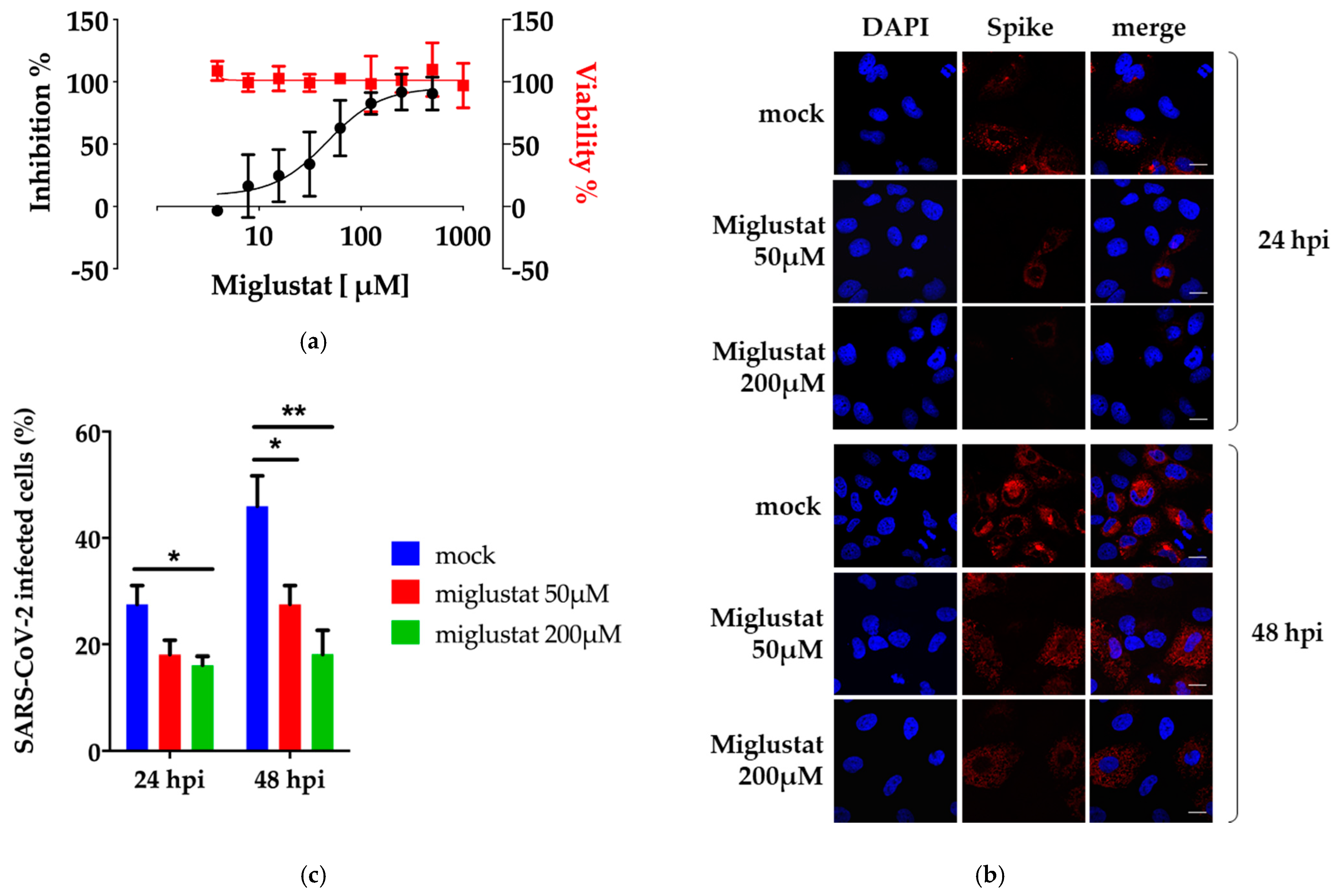
3.1. Anti-SARS-CoV-2 Activity of Miglustat in Vero E6 and Huh7 Cells
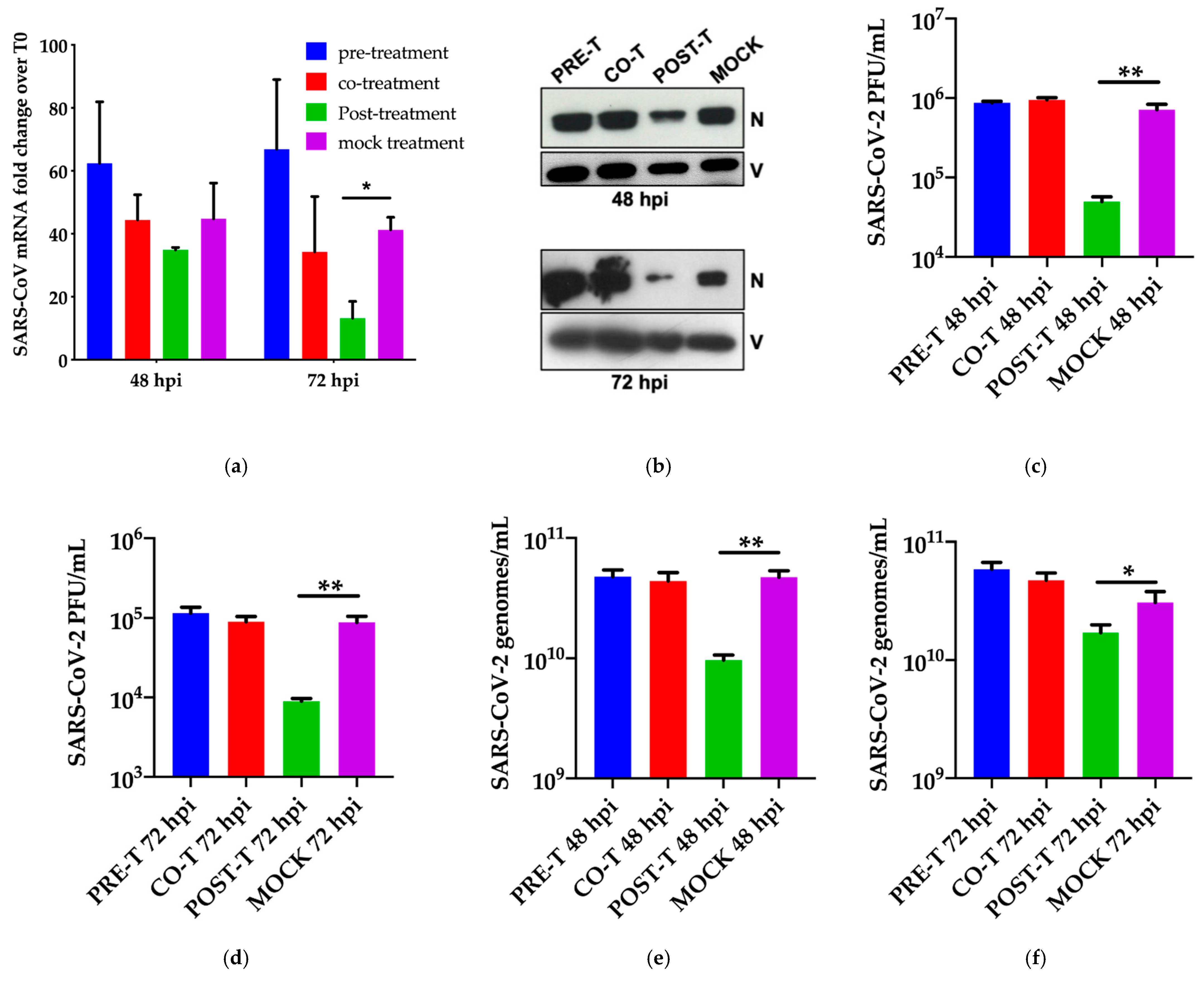
3.2. Dissection of Miglustat Antiviral Activity by Time of Addition Experiments
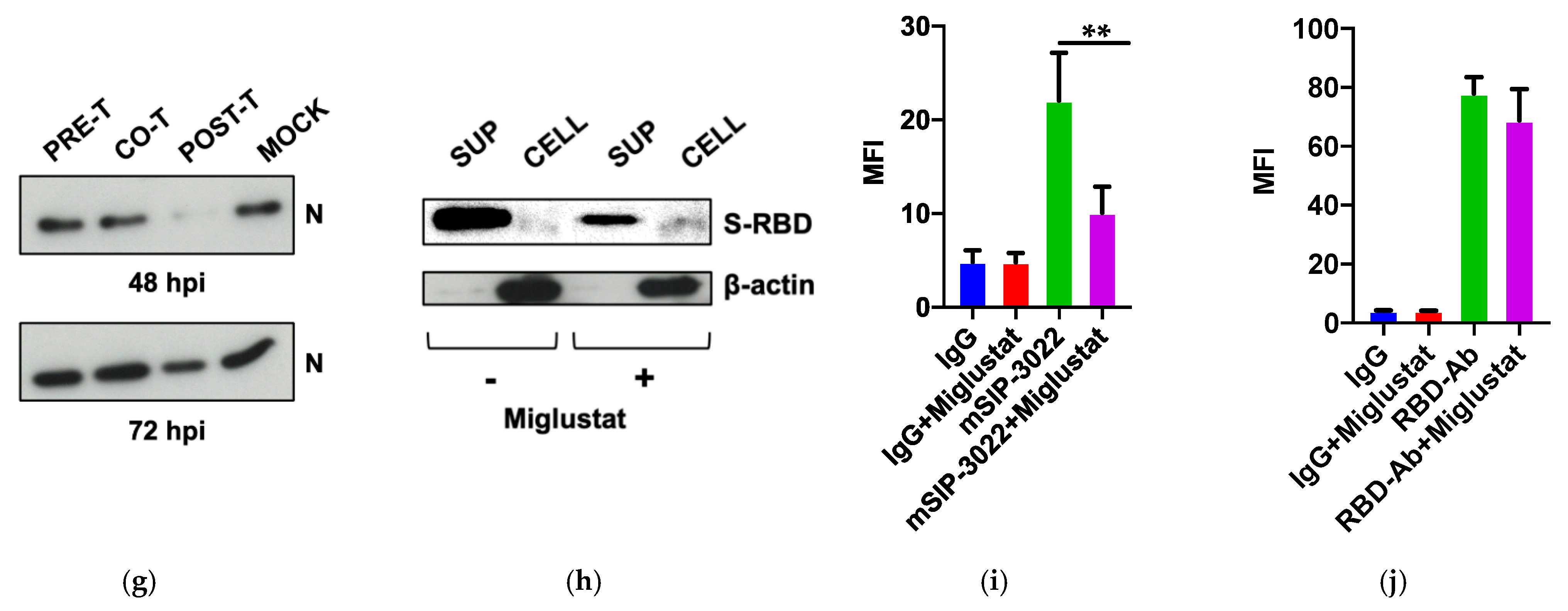
3.3. Effect of Miglustat on the Spike Protein
3.4. Anti-SARS-CoV-2 Activity of Miglustat and Celgosivir
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef]
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.M.; Wang, W.; Song, Z.G.; Hu, Y.; Tao, Z.W.; Tian, J.H.; Pei, Y.Y.; et al. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef] [PubMed]
- WHO. World Health Organization-Coronavirus Disease 2019 (COVID-19). Weekly Epidemiological Update. Available online: https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19---23-march-2021 (accessed on 29 April 2021).
- V’Kovski, P.; Kratzel, A.; Steiner, S.; Stalder, H.; Thiel, V. Coronavirus biology and replication: Implications for SARS-CoV-2. Nat. Rev. Microbiol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Xie, M.; Chen, Q. Insight into 2019 novel coronavirus-An updated interim review and lessons from SARS-CoV and MERS-CoV. Int. J. Infect. Dis. 2020, 94, 119–124. [Google Scholar] [CrossRef] [PubMed]
- Tay, M.Z.; Poh, C.M.; Renia, L.; MacAry, P.A.; Ng, L.F.P. The trinity of COVID-19: Immunity, inflammation and intervention. Nat. Rev. Immunol. 2020. [Google Scholar] [CrossRef]
- Sanders, J.M.; Monogue, M.L.; Jodlowski, T.Z.; Cutrell, J.B. Pharmacologic Treatments for Coronavirus Disease 2019 (COVID-19): A Review. JAMA 2020. [Google Scholar] [CrossRef]
- Liu, Z.; VanBlargan, L.A.; Bloyet, L.M.; Rothlauf, P.W.; Chen, R.E.; Stumpf, S.; Zhao, H.; Errico, J.M.; Theel, E.S.; Liebeskind, M.J.; et al. Identification of SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. Cell Host. Microbe 2021, 29, 477–488. [Google Scholar] [CrossRef]
