Xeno-Nucleic Acid (XNA) 2’-Fluoro-Arabino Nucleic Acid (FANA) Aptamers to the Receptor-Binding Domain of SARS-CoV-2 S Protein Block ACE2 Binding
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Methods
2.2.1. End-Labeling of Oligonucleotides with T4 Polynucleotide Kinase
2.2.2. Selection of FANA Aptamers with SARS-CoV-2 RBD Using Magnetic Dynabeads™
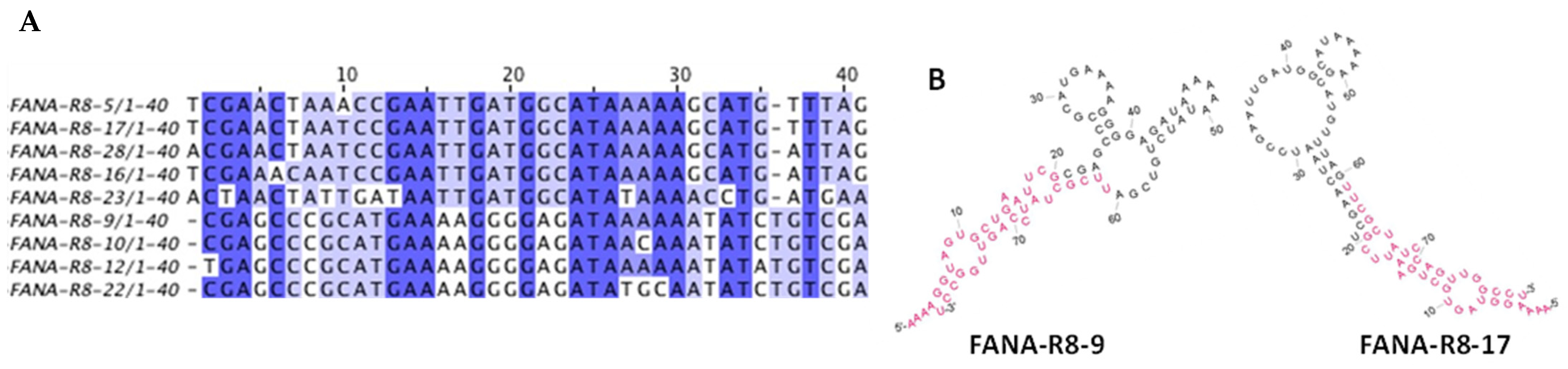
2.2.3. Sequence Analysis of FANA Products Recovered from Round 8
2.2.4. Determination of Apparent Equilibrium Dissociation Constant (KD,app) Using Nitrocellulose Filter Binding Assays
2.2.5. Competition Binding Assays
2.2.6. Dissociation Constant (koff) and Half-Life (t1/2) Determinations
2.2.7. Binding Inhibitions Analysis
3. Results
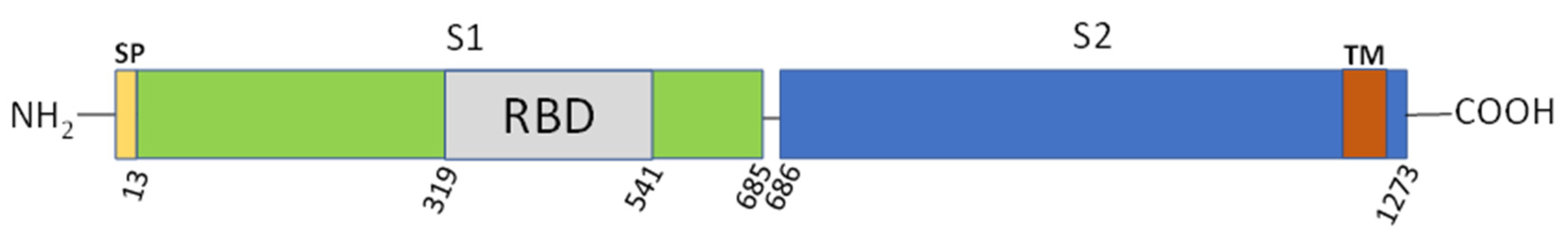
3.1. Selection of FANA Aptamers against the Spike Receptor Binding Domain (RBD)
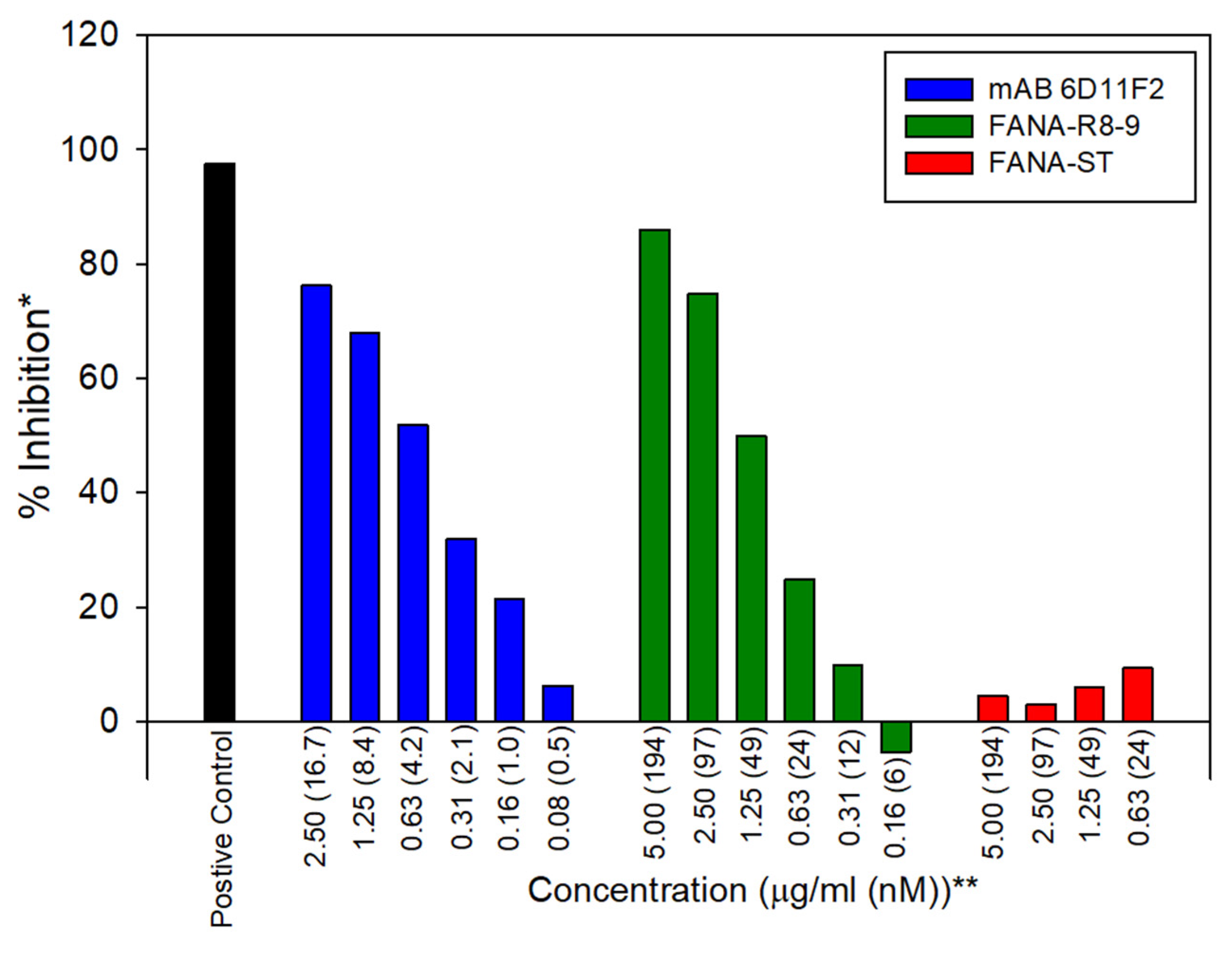
3.2. Functional Assessment of RBD-Binding Aptamers
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Perlman, S. Another Decade, Another Coronavirus. N. Engl. J. Med. 2020, 382, 760–762. [Google Scholar] [CrossRef]
- Civljak, R.; Markotic, A.; Kuzman, I. The third coronavirus epidemic in the third millennium: Whats next? Croat. Med. J. 2020, 61, 1–4. [Google Scholar] [CrossRef]
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed]
- Hamming, I.; Timens, W.; Bulthuis, M.L.; Lely, A.T.; Navis, G.; van Goor, H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 2004, 203, 631–637. [Google Scholar] [CrossRef]
- Sims, A.C.; Baric, R.S.; Yount, B.; Burkett, S.E.; Collins, P.L.; Pickles, R.J. Severe acute respiratory syndrome coronavirus infection of human ciliated airway epithelia: Role of ciliated cells in viral spread in the conducting airways of the lungs. J. Virol. 2005, 79, 15511–15524. [Google Scholar] [CrossRef]
- Taylor, P.C.; Adams, A.C.; Hufford, M.M.; de la Torre, I.; Winthrop, K.; Gottlieb, R.L. Neutralizing monoclonal antibodies for treatment of COVID-19. Nat. Rev. Immunol. 2021, 21, 382–393. [Google Scholar] [CrossRef]
