The Immune Response to SARS-CoV-2 and Variants of Concern
Abstract
:1. Background about COVID-19
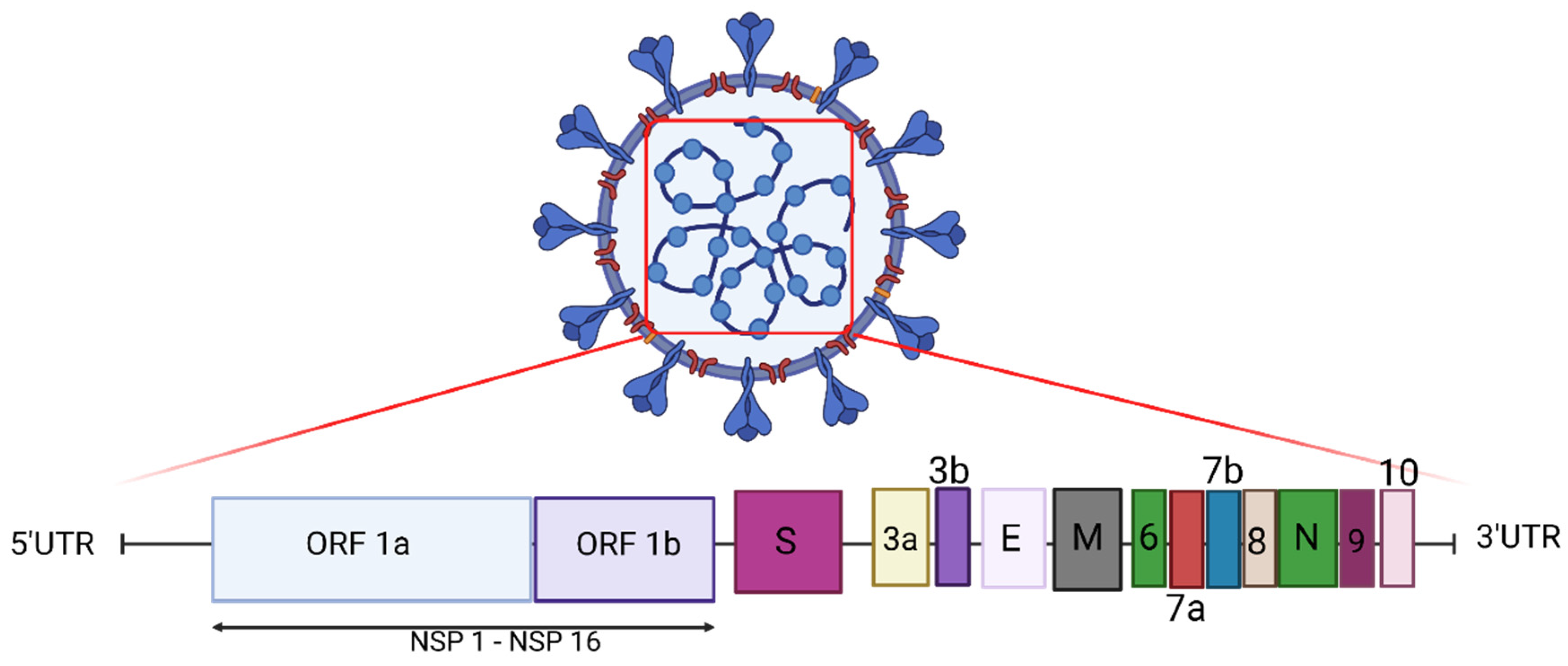
2. Genome of SARS-CoV-2
3. Entry of Virus into Host Cells
4. Adaptive Immune Response: Protection vs. Severe Disease
5. Adaptive Immune Responses in Asymptomatic, Mild, and Severe Cases of COVID-19
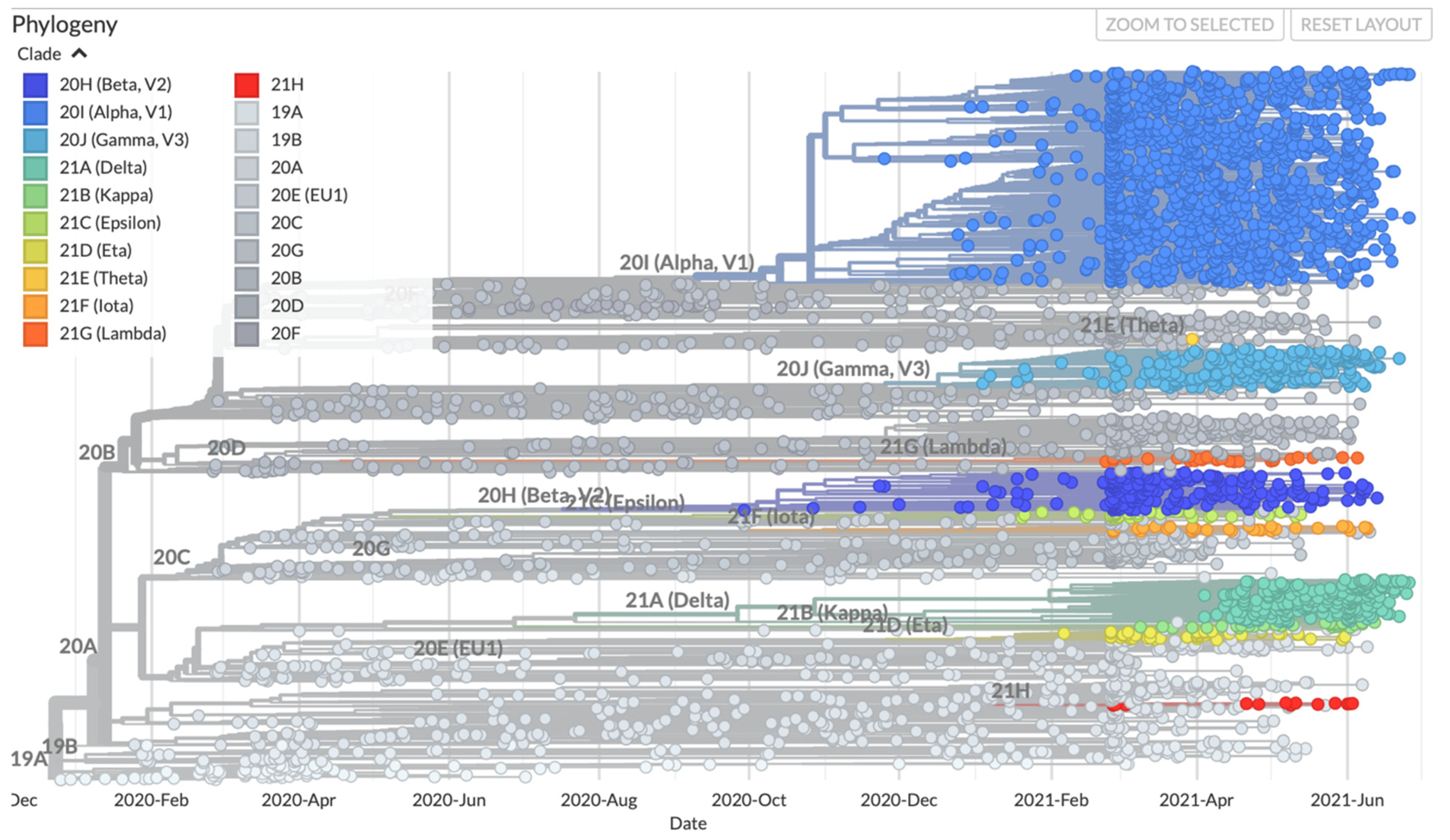
6. Recent Emergence of Variants and the Theories around the Emergence of New Variants
7. SARS-CoV-2 Variants of Concern (VOC)
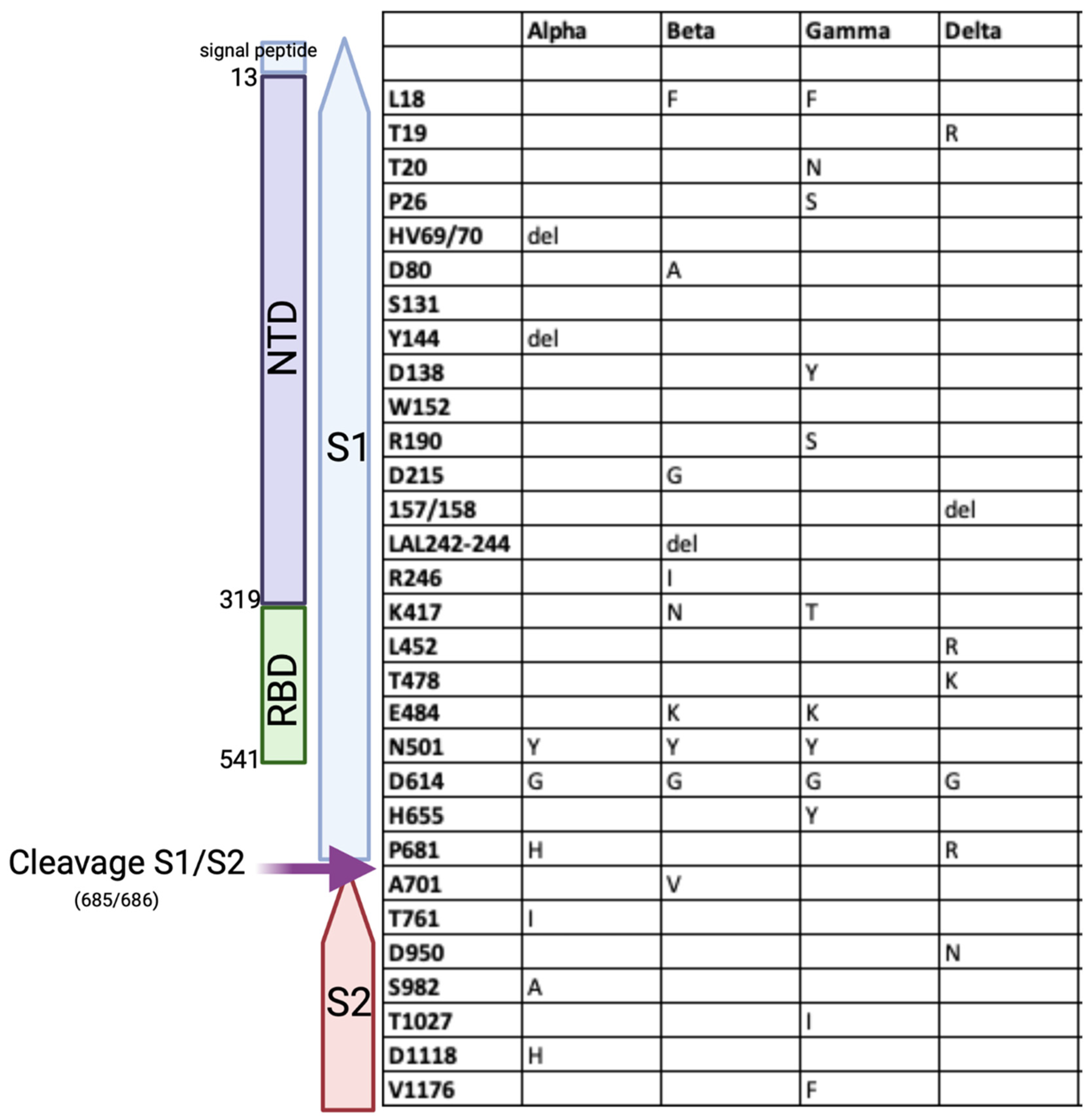
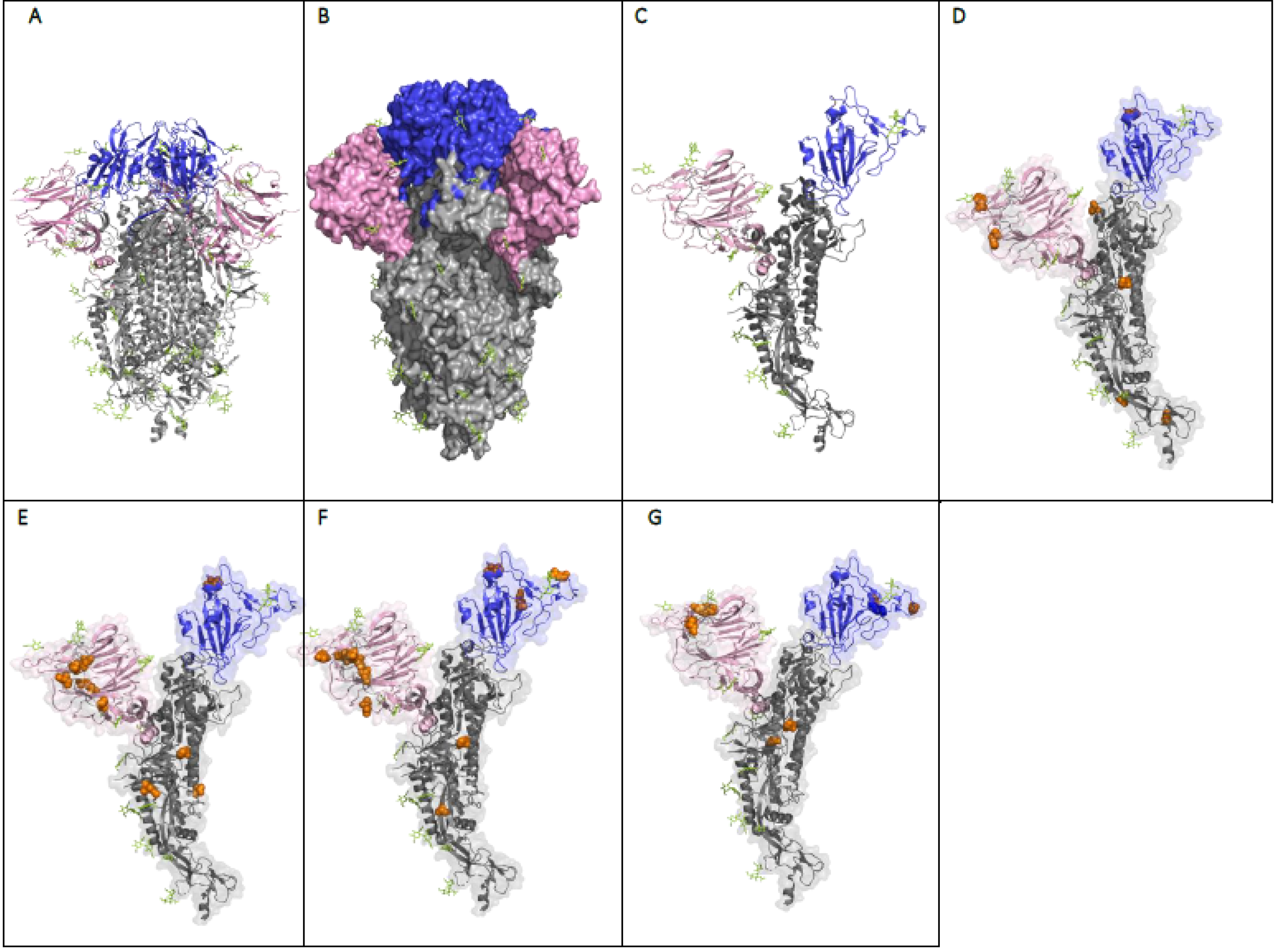
8. The Effect of the Mutations on the Structure and Function of the SARS-CoV-2 Spike Protein
9. Efficacy of Immunity from Prior Infection against SARS-CoV-2 Variants of Concern
10. Currently Available Vaccines and Their Efficacy
11. The Efficacy and Effectiveness of the Vaccines against Variants of Concern
12. The Neutralising Activity of Post-Vaccine Sera against VOCs
13. Heterologous Prime-Boost Vaccination and Variants of Concern
14. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Muralidar, S.; Ambi, S.V.; Sekaran, S.; Krishnan, U.M. The emergence of COVID-19 as a global pandemic: Understanding the epidemiology, immune response and potential therapeutic targets of SARS-CoV-2. Biochimie 2020, 179, 85–100. [Google Scholar] [CrossRef]
- Dos Santos, W.G. Natural history of COVID-19 and current knowledge on treatment therapeutic options. Biomed. Pharmacother. 2020, 129, 110493. [Google Scholar] [CrossRef]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef] [PubMed]
- WHO. Report of the WHO-China Joint Mission on Coronavirus Disease 2019 (COVID-19). Available online: https://www.who.int/docs/default-source/coronaviruse/who-china-joint-mission-on-covid-19-final-report.pdf (accessed on 12 May 2021).
- WHO Director-General’s Opening Remarks at the Media Briefing on COVID-19—11. Available online: https://www.who.int/director-general/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---11-march-2020 (accessed on 30 March 2021).
- Alanagreh, L.; Alzoughool, F.; Atoum, M. The Human Coronavirus Disease COVID-19: Its Origin, Characteristics, and Insights into Potential Drugs and Its Mechanisms. Pathogens 2020, 9, 331. [Google Scholar] [CrossRef] [PubMed]
- Bulut, C.; Kato, Y. Epidemiology of COVID-19. Turk. J. Med. Sci. 2020, 50, 563–570. [Google Scholar] [CrossRef] [PubMed]
- Casalino, L.; Gaieb, Z.; Goldsmith, J.A.; Hjorth, C.K.; Dommer, A.C.; Harbison, A.M.; Fogarty, C.A.; Barros, E.P.; Taylor, B.C.; McLellan, J.S.; et al. Shielding and Beyond: The Roles of Glycans in SARS-CoV-2 Spike Protein. ACS Cent. Sci. 2020, 6, 1722–1734. [Google Scholar] [CrossRef]
- Jaimes, J.A.; Andre, N.M.; Chappie, J.S.; Millet, J.K.; Whittaker, G.R. Phylogenetic Analysis and Structural Modeling of SARS-CoV-2 Spike Protein Reveals an Evolutionary Distinct and Proteolytically Sensitive Activation Loop. J. Mol. Biol. 2020, 432, 3309–3325. [Google Scholar] [CrossRef]