- Elbein, A.D. Glycosidase inhibitors: Inhibitors of N-linked oligosaccharide processing. FASEB J. 1991, 5, 3055–3063. [Google Scholar] [CrossRef]
- Fischl, M.A.; Resnick, L.; Coombs, R.; Kremer, A.B.; Pottage, J.C., Jr.; Fass, R.J.; Fife, K.H.; Powderly, W.G.; Collier, A.C.; Aspinall, R.L.; et al. The safety and efficacy of combination N-butyl-deoxynojirimycin (SC-48334) and zidovudine in patients with HIV-1 infection and 200-500 CD4 cells/mm3. J. Acquir. Immune Defic. Syndr. 1994, 7, 139–147. [Google Scholar]
- Tierney, M.; Pottage, J.; Kessler, H.; Fischl, M.; Richman, D.; Merigan, T.; Powderly, W.; Smith, S.; Karim, A.; Sherman, J.; et al. The tolerability and pharmacokinetics of N-butyl-deoxynojirimycin in patients with advanced HIV disease (ACTG 100). The AIDS Clinical Trials Group (ACTG) of the National Institute of Allergy and Infectious Diseases. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 1995, 10, 549–553. [Google Scholar] [CrossRef]
- Dwek, R.A.; Butters, T.D.; Platt, F.M.; Zitzmann, N. Targeting glycosylation as a therapeutic approach. Nat. Rev. Drug Discov 2002, 1, 65–75. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.; Warren, T.K.; Zhao, X.; Gill, T.; Guo, F.; Wang, L.; Comunale, M.A.; Du, Y.; Alonzi, D.S.; Yu, W.; et al. Small molecule inhibitors of ER alpha-glucosidases are active against multiple hemorrhagic fever viruses. Antiviral. Res. 2013, 98, 432–440. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.F.; Lee, C.J.; Liao, C.L.; Dwek, R.A.; Zitzmann, N.; Lin, Y.L. Antiviral effects of an iminosugar derivative on flavivirus infections. J. Virol. 2002, 76, 3596–3604. [Google Scholar] [CrossRef]
- Platt, F.M.; d’Azzo, A.; Davidson, B.L.; Neufeld, E.F.; Tifft, C.J. Lysosomal storage diseases. Nat. Rev. Dis. Primers 2018, 4, 27. [Google Scholar] [CrossRef] [PubMed]
- Sunkara, P.S.; Taylor, D.L.; Kang, M.S.; Bowlin, T.L.; Liu, P.S.; Tyms, A.S.; Sjoerdsma, A. Anti-HIV activity of castanospermine analogues. Lancet 1989, 1, 1206. [Google Scholar] [CrossRef]