- Jaworski, J.P. Neutralizing monoclonal antibodies for COVID-19 treatment and prevention. Biomed. J. 2021, 44, 7–17. [Google Scholar] [CrossRef]
- Marovich, M.; Mascola, J.R.; Cohen, M.S. Monoclonal Antibodies for Prevention and Treatment of COVID-19. JAMA 2020, 324, 131–132. [Google Scholar] [CrossRef] [PubMed]
- Prasad, R. Aptamer-based assay developed for coronavirus detection. The Hindu, 13 July 2020. [Google Scholar]
- Brody, E.N.; Gold, L. Aptamers as therapeutic and diagnostic agents. J. Biotechnol. 2000, 74, 5–13. [Google Scholar] [CrossRef]
- Deisingh, A.K. Aptamer-based biosensors: Biomedical applications. Handb. Exp. Pharmacol. 2006, 173, 341–357. [Google Scholar]
- Gold, L. The SELEX process: A surprising source of therapeutic and diagnostic compounds. Harvey Lect. 1995, 91, 47–57. [Google Scholar] [PubMed]
- James, W. Aptamers in the virologists’ toolkit. J. Gen. Virol. 2007, 88, 351–364. [Google Scholar] [CrossRef] [PubMed]
- Joshi, P.J.; Fisher, T.S.; Prasad, V.R. Anti-HIV inhibitors based on nucleic acids: Emergence of aptamers as potent antivirals. Curr. Drug Targets Infect. Disord. 2003, 3, 383–400. [Google Scholar] [CrossRef]
- Nimjee, S.M.; Rusconi, C.P.; Sullenger, B.A. Aptamers: An emerging class of therapeutics. Annu. Rev. Med. 2005, 56, 555–583. [Google Scholar] [CrossRef]
- Porschewski, P.; Grattinger, M.A.; Klenzke, K.; Erpenbach, A.; Blind, M.R.; Schafer, F. Using aptamers as capture reagents in bead-based assay systems for diagnostics and hit identification. J. Biomol. Screen. 2006, 11, 773–781. [Google Scholar] [CrossRef][Green Version]
- Zhang, Z.; Blank, M.; Schluesener, H.J. Nucleic acid aptamers in human viral disease. Arch. Immunol. Ther. Exp. (Warsz.) 2004, 52, 307–315. [Google Scholar]
- Mok, W.; Li, Y. Recent Progress in Nucleic Acid Aptamer-Based Biosensors and Bioassays. Sensors 2008, 8, 7050–7084. [Google Scholar] [CrossRef]