- Shang, J.; Wan, Y.; Luo, C.; Ye, G.; Geng, Q.; Auerbach, A.; Li, F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2020, 117, 11727–11734. [Google Scholar] [CrossRef]
- Taefehshokr, N.; Taefehshokr, S.; Hemmat, N.; Heit, B. Covid-19: Perspectives on Innate Immune Evasion. Front. Immunol. 2020, 11, 580641. [Google Scholar] [CrossRef]
- Ke, Z.; Oton, J.; Qu, K.; Cortese, M.; Zila, V.; McKeane, L.; Nakane, T.; Zivanov, J.; Neufeldt, C.J.; Cerikan, B.; et al. Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Nature 2020, 588, 498–502. [Google Scholar] [CrossRef]
- Chowdhury, M.A.; Hossain, N.; Kashem, M.A.; Shahid, M.A.; Alam, A. Immune response in COVID-19: A review. J. Infect. Public Health 2020, 13, 1619–1629. [Google Scholar] [CrossRef]
- Yang, X.; Dai, T.; Zhou, X.; Qian, H.; Guo, R.; Lei, L.; Zhang, X.; Zhang, D.; Shi, L.; Cheng, Y.; et al. Naturally activated adaptive immunity in COVID-19 patients. J. Cell Mol. Med. 2020, 24, 12457–12463. [Google Scholar] [CrossRef]
- Mazzoni, A.; Maggi, L.; Capone, M.; Spinicci, M.; Salvati, L.; Colao, M.G.; Vanni, A.; Kiros, S.T.; Mencarini, J.; Zammarchi, L.; et al. Cell-mediated and humoral adaptive immune responses to SARS-CoV-2 are lower in asymptomatic than symptomatic COVID-19 patients. Eur. J. Immunol. 2020, 50, 2013–2024. [Google Scholar] [CrossRef] [PubMed]
- Sette, A.; Crotty, S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell 2021, 184, 861–880. [Google Scholar] [CrossRef] [PubMed]
- Cevik, M.; Kuppalli, K.; Kindrachuk, J.; Peiris, M. Virology, transmission, and pathogenesis of SARS-CoV-2. BMJ 2020, 371, m3862. [Google Scholar] [CrossRef] [PubMed]
- Dhochak, N.; Singhal, T.; Kabra, S.K.; Lodha, R. Pathophysiology of COVID-19: Why Children Fare Better than Adults? Indian J. Pediatr. 2020, 87, 537–546. [Google Scholar] [CrossRef]
- Cohen, M.S.; Nirula, A.; Mulligan, M.J.; Novak, R.M.; Marovich, M.; Yen, C.; Stemer, A.; Mayer, S.M.; Wohl, D.; Brengle, B.; et al. Effect of Bamlanivimab vs Placebo on Incidence of COVID-19 Among Residents and Staff of Skilled Nursing and Assisted Living Facilities: A Randomized Clinical Trial. JAMA 2021, 326, 46–55. [Google Scholar] [CrossRef]
- Quast, I.; Tarlinton, D. B cell memory: Understanding COVID-19. Immunity 2021, 54, 205–210. [Google Scholar] [CrossRef]
- Brouwer, P.J.M.; Caniels, T.G.; van der Straten, K.; Snitselaar, J.L.; Aldon, Y.; Bangaru, S.; Torres, J.L.; Okba, N.M.A.; Claireaux, M.; Kerster, G.; et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science 2020, 369, 643–650. [Google Scholar] [CrossRef]
- Yuan, M.; Huang, D.; Lee, C.D.; Wu, N.C.; Jackson, A.M.; Zhu, X.; Liu, H.; Peng, L.; van Gils, M.J.; Sanders, R.W.; et al. Structural and functional ramifications of antigenic drift in recent SARS-CoV-2 variants. Science 2021, 373, 818–823. [Google Scholar] [CrossRef]