- Taylor, D.L.; Sunkara, P.S.; Liu, P.S.; Kang, M.S.; Bowlin, T.L.; Tyms, A.S. 6-0-butanoylcastanospermine (MDL 28,574) inhibits glycoprotein processing and the growth of HIVs. AIDS 1991, 5, 693–698. [Google Scholar] [CrossRef]
- Durantel, D. Celgosivir, an alpha-glucosidase I inhibitor for the potential treatment of HCV infection. Curr. Opin. Investig. Drugs 2009, 10, 860–870. [Google Scholar] [PubMed]
- Low, J.G.; Sung, C.; Wijaya, L.; Wei, Y.; Rathore, A.P.S.; Watanabe, S.; Tan, B.H.; Toh, L.; Chua, L.T.; Hou, Y.; et al. Efficacy and safety of celgosivir in patients with dengue fever (CELADEN): A phase 1b, randomised, double-blind, placebo-controlled, proof-of-concept trial. Lancet Infect. Dis. 2014, 14, 706–715. [Google Scholar] [CrossRef]
- Watanabe, S.; Chan, K.W.; Dow, G.; Ooi, E.E.; Low, J.G.; Vasudevan, S.G. Optimizing celgosivir therapy in mouse models of dengue virus infection of serotypes 1 and 2: The search for a window for potential therapeutic efficacy. Antiviral. Res. 2016, 127, 10–19. [Google Scholar] [CrossRef]
- Sung, C.; Wei, Y.; Watanabe, S.; Lee, H.S.; Khoo, Y.M.; Fan, L.; Rathore, A.P.; Chan, K.W.; Choy, M.M.; Kamaraj, U.S.; et al. Extended Evaluation of Virological, Immunological and Pharmacokinetic Endpoints of CELADEN: A Randomized, Placebo-Controlled Trial of Celgosivir in Dengue Fever Patients. PLoS Negl. Trop. Dis. 2016, 10, e0004851. [Google Scholar] [CrossRef]
- Milani, M.; Donalisio, M.; Milan Bonotto, R.; Scheneider, E.; Arduino, I.; Boni, F.; Lembo, D.; Marcello, A.; Mastrangelo, E. Combined in silico docking and in vitro antiviral testing for drug repurposing identified lurasidone and elbasvir as SARS-CoV-2 and HCoV-OC43 inhibitors. BioRxiv 2020. [Google Scholar] [CrossRef]
- Licastro, D.; Rajasekharan, S.; Dal Monego, S.; Segat, L.; D’Agaro, P.; Marcello, A. Isolation and Full-Length Genome Characterization of SARS-CoV-2 from COVID-19 Cases in Northern Italy. J. Virol. 2020, 94. [Google Scholar] [CrossRef] [PubMed]