- Andreola, M.L.; Pileur, F.; Calmels, C.; Ventura, M.; Tarrago-Litvak, L.; Toulme, J.J.; Litvak, S. DNA aptamers selected against the HIV-1 RNase H display in vitro antiviral activity. Biochemistry 2001, 40, 10087–10094. [Google Scholar] [CrossRef]
- de Soultrait, V.R.; Lozach, P.Y.; Altmeyer, R.; Tarrago-Litvak, L.; Litvak, S.; Andreola, M.L. DNA aptamers derived from HIV-1 RNase H inhibitors are strong anti-integrase agents. J. Mol. Biol. 2002, 324, 195–203. [Google Scholar] [CrossRef]
- Ferguson, M.R.; Rojo, D.R.; Somasunderam, A.; Thiviyanathan, V.; Ridley, B.D.; Yang, X.; Gorenstein, D.G. Delivery of double-stranded DNA thioaptamers into HIV-1 infected cells for antiviral activity. Biochem. Biophys. Res. Commun. 2006, 344, 792–797. [Google Scholar] [CrossRef] [PubMed]
- Fisher, T.S.; Joshi, P.; Prasad, V.R. HIV-1 reverse transcriptase mutations that confer decreased in vitro susceptibility to anti-RT DNA aptamer RT1t49 confer cross resistance to other anti-RT aptamers but not to standard RT inhibitors. AIDS Res. Ther. 2005, 2, 8. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hotoda, H.; Koizumi, M.; Koga, R.; Kaneko, M.; Momota, K.; Ohmine, T.; Furukawa, H.; Agatsuma, T.; Nishigaki, T.; Sone, J.; et al. Biologically active oligodeoxyribonucleotides. 5. 5’-End-substituted d(TGGGAG) possesses anti-human immunodeficiency virus type 1 activity by forming a G-quadruplex structure. J. Med. Chem. 1998, 41, 3655–3663. [Google Scholar] [CrossRef] [PubMed]
- Joshi, P.J.; North, T.W.; Prasad, V.R. Aptamers directed to HIV-1 reverse transcriptase display greater efficacy over small hairpin RNAs targeted to viral RNA in blocking HIV-1 replication. Mol. Ther. 2005, 11, 677–686. [Google Scholar] [CrossRef]
- Kolb, G.; Reigadas, S.; Castanotto, D.; Faure, A.; Ventura, M.; Rossi, J.J.; Toulme, J.-J. Endogenous expression of an anti-TAR aptamer reduces HIV-1 replication. RNA Biol. 2006, 3, 150–156. [Google Scholar] [CrossRef] [PubMed]
- Matzen, K.; Elzaouk, L.; Matskevich, A.A.; Nitzsche, A.; Heinrich, J.; Moelling, K. RNase H-mediated retrovirus destruction in vivo triggered by oligodeoxynucleotides. Nat. Biotechnol. 2007, 25, 669–674. [Google Scholar] [CrossRef]