- Deshpande, A.; Harris, B.D.; Martinez-Sobrido, L.; Kobie, J.J.; Walter, M.R. Epitope Classification and RBD Binding Properties of Neutralizing Antibodies Against SARS-CoV-2 Variants of Concern. Front. Immunol. 2021, 12, 691715. [Google Scholar] [CrossRef] [PubMed]
- Barnes, C.O.; Jette, C.A.; Abernathy, M.E.; Dam, K.A.; Esswein, S.R.; Gristick, H.B.; Malyutin, A.G.; Sharaf, N.G.; Huey-Tubman, K.E.; Lee, Y.E.; et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature 2020, 588, 682–687. [Google Scholar] [CrossRef] [PubMed]
- Grifoni, A.; Weiskopf, D.; Ramirez, S.I.; Mateus, J.; Dan, J.M.; Moderbacher, C.R.; Rawlings, S.A.; Sutherland, A.; Premkumar, L.; Jadi, R.S.; et al. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell 2020, 181, 1489–1501. [Google Scholar] [CrossRef] [PubMed]
- Koutsakos, M.; Nguyen, T.H.O.; Kedzierska, K. With a Little Help from T Follicular Helper Friends: Humoral Immunity to Influenza Vaccination. J. Immunol. 2019, 202, 360–367. [Google Scholar] [CrossRef] [Green Version]
- Zhang, F.; Gan, R.; Zhen, Z.; Hu, X.; Li, X.; Zhou, F.; Liu, Y.; Chen, C.; Xie, S.; Zhang, B.; et al. Adaptive immune responses to SARS-CoV-2 infection in severe versus mild individuals. Signal Transduct. Target. Ther. 2020, 5, 156. [Google Scholar] [CrossRef]
- Ogbe, A.; Kronsteiner, B.; Skelly, D.T.; Pace, M.; Brown, A.; Adland, E.; Adair, K.; Akhter, H.D.; Ali, M.; Ali, S.E.; et al. T cell assays differentiate clinical and subclinical SARS-CoV-2 infections from cross-reactive antiviral responses. Nat. Commun. 2021, 12, 2055. [Google Scholar] [CrossRef]
- Sekine, T.; Perez-Potti, A.; Rivera-Ballesteros, O.; Stralin, K.; Gorin, J.B.; Olsson, A.; Llewellyn-Lacey, S.; Kamal, H.; Bogdanovic, G.; Muschiol, S.; et al. Robust T Cell Immunity in Convalescent Individuals with Asymptomatic or Mild COVID-19. Cell 2020, 183, 158–168. [Google Scholar] [CrossRef]
- Koblischke, M.; Traugott, M.T.; Medits, I.; Spitzer, F.S.; Zoufaly, A.; Weseslindtner, L.; Simonitsch, C.; Seitz, T.; Hoepler, W.; Puchhammer-Stockl, E.; et al. Dynamics of CD4 T Cell and Antibody Responses in COVID-19 Patients with Different Disease Severity. Front. Med. 2020, 7, 592629. [Google Scholar] [CrossRef]
- Ni, L.; Ye, F.; Cheng, M.L.; Feng, Y.; Deng, Y.Q.; Zhao, H.; Wei, P.; Ge, J.; Gou, M.; Li, X.; et al. Detection of SARS-CoV-2-Specific Humoral and Cellular Immunity in COVID-19 Convalescent Individuals. Immunity 2020, 52, 971–977. [Google Scholar] [CrossRef]
- Liu, C.; Yu, X.; Gao, C.; Zhang, L.; Zhai, H.; Hu, Y.; Liu, E.; Wang, Q.; Gao, Y.; Wei, D.; et al. Characterization of antibody responses to SARS-CoV-2 in convalescent COVID-19 patients. J. Med. Virol. 2021, 93, 2227–2233. [Google Scholar] [CrossRef]
- Whitcombe, A.L.; McGregor, R.; Craigie, A.; James, A.; Charlewood, R.; Lorenz, N.; Dickson, J.M.; Sheen, C.R.; Koch, B.; Fox-Lewis, S.; et al. Comprehensive analysis of SARS-CoV-2 antibody dynamics in New Zealand. Clin. Transl. Immunol. 2021, 10, e1261. [Google Scholar] [CrossRef]