- Stadlbauer, D.; Amanat, F.; Chromikova, V.; Jiang, K.; Strohmeier, S.; Arunkumar, G.A.; Tan, J.; Bhavsar, D.; Capuano, C.; Kirkpatrick, E.; et al. SARS-CoV-2 Seroconversion in Humans: A Detailed Protocol for a Serological Assay, Antigen Production, and Test Setup. Curr. Protoc. Microbiol. 2020, 57, e100. [Google Scholar] [CrossRef] [PubMed]
- Marcello, A.; Civra, A.; Milan Bonotto, R.; Nascimento Alves, L.; Rajasekharan, S.; Giacobone, C.; Caccia, C.; Cavalli, R.; Adami, M.; Brambilla, P.; et al. The cholesterol metabolite 27-hydroxycholesterol inhibits SARS-CoV-2 and is markedly decreased in COVID-19 patients. Redox. Biol. 2020, 36, 101682. [Google Scholar] [CrossRef]
- ter Meulen, J.; van den Brink, E.N.; Poon, L.L.; Marissen, W.E.; Leung, C.S.; Cox, F.; Cheung, C.Y.; Bakker, A.Q.; Bogaards, J.A.; van Deventer, E.; et al. Human monoclonal antibody combination against SARS coronavirus: Synergy and coverage of escape mutants. PLoS Med. 2006, 3, e237. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Li, C.; Huang, A.; Xia, S.; Lu, S.; Shi, Z.; Lu, L.; Jiang, S.; Yang, Z.; Wu, Y.; et al. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg. Microbes Infect. 2020, 9, 382–385. [Google Scholar] [CrossRef]
- Petris, G.; Bestagno, M.; Arnoldi, F.; Burrone, O.R. New tags for recombinant protein detection and O-glycosylation reporters. PLoS ONE 2014, 9, e96700. [Google Scholar] [CrossRef]
- Carletti, T.; Zakaria, M.K.; Faoro, V.; Reale, L.; Kazungu, Y.; Licastro, D.; Marcello, A. Viral priming of cell intrinsic innate antiviral signaling by the unfolded protein response. Nat. Commun. 2019, 10, 3889. [Google Scholar] [CrossRef]
- Milani, M.; Donalisio, M.; Bonotto, R.M.; Schneider, E.; Arduino, I.; Boni, F.; Lembo, D.; Marcello, A.; Mastrangelo, E. Combined in silico and in vitro approaches identified the antipsychotic drug lurasidone and the antiviral drug elbasvir as SARS-CoV2 and HCoV-OC43 inhibitors. Antiviral. Res. 2021, 189, 105055. [Google Scholar] [CrossRef]