- Moelling, K.; Matskevich, A.; Jung, J.S. Relationship between retroviral replication and RNA interference machineries. Cold Spring Harb. Symp. Quant. Biol. 2006, 71, 365–368. [Google Scholar] [CrossRef]
- Neff, C.P.; Zhou, J.; Remling, L.; Kuruvilla, J.; Zhang, J.; Li, H.; Smith, D.D.; Swiderski, P.; Rossi, J.J.; Akkina, R. An aptamer-siRNA chimera suppresses HIV-1 viral loads and protects from helper CD4(+) T cell decline in humanized mice. Sci. Transl. Med. 2011, 3, 66ra66. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Neff, C.P.; Liu, X.; Zhang, J.; Li, H.; Smith, D.D.; Swiderski, P.; Aboellail, T.; Huang, Y.; Du, Q.; et al. Systemic administration of combinatorial dsiRNAs via nanoparticles efficiently suppresses HIV-1 infection in humanized mice. Mol. Ther. 2011, 19, 2228–2238. [Google Scholar] [CrossRef] [PubMed]
- DeStefano, J.J.; Nair, G.R. Novel aptamer inhibitors of human immunodeficiency virus reverse transcriptase. Oligonucleotides 2008, 18, 133–144. [Google Scholar] [CrossRef]
- Lai, Y.T.; DeStefano, J.J. DNA aptamers to human immunodeficiency virus reverse transcriptase selected by a primer-free SELEX method: Characterization and comparison with other aptamers. Nucleic Acid Ther. 2012, 22, 162–176. [Google Scholar] [CrossRef] [PubMed]
- Michalowski, D.; Chitima-Matsiga, R.; Held, D.M.; Burke, D.H. Novel bimodular DNA aptamers with guanosine quadruplexes inhibit phylogenetically diverse HIV-1 reverse transcriptases. Nucleic Acids Res. 2008, 36, 7124–7135. [Google Scholar] [CrossRef] [PubMed]
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Pinheiro, V.B.; Holliger, P. The XNA world: Progress towards replication and evolution of synthetic genetic polymers. Curr. Opin. Chem. Biol. 2012, 16, 245–252. [Google Scholar] [CrossRef]
- Pinheiro, V.B.; Taylor, A.I.; Cozens, C.; Abramov, M.; Renders, M.; Zhang, S.; Chaput, J.C.; Wengel, J.; Peak-Chew, S.Y.; McLaughlin, S.H.; et al. Synthetic genetic polymers capable of heredity and evolution. Science 2012, 336, 341–344. [Google Scholar] [CrossRef]