- Lee, W.T.; Girardin, R.C.; Dupuis, A.P.; Kulas, K.E.; Payne, A.F.; Wong, S.J.; Arinsburg, S.; Nguyen, F.T.; Mendu, D.R.; Firpo-Betancourt, A.; et al. Neutralizing Antibody Responses in COVID-19 Convalescent Sera. J. Infect. Dis. 2021, 223, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Mentzer, A.J.; Liu, G.; Yao, X.; Yin, Z.; Dong, D.; Dejnirattisai, W.; Rostron, T.; Supasa, P.; Liu, C.; et al. Broad and strong memory CD4(+) and CD8(+) T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat. Immunol. 2020, 21, 1336–1345. [Google Scholar] [CrossRef]
- Malani, A.N.; Sherbeck, J.P.; Malani, P.N. Convalescent Plasma and COVID-19. JAMA 2020, 324, 524. [Google Scholar] [CrossRef] [PubMed]
- Tan, A.T.; Linster, M.; Tan, C.W.; Le Bert, N.; Chia, W.N.; Kunasegaran, K.; Zhuang, Y.; Tham, C.Y.L.; Chia, A.; Smith, G.J.D.; et al. Early induction of functional SARS-CoV-2-specific T cells associates with rapid viral clearance and mild disease in COVID-19 patients. Cell Rep. 2021, 34, 108728. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.H.; Rha, M.S.; Sa, M.; Choi, H.K.; Jeon, J.H.; Seok, H.; Park, D.W.; Park, S.H.; Jeong, H.W.; Choi, W.S.; et al. SARS-CoV-2-specific T cell memory is sustained in COVID-19 convalescent patients for 10 months with successful development of stem cell-like memory T cells. Nat. Commun. 2021, 12, 4043. [Google Scholar] [CrossRef]
- Moderbacher, C.R.; Ramirez, S.I.; Dan, J.M.; Grifoni, A.; Hastie, K.M.; Weiskopf, D.; Belanger, S.; Abbott, R.K.; Kim, C.; Choi, J.; et al. Antigen-Specific Adaptive Immunity to SARS-CoV-2 in Acute COVID-19 and Associations with Age and Disease Severity. Cell 2020, 183, 996–1012. [Google Scholar] [CrossRef] [PubMed]
- Braun, J.; Loyal, L.; Frentsch, M.; Wendisch, D.; Georg, P.; Kurth, F.; Hippenstiel, S.; Dingeldey, M.; Kruse, B.; Fauchere, F.; et al. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature 2020, 587, 270–274. [Google Scholar] [CrossRef]
- Lauring, A.S.; Hodcroft, E.B. Genetic Variants of SARS-CoV-2-What Do They Mean? JAMA 2021, 325, 529–531. [Google Scholar] [CrossRef]
- Dimonte, S.; Babakir-Mina, M.; Hama-Soor, T.; Ali, S. Genetic Variation and Evolution of the 2019 Novel Coronavirus. Public Health Genom. 2021, 24, 54–66. [Google Scholar] [CrossRef]
- Koma, T.; Adachi, S.; Doi, N.; Adachi, A.; Nomaguchi, M. Toward Understanding Molecular Bases for Biological Diversification of Human Coronaviruses: Present Status and Future Perspectives. Front. Microbiol. 2020, 11, 2016. [Google Scholar] [CrossRef] [PubMed]
- Khatamzas, E.; Rehn, A.; Muenchhoff, M.; Hellmuth, J.; Gaitzsch, E.; Weiglein, T.; Georgi, E.; Scherer, C.; Stecher, S.; Weigert, O.; et al. Emergence of multiple SARS-CoV-2 mutations in an immunocompromised host. medRxiv 2021. [Google Scholar] [CrossRef]
- Avanzato, V.A.; Matson, M.J.; Seifert, S.N.; Pryce, R.; Williamson, B.N.; Anzick, S.L.; Barbian, K.; Judson, S.D.; Fischer, E.R.; Martens, C.; et al. Case Study: Prolonged Infectious SARS-CoV-2 Shedding from an Asymptomatic Immunocompromised Individual with Cancer. Cell 2020, 183, 1901–1912. [Google Scholar] [CrossRef]