- Ritchie, G.; Harvey, D.J.; Feldmann, F.; Stroeher, U.; Feldmann, H.; Royle, L.; Dwek, R.A.; Rudd, P.M. Identification of N-linked carbohydrates from severe acute respiratory syndrome (SARS) spike glycoprotein. Virology 2010, 399, 257–269. [Google Scholar] [CrossRef]
- Fukushi, M.; Yoshinaka, Y.; Matsuoka, Y.; Hatakeyama, S.; Ishizaka, Y.; Kirikae, T.; Sasazuki, T.; Miyoshi-Akiyama, T. Monitoring of S protein maturation in the endoplasmic reticulum by calnexin is important for the infectivity of severe acute respiratory syndrome coronavirus. J. Virol. 2012, 86, 11745–11753. [Google Scholar] [CrossRef] [PubMed]
- Yuan, M.; Wu, N.C.; Zhu, X.; Lee, C.D.; So, R.T.Y.; Lv, H.; Mok, C.K.P.; Wilson, I.A. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science 2020, 368, 630–633. [Google Scholar] [CrossRef] [PubMed]
- Zakaria, M.K.; Carletti, T.; Marcello, A. Cellular Targets for the Treatment of Flavivirus Infections. Front. Cell Infect. Microbiol. 2018, 8, 398. [Google Scholar] [CrossRef]
- Williams, S.J.; Goddard-Borger, E.D. alpha-glucosidase inhibitors as host-directed antiviral agents with potential for the treatment of COVID-19. Biochem Soc. Trans. 2020, 48, 1287–1295. [Google Scholar] [CrossRef]
- van Giersbergen, P.L.; Dingemanse, J. Influence of food intake on the pharmacokinetics of miglustat, an inhibitor of glucosylceramide synthase. J. Clin. Pharmacol. 2007, 47, 1277–1282. [Google Scholar] [CrossRef] [PubMed]
- Nunes-Santos, C.J.; Kuehn, H.S.; Rosenzweig, S.D. N-Glycan Modification in Covid-19 Pathophysiology: In vitro Structural Changes with Limited Functional Effects. J. Clin. Immunol. 2021, 41, 335–344. [Google Scholar] [CrossRef] [PubMed]
- Helenius, A.; Aebi, M. Intracellular functions of N-linked glycans. Science 2001, 291, 2364–2369. [Google Scholar] [CrossRef]