- Schmidt, M.; Pei, L. Synthetic toxicology: Where engineering meets biology and toxicology. Toxicol. Sci. 2011, 120, S204–S224. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, Z.; Gemeinhart, R.A. Progress in microRNA delivery. J. Control. Release 2013, 172, 962–974. [Google Scholar] [CrossRef]
- Watts, J.K.; Katolik, A.; Viladoms, J.; Damha, M.J. Studies on the hydrolytic stability of 2’-fluoroarabinonucleic acid (2’F-ANA). Org. Biomol. Chem. 2009, 7, 1904–1910. [Google Scholar] [CrossRef] [PubMed]
- Watts, J.K.; Damha, M.J. 2 ‘ F-arabinonucleic acids (2 ‘ F-ANA)—History, properties, and new frontiers. Can. J. Chem. 2008, 86, 641–656. [Google Scholar] [CrossRef]
- Alves Ferreira-Bravo, I.; Cozens, C.; Holliger, P.; DeStefano, J.J. Selection of 2’-deoxy-2’-fluoroarabinonucleotide (FANA) aptamers that bind HIV-1 reverse transcriptase with picomolar affinity. Nucleic Acids Res. 2015, 43, 9587–9599. [Google Scholar] [CrossRef]
- Rose, K.M.; Alves Ferreira-Bravo, I.; Li, M.; Craigie, R.; Ditzler, M.A.; Holliger, P.; DeStefano, J.J. Selection of 2’-Deoxy-2’-Fluoroarabino Nucleic Acid (FANA) Aptamers That Bind HIV-1 Integrase with Picomolar Affinity. ACS Chem. Biol. 2019, 14, 2166–2175. [Google Scholar] [CrossRef]
- Huang, Y.; Yang, C.; Xu, X.F.; Xu, W.; Liu, S.W. Structural and functional properties of SARS-CoV-2 spike protein: Potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020, 41, 1141–1149. [Google Scholar] [CrossRef]
- Xia, X. Domains and Functions of Spike Protein in SARS-CoV-2 in the Context of Vaccine Design. Viruses 2021, 13, 109. [Google Scholar]
- Rozewicki, J.; Li, S.; Amada, K.M.; Standley, D.M.; Katoh, K. MAFFT-DASH: Integrated protein sequence and structural alignment. Nucleic Acids Res. 2019, 47, W5–W10. [Google Scholar] [CrossRef] [PubMed]
- RNAfold. Available online: http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi (accessed on 1 July 2021).