- Borges, V.; Isidro, J.; Cunha, M.; Cochicho, D.; Martins, L.; Banha, L.; Figueiredo, M.; Rebelo, L.; Trindade, M.C.; Duarte, S. Long-term evolution of SARS-CoV-2 in an immunocompromised patient with non-Hodgkin lymphoma. mSphere 2021, 28, e0024421. [Google Scholar]
- Choi, B.; Choudhary, M.C.; Regan, J.; Sparks, J.A.; Padera, R.F.; Qiu, X.; Solomon, I.H.; Kuo, H.H.; Boucau, J.; Bowman, K.; et al. Persistence and Evolution of SARS-CoV-2 in an Immunocompromised Host. N. Engl. J. Med. 2020, 383, 2291–2293. [Google Scholar] [CrossRef]
- Jungreis, I.; Sealfon, R.; Kellis, M. SARS-CoV-2 gene content and COVID-19 mutation impact by comparing 44 Sarbecovirus genomes. Res. Sq. 2020. [Google Scholar] [CrossRef]
- Bazykin, G.A.; Stanevich, O.; Danilenko, D.; Fadeev, A.; Komissarova, K.; Ivanova, A.; Sergeeva, M.; Safina, K.; Nabieva, E.; Klink, G.; et al. Emergence of Y453F and Δ69-70HV Mutations in a Lymphoma Patient with Long-Term COVID-19. Available online: https://virological.org/t/emergence-of-y453f-and-469-470hv-mutations-in-a-lymphoma-patient-with-long-term-covid-419/580 (accessed on 20 April 2021).
- Dearlove, B.; Lewitus, E.; Bai, H.; Li, Y.; Reeves, D.B.; Joyce, M.G.; Scott, P.T.; Amare, M.F.; Vasan, S.; Michael, N.L.; et al. A SARS-CoV-2 vaccine candidate would likely match all currently circulating variants. Proc. Natl. Acad. Sci. USA 2020, 117, 23652–23662. [Google Scholar] [CrossRef]
- Goldstein, S.A.; Brown, J.; Pedersen, B.S.; Quinlan, A.R.; Elde, N.C. Extensive recombination-driven coronavirus diversification expands the pool of potential pandemic pathogens. bioRxiv 2021. [Google Scholar] [CrossRef]
- Li, X.; Giorgi, E.E.; Marichannegowda, M.H.; Foley, B.; Xiao, C.; Kong, X.P.; Chen, Y.; Gnanakaran, S.; Korber, B.; Gao, F. Emergence of SARS-CoV-2 through recombination and strong purifying selection. Sci. Adv. 2020, 6, eabb9153. [Google Scholar] [CrossRef] [PubMed]
- Graham, R.L.; Baric, R.S. Recombination, reservoirs, and the modular spike: Mechanisms of coronavirus cross-species transmission. J. Virol. 2010, 84, 3134–3146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, Z.; Meng, K.; Meng, G. Genomic recombination events may reveal the evolution of coronavirus and the origin of SARS-CoV-2. Sci. Rep. 2020, 10, 21617. [Google Scholar] [CrossRef]
- Haddad, D.; John, S.E.; Mohammad, A.; Hammad, M.M.; Hebbar, P.; Channanath, A.; Nizam, R.; Al-Qabandi, S.; Al Madhoun, A.; Alshukry, A.; et al. SARS-CoV-2: Possible recombination and emergence of potentially more virulent strains. PLoS ONE 2021, 16, e0251368. [Google Scholar] [CrossRef] [PubMed]
- Latif, A.A.; Mullen, J.L.; Alkuzweny, M.; Tsueng, G.; Cano, M.; Haag, E.; Zhou, J.; Zeller, M.; Hufbauer, E.; Matteson, N.; et al. Center for Viral Systems Biology. Available online: https://outbreak.info/compare-lineages (accessed on 2 August 2021).