- Zhao, X.; Guo, F.; Comunale, M.A.; Mehta, A.; Sehgal, M.; Jain, P.; Cuconati, A.; Lin, H.; Block, T.M.; Chang, J.; et al. Inhibition of endoplasmic reticulum-resident glucosidases impairs severe acute respiratory syndrome coronavirus and human coronavirus NL63 spike protein-mediated entry by altering the glycan processing of angiotensin I-converting enzyme 2. Antimicrob. Agents Chemother. 2015, 59, 206–216. [Google Scholar] [CrossRef] [PubMed]
- Clarke, E.C.; Nofchissey, R.A.; Ye, C.; Bradfute, S.B. The iminosugars celgosivir, castanospermine and UV-4 inhibit SARS-CoV-2 replication. Glycobiology 2020. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rajasekharan, S.; Milan Bonotto, R.; Nascimento Alves, L.; Kazungu, Y.; Poggianella, M.; Martinez-Orellana, P.; Skoko, N.; Polez, S.; Marcello, A. Inhibitors of Protein Glycosylation Are Active against the Coronavirus Severe Acute Respiratory Syndrome Coronavirus SARS-CoV-2. Viruses 2021, 13, 808. https://doi.org/10.3390/v13050808
Rajasekharan S, Milan Bonotto R, Nascimento Alves L, Kazungu Y, Poggianella M, Martinez-Orellana P, Skoko N, Polez S, Marcello A. Inhibitors of Protein Glycosylation Are Active against the Coronavirus Severe Acute Respiratory Syndrome Coronavirus SARS-CoV-2. Viruses. 2021; 13(5):808. https://doi.org/10.3390/v13050808
Chicago/Turabian StyleRajasekharan, Sreejith, Rafaela Milan Bonotto, Lais Nascimento Alves, Yvette Kazungu, Monica Poggianella, Pamela Martinez-Orellana, Natasa Skoko, Sulena Polez, and Alessandro Marcello. 2021. "Inhibitors of Protein Glycosylation Are Active against the Coronavirus Severe Acute Respiratory Syndrome Coronavirus SARS-CoV-2" Viruses 13, no. 5: 808. https://doi.org/10.3390/v13050808
APA StyleRajasekharan, S., Milan Bonotto, R., Nascimento Alves, L., Kazungu, Y., Poggianella, M., Martinez-Orellana, P., Skoko, N., Polez, S., & Marcello, A. (2021). Inhibitors of Protein Glycosylation Are Active against the Coronavirus Severe Acute Respiratory Syndrome Coronavirus SARS-CoV-2. Viruses, 13(5), 808. https://doi.org/10.3390/v13050808