- Li, J.; Zhang, Z.; Gu, J.; Stacey, H.D.; Ang, J.C.; Capretta, A.; Filipe, C.D.M.; Mossman, K.L.; Balion, C.; Salena, B.J.; et al. Diverse high-affinity DNA aptamers for wild-type and B.1.1.7 SARS-CoV-2 spike proteins from a pre-structured DNA library. Nucleic Acids Res. 2021, 49, 7267–7279. [Google Scholar] [CrossRef]
- Liu, Z.; Xiao, X.; Wei, X.; Li, J.; Yang, J.; Tan, H.; Zhu, J.; Zhang, Q.; Wu, J.; Liu, L. Composition and divergence of coronavirus spike proteins and host ACE2 receptors predict potential intermediate hosts of SARS-CoV-2. J. Med. Virol. 2020, 92, 595–601. [Google Scholar] [CrossRef]
- Song, Y.; Song, J.; Wei, X.; Huang, M.; Sun, M.; Zhu, L.; Lin, B.; Shen, H.; Zhu, Z.; Yang, C. Discovery of Aptamers Targeting the Receptor-Binding Domain of the SARS-CoV-2 Spike Glycoprotein. Anal. Chem. 2020, 92, 9895–9900. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, A.; Weber, A.; Bayin, M.; Breuers, S.; Fieberg, V.; Famulok, M.; Mayer, G. A SARS-CoV-2 Spike Binding DNA Aptamer that Inhibits Pseudovirus Infection by an RBD-Independent Mechanism. Angew. Chem. Weinheim Bergstr. Ger. 2021, 133, 10367–10373. [Google Scholar] [CrossRef]
- Yang, G.; Li, Z.; Mohammed, I.; Zhao, L.; Wei, W.; Xiao, H.; Guo, W.; Zhao, Y.; Qu, F.; Huang, Y. Identification of SARS-CoV-2-against aptamer with high neutralization activity by blocking the RBD domain of spike protein 1. Signal. Transduct. Target. Ther. 2021, 6, 227. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Liu, S.; Wei, X.; Wan, S.; Huang, M.; Song, T.; Lu, Y.; Weng, X.; Lin, Z.; Chen, H.; et al. Aptamer Blocking Strategy Inhibits SARS-CoV-2 Virus Infection. Angew. Chem. Int. Ed. Engl. 2021, 60, 10266–10272. [Google Scholar] [CrossRef]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef]
- Eremeeva, E.; Herdewijn, P. Non canonical genetic material. Curr. Opin. Biotechnol. 2019, 57, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Anosova, I.; Kowal, E.A.; Dunn, M.R.; Chaput, J.C.; Van Horn, W.D.; Egli, M. The structural diversity of artificial genetic polymers. Nucleic Acids Res. 2016, 44, 1007–1021. [Google Scholar] [CrossRef] [PubMed]
- Kasahara, Y.; Irisawa, Y.; Fujita, H.; Yahara, A.; Ozaki, H.; Obika, S.; Kuwahara, M. Capillary electrophoresis-systematic evolution of ligands by exponential enrichment selection of base- and sugar-modified DNA aptamers: Target binding dominated by 2’-O,4’-C-methylene-bridged/locked nucleic acid primer. Anal. Chem. 2013, 85, 4961–4967. [Google Scholar] [CrossRef] [PubMed]
- Trujillo, C.A.; Nery, A.A.; Alves, J.M.; Martins, A.H.; Ulrich, H. Development of the anti-VEGF aptamer to a therapeutic agent for clinical ophthalmology. Clin. Ophthalmol. 2007, 1, 393–402. [Google Scholar]