- Leung, K.; Shum, M.H.; Leung, G.M.; Lam, T.T.; Wu, J.T. Early transmissibility assessment of the N501Y mutant strains of SARS-CoV-2 in the United Kingdom, October to November 2020. Euro. Surveill. 2021, 26, 2002106. [Google Scholar] [CrossRef]
- Campbell, F.; Archer, B.; Laurenson-Schafer, H.; Jinnai, Y.; Konings, F.; Batra, N.; Pavlin, B.; Vandemaele, K.; Van Kerkhove, M.D.; Jombart, T.; et al. Increased transmissibility and global spread of SARS-CoV-2 variants of concern as at June 2021. Eurosurveillance 2021, 26, 2100509. [Google Scholar] [CrossRef] [PubMed]
- Washington, N.L.; Gangavarapu, K.; Zeller, M.; Bolze, A.; Cirulli, E.T.; Schiabor Barrett, K.M.; Larsen, B.B.; Anderson, C.; White, S.; Cassens, T.; et al. Emergence and rapid transmission of SARS-CoV-2 B.1.1.7 in the United States. Cell 2021, 184, 2587–2594. [Google Scholar] [CrossRef] [PubMed]
- Thorne, L.G.; Bouhaddou, M.; Reuschl, A.K.; Zuliani-Alvarez, L.; Polacco, B.; Pelin, A.; Batra, J.; Whelan, M.V.X.; Ummadi, M.; Rojc, A.; et al. Evolution of enhanced innate immune evasion by the SARS-CoV-2 B.1.1.7 UK variant. bioRxiv 2021. [Google Scholar] [CrossRef]
- Hsu, J.C.; Laurent-Rolle, M.; Pawlak, J.B.; Wilen, C.B.; Cresswell, P. Translational shutdown and evasion of the innate immune response by SARS-CoV-2 NSP14 protein. Proc. Natl. Acad. Sci. USA 2021, 118, e2101161118. [Google Scholar] [CrossRef] [PubMed]
- Harvey, W.T.; Carabelli, A.M.; Jackson, B.; Gupta, R.K.; Thomson, E.C.; Harrison, E.M.; Ludden, C.; Reeve, R.; Rambaut, A.; Consortium, C.-G.U.; et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021, 19, 409–424. [Google Scholar] [CrossRef]
- Supasa, P.; Zhou, D.; Dejnirattisai, W.; Liu, C.; Mentzer, A.J.; Ginn, H.M.; Zhao, Y.; Duyvesteyn, H.M.E.; Nutalai, R.; Tuekprakhon, A.; et al. Reduced neutralization of SARS-CoV-2 B.1.1.7 variant by convalescent and vaccine sera. Cell 2021, 184, 2201–2211. [Google Scholar] [CrossRef] [PubMed]
- Planas, D.; Veyer, D.; Baidaliuk, A.; Staropoli, I.; Guivel-Benhassine, F.; Rajah, M.M.; Planchais, C.; Porrot, F.; Robillard, N.; Puech, J.; et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature 2021, 596, 276–280. [Google Scholar] [CrossRef]
- Wang, P.; Nair, M.S.; Liu, L.; Iketani, S.; Luo, Y.; Guo, Y.; Wang, M.; Yu, J.; Zhang, B.; Kwong, P.D.; et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature 2021, 593, 130–135. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Li, X.; Li, T.; Zhang, S.; Wang, L.; Wu, X.; Liu, J. The genetic sequence, origin, and diagnosis of SARS-CoV-2. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 1629–1635. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Arora, P.; Gross, R.; Seidel, A.; Hornich, B.F.; Hahn, A.S.; Kruger, N.; Graichen, L.; Hofmann-Winkler, H.; Kempf, A.; et al. SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell 2021, 184, 2384–2393. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Ginn, H.M.; Dejnirattisai, W.; Supasa, P.; Wang, B.; Tuekprakhon, A.; Nutalai, R.; Zhou, D.; Mentzer, A.J.; Zhao, Y.; et al. Reduced neutralization of SARS-CoV-2 B.1.617 by vaccine and convalescent serum. Cell 2021, 184, 4220–4236. [Google Scholar] [CrossRef] [PubMed]
- Collier, D.A.; De Marco, A.; Ferreira, I.; Meng, B.; Datir, R.P.; Walls, A.C.; Kemp, S.A.; Bassi, J.; Pinto, D.; Silacci-Fregni, C.; et al. Sensitivity of SARS-CoV-2 B.1.1.7 to mRNA vaccine-elicited antibodies. Nature 2021, 593, 136–141. [Google Scholar] [CrossRef] [PubMed]
- Edara, V.V.; Hudson, W.H.; Xie, X.; Ahmed, R.; Suthar, M.S. Neutralizing Antibodies Against SARS-CoV-2 Variants After Infection and Vaccination. JAMA 2021, 325, 1896–1898. [Google Scholar] [CrossRef]
- Muik, A.; Wallisch, A.K.; Sanger, B.; Swanson, K.A.; Muhl, J.; Chen, W.; Cai, H.; Maurus, D.; Sarkar, R.; Tureci, O.; et al. Neutralization of SARS-CoV-2 lineage B.1.1.7 pseudovirus by BNT162b2 vaccine-elicited human sera. Science 2021, 371, 1152–1153. [Google Scholar] [CrossRef]
- Wall, E.C.; Wu, M.; Harvey, R.; Kelly, G.; Warchal, S.; Sawyer, C.; Daniels, R.; Hobson, P.; Hatipoglu, E.; Ngai, Y.; et al. Neutralising antibody activity against SARS-CoV-2 VOCs B.1.617.2 and B.1.351 by BNT162b2 vaccination. Lancet 2021, 397, 2331–2333. [Google Scholar] [CrossRef]
- Wu, K.; Werner, A.P.; Koch, M.; Choi, A.; Narayanan, E.; Stewart-Jones, G.B.E.; Colpitts, T.; Bennett, H.; Boyoglu-Barnum, S.; Shi, W.; et al. Serum Neutralizing Activity Elicited by mRNA-1273 Vaccine. N. Engl. J. Med. 2021, 384, 1468–1470. [Google Scholar] [CrossRef]
- Emary, K.R.W.; Golubchik, T.; Aley, P.K.; Ariani, C.V.; Angus, B.; Bibi, S.; Blane, B.; Bonsall, D.; Cicconi, P.; Charlton, S.; et al. Efficacy of ChAdOx1 nCoV-19 (AZD1222) vaccine against SARS-CoV-2 variant of concern 202012/01 (B.1.1.7): An exploratory analysis of a randomised controlled trial. Lancet 2021, 397, 1351–1362. [Google Scholar] [CrossRef]
- Pearson, C.A.B.; Davies, N.G.; Kucharski, A.J.; Edmunds, W.J.; Eggo, R.M. Estimates of severity and transmissibility of novel South Africa SARS-CoV-2 variant501Y. Cent. Mathem. Model. Infect. Dis. 2021. Available online: https://cmmid.github.io/topics/covid19/reports/sa-novel-variant/2021_01_11_Transmissibility_and_severity_of_501Y_V2_in_SA.pdf (accessed on 8 April 2021).
- Lazarevic, I.; Pravica, V.; Miljanovic, D.; Cupic, M. Immune Evasion of SARS-CoV-2 Emerging Variants: What Have We Learnt So Far? Viruses 2021, 13, 1192. [Google Scholar] [CrossRef]
- Cele, S.; Gazy, I.; Jackson, L.; Hwa, S.H.; Tegally, H.; Lustig, G.; Giandhari, J.; Pillay, S.; Wilkinson, E.; Naidoo, Y.; et al. Escape of SARS-CoV-2 501Y.V2 from neutralization by convalescent plasma. Nature 2021, 593, 142–146. [Google Scholar] [CrossRef] [PubMed]
- Wibmer, C.K.; Ayres, F.; Hermanus, T.; Madzivhandila, M.; Kgagudi, P.; Oosthuysen, B.; Lambson, B.E.; de Oliveira, T.; Vermeulen, M.; van der Berg, K.; et al. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat. Med. 2021, 27, 622–625. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.; Dejnirattisai, W.; Supasa, P.; Liu, C.; Mentzer, A.J.; Ginn, H.M.; Zhao, Y.; Duyvesteyn, H.M.E.; Tuekprakhon, A.; Nutalai, R.; et al. Evidence of escape of SARS-CoV-2 variant B.1.351 from natural and vaccine-induced sera. Cell 2021, 184, 2348–2361. [Google Scholar] [CrossRef]
- Davis, C.; Logan, N.; Tyson, G.; Orton, R.; Harvey, W.; Haughney, J.; Perkins, J.; Consortium, T.C.-G.U.; Peacock, T.P.; Barclay, W.S.; et al. Reduced neutralisation of the Delta (B.1.617.2) SARS-CoV-2 variant of concern following vaccination. medRxiv 2021. [Google Scholar] [CrossRef]
- Madhi, S.A.; Baillie, V.; Cutland, C.L.; Voysey, M.; Koen, A.L.; Fairlie, L.; Padayachee, S.D.; Dheda, K.; Barnabas, S.L.; Bhorat, Q.E.; et al. Efficacy of the ChAdOx1 nCoV-19 Covid-19 Vaccine against the B.1.351 Variant. N. Engl. J. Med. 2021, 384, 1885–1898. [Google Scholar] [CrossRef]
- Shen, X.; Tang, H.; Pajon, R.; Smith, G.; Glenn, G.M.; Shi, W.; Korber, B.; Montefiori, D.C. Neutralization of SARS-CoV-2 Variants B.1.429 and B.1.351. N. Engl. J. Med. 2021, 384, 2352–2354. [Google Scholar] [CrossRef]
- Faria, N.R.; Mellan, T.A.; Whittaker, C.; Claro, I.M.; Candido, D.D.S.; Mishra, S.; Crispim, M.A.E.; Sales, F.C.; Hawryluk, I.; McCrone, J.T.; et al. Genomics and epidemiology of a novel SARS-CoV-2 lineage in Manaus, Brazil. medRxiv 2021, 372, 815–821. [Google Scholar] [CrossRef]
- Naveca, F.G.; Nascimento, V.; de Souza, V.C.; Corado, A.L.; Nascimento, F.; Silva, G.; Costa, A.; Duarte, D.; Pessoa, K.; Mejia, M.; et al. COVID-19 in Amazonas, Brazil, was driven by the persistence of endemic lineages and P.1 emergence. Nat. Med. 2021, 27, 1230–1238. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Kruger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Casner, R.G.; Nair, M.S.; Wang, M.; Yu, J.; Cerutti, G.; Liu, L.; Kwong, P.D.; Huang, Y.; Shapiro, L.; et al. Increased Resistance of SARS-CoV-2 Variant P.1 to Antibody Neutralization. Cell Host Microbe 2021, 29, 747–751. [Google Scholar] [CrossRef] [PubMed]
- Dejnirattisai, W.; Zhou, D.; Supasa, P.; Liu, C.; Mentzer, A.J.; Ginn, H.M.; Zhao, Y.; Duyvesteyn, H.M.E.; Tuekprakhon, A.; Nutalai, R.; et al. Antibody evasion by the P.1 strain of SARS-CoV-2. Cell 2021, 184, 2939–2954. [Google Scholar] [CrossRef] [PubMed]
- Edara, V.V.; Pinsky, B.A.; Suthar, M.S.; Lai, L.; Davis-Gardner, M.E.; Floyd, K.; Flowers, M.W.; Wrammert, J.; Hussaini, L.; Ciric, C.R.; et al. Infection and Vaccine-Induced Neutralizing-Antibody Responses to the SARS-CoV-2 B.1.617 Variants. N. Engl. J. Med. 2021, 385, 664–666. [Google Scholar] [CrossRef]
- Kidd, M.; Richter, A.; Best, A.; Cumley, N.; Mirza, J.; Percival, B.; Mayhew, M.; Megram, O.; Ashford, F.; White, T.; et al. S-variant SARS-CoV-2 lineage B1.1.7 is associated with significantly higher viral loads in samples tested by ThermoFisher TaqPath RT-qPCR. J. Infect. Dis. 2021, 223, 1666–1670. [Google Scholar] [CrossRef]
- Mahase, E. Covid-19: What have we learnt about the new variant in the UK? BMJ 2020, 371, m4944. [Google Scholar] [CrossRef] [PubMed]
- Tegally, H.; Wilkinson, E.; Giovanetti, M.; Iranzadeh, A.; Fonseca, V.; Giandhari, J.; Doolabh, D.; Pillay, S.; San, E.J.; Msomi, N.; et al. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. medRxiv 2020. [Google Scholar] [CrossRef]
- Kemp, S.A.; Collier, D.A.; Datir, R.; Ferreira, I.; Gayed, S.; Jahun, A.; Hosmillo, M.; Rees-Spear, C.; Mlcochova, P.; Lumb, I.U.; et al. Neutralising antibodies in Spike mediated SARS-CoV-2 adaptation. medRxiv 2020. [Google Scholar] [CrossRef]