- Lee, J.H.; Canny, M.D.; De Erkenez, A.; Krilleke, D.; Ng, Y.S.; Shima, D.T.; Pardi, A.; Jucker, F. A therapeutic aptamer inhibits angiogenesis by specifically targeting the heparin binding domain of VEGF165. Proc. Natl. Acad. Sci. USA 2005, 102, 18902–18907. [Google Scholar] [CrossRef] [PubMed]
- Rohloff, J.C.; Gelinas, A.D.; Jarvis, T.C.; Ochsner, U.A.; Schneider, D.J.; Gold, L.; Janjic, N. Nucleic Acid Ligands With Protein-like Side Chains: Modified Aptamers and Their Use as Diagnostic and Therapeutic Agents. Mol. Ther. Nucleic Acids 2014, 3, e201. [Google Scholar] [CrossRef]
- Bernetti, M.; Cavalli, A.; Mollica, L. Protein-ligand (un)binding kinetics as a new paradigm for drug discovery at the crossroad between experiments and modelling. MedChemComm 2017, 8, 534–550. [Google Scholar] [CrossRef]



| a Aptamer | b RBD KD,app (nM) | c S1 KD.app (nM) | S1 (No His Tag) KD,app (nM) | d S Protein Timer KD,app (nM) |
|---|---|---|---|---|
| e FANA-ST | f 1695 ± 754 | |||
| FANA-R8–3 | 68.1 ± 16.9 | 46.4 ± 2.8 | ||
| FANA-R8–5 | 26.4 | 16.6 | ||
| FANA-R8–7 | 62.8 ± 8.4 | 31.2 ± 11.7 | ||
| FANA-R8–9 | 23.5 ± 4.6 | 14.7 ± 3.9 | 29.6 ± 3.9 | 14.4 ± 4.6 |
| FANA-R8–9 | 1.4 ± 0.4 (Delta Variant) g | |||
| h FANA-R8–9 (D-MEM+10% FBS) | 19.6 ± 6.2 | |||
| FANA-R8–15 | 43.3 ± 0.2 | 27.7 ± 5.3 | ||
| FANA-R8–17 | 29.5 ± 6.5 | 19.1 ± 7.5 | ||
| FANA-R8–22 | 21.3 | 13.3 | ||
| i S1-SARS-CoV-1 | 259 ± 161 | |||
| j CoV2-RBD-1C | 872 ± 359 | 105 ± 34 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alves Ferreira-Bravo, I.; DeStefano, J.J. Xeno-Nucleic Acid (XNA) 2’-Fluoro-Arabino Nucleic Acid (FANA) Aptamers to the Receptor-Binding Domain of SARS-CoV-2 S Protein Block ACE2 Binding. Viruses 2021, 13, 1983. https://doi.org/10.3390/v13101983
Alves Ferreira-Bravo I, DeStefano JJ. Xeno-Nucleic Acid (XNA) 2’-Fluoro-Arabino Nucleic Acid (FANA) Aptamers to the Receptor-Binding Domain of SARS-CoV-2 S Protein Block ACE2 Binding. Viruses. 2021; 13(10):1983. https://doi.org/10.3390/v13101983
Chicago/Turabian StyleAlves Ferreira-Bravo, Irani, and Jeffrey J. DeStefano. 2021. "Xeno-Nucleic Acid (XNA) 2’-Fluoro-Arabino Nucleic Acid (FANA) Aptamers to the Receptor-Binding Domain of SARS-CoV-2 S Protein Block ACE2 Binding" Viruses 13, no. 10: 1983. https://doi.org/10.3390/v13101983
APA StyleAlves Ferreira-Bravo, I., & DeStefano, J. J. (2021). Xeno-Nucleic Acid (XNA) 2’-Fluoro-Arabino Nucleic Acid (FANA) Aptamers to the Receptor-Binding Domain of SARS-CoV-2 S Protein Block ACE2 Binding. Viruses, 13(10), 1983. https://doi.org/10.3390/v13101983