- Jangra, S.; Ye, C.; Rathnasinghe, R.; Stadlbauer, D.; Krammer, F.; Simon, V.; Martinez-Sobrido, L.; Garcia-Sastre, A.; Schotsaert, M. The E484K mutation in the SARS-CoV-2 spike protein reduces but does not abolish neutralizing activity of human convalescent and post-vaccination sera. medRxiv 2021. [Google Scholar] [CrossRef]
- Starr, T.N.; Greaney, A.J.; Hilton, S.K.; Ellis, D.; Crawford, K.H.D.; Dingens, A.S.; Navarro, M.J.; Bowen, J.E.; Tortorici, M.A.; Walls, A.C.; et al. Deep Mutational Scanning of SARS-CoV-2 Receptor Binding Domain Reveals Constraints on Folding and ACE2 Binding. Cell 2020, 182, 1295–1310. [Google Scholar] [CrossRef]
- Wise, J. Covid-19: The E484K mutation and the risks it poses. BMJ 2021, 372, n359. [Google Scholar] [CrossRef] [PubMed]
- Resende, P.C.; Bezerra, J.F.; Vasconcelos, R.; Arantes, I.; Appolinario, L.; Mendonça, A.C.; Paixao, A.C.; Rodrigues, A.C.D.; Silva, T.; Rocha, A.S. Spike E484K Mutation in the First SARS-CoV-2 Reinfection Case Confirmed in Brazil. 2020. Available online: https://virological.org/t/spike-e484k-mutation-in-the-first-sars-cov-482-reinfection-case-confirmed-in-brazil-2020/2584 (accessed on 18 May 2021).
- Nonaka, C.K.V.; Franco, M.M.; Graf, T.; de Lorenzo Barcia, C.A.; de Avila Mendonca, R.N.; de Sousa, K.A.F.; Neiva, L.M.C.; Fosenca, V.; Mendes, A.V.A.; de Aguiar, R.S.; et al. Genomic Evidence of SARS-CoV-2 Reinfection Involving E484K Spike Mutation, Brazil. Emerg. Infect. Dis. 2021, 27, 1522–1524. [Google Scholar] [CrossRef] [PubMed]
- Erol, A. Are the emerging SARS-COV-2 mutations friend or foe? Immunol. Lett. 2021, 230, 63–64. [Google Scholar] [CrossRef]
- Fiorentini, S.; Messali, S.; Zani, A.; Caccuri, F.; Giovanetti, M.; Ciccozzi, M.; Caruso, A. First detection of SARS-CoV-2 spike protein N501 mutation in Italy in August, 2020. Lancet Infect. Dis. 2021, 21, e147. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, J.; Plante, K.S.; Plante, J.A.; Xie, X.; Zhang, X.; Ku, Z.; An, Z.; Scharton, D.; Schindewolf, C.; et al. The N501Y spike substitution enhances SARS-CoV-2 transmission. bioRxiv 2021. [Google Scholar] [CrossRef]
- Garry, R.F. Mutations arising in SARS-CoV-2 spike on sustained human-to-human transmission and human-to-animal passage. Image 2021, 908, 292. [Google Scholar]
- Meng, B.; Kemp, S.A.; Papa, G.; Datir, R.; Ferreira, I.; Marelli, S.; Harvey, W.T.; Lytras, S.; Mohamed, A.; Gallo, G.; et al. Recurrent emergence of SARS-CoV-2 spike deletion H69/V70 and its role in the Alpha variant B.1.1.7. Cell Rep. 2021, 35, 109292. [Google Scholar] [CrossRef]
- McCallum, M.; Marco, A.; Lempp, F.; Tortorici, M.A.; Pinto, D.; Walls, A.C.; Beltramello, M.; Chen, A.; Liu, Z.; Zatta, F.; et al. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell 2021, 184, 2332–2347. [Google Scholar] [CrossRef]
- Cerutti, G.; Guo, Y.; Zhou, T.; Gorman, J.; Lee, M.; Rapp, M.; Reddem, E.R.; Yu, J.; Bahna, F.; Bimela, J.; et al. Potent SARS-CoV-2 neutralizing antibodies directed against spike N-terminal domain target a single supersite. Cell Host Microbe 2021, 29, 819–833. [Google Scholar] [CrossRef]
- Coutard, B.; Valle, C.; de Lamballerie, X.; Canard, B.; Seidah, N.G.; Decroly, E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antivir. Res. 2020, 176, 104742. [Google Scholar] [CrossRef]
- Stokel-Walker, C. What we know about covid-19 reinfection so far. BMJ 2021, 372, n99. [Google Scholar] [CrossRef] [PubMed]
- Hall, V.; Foulkes, S.; Charlett, A.; Atti, A.; Monk, E.; Simmons, R.; Wellington, E.; Cole, M.; Saei, A.; Oguti, B.; et al. Do antibody positive healthcare workers have lower SARS-CoV-2 infection rates than antibody negative healthcare workers? Large multi-centre prospective cohort study (the SIREN study), England: June to November 2020. medRxiv 2021. [Google Scholar] [CrossRef]
- Wilkinson, E. Covid-19 reinfection “rare” says NHS study but some may still pass the virus on. Pulse 2021. Available online: https://www.pulsetoday.co.uk/news/coronavirus/covid-19-reinfection-rare-says-nhs-study-but-some-may-still-pass-the-virus-on/ (accessed on 14 April 2021).
- Graham, M.S.; Sudre, C.H.; May, A.; Antonelli, M.; Murray, B.; Varsavsky, T.; Klaser, K.; Canas, L.S.; Molteni, E.; Modat, M.; et al. Changes in symptomatology, reinfection, and transmissibility associated with the SARS-CoV-2 variant B.1.1.7: An ecological study. Lancet Public Health 2021, 6, e335–e345. [Google Scholar] [CrossRef]
- Sabino, E.C.; Buss, L.F.; Carvalho, M.P.S.; Prete, C.A., Jr.; Crispim, M.A.E.; Fraiji, N.A.; Pereira, R.H.M.; Parag, K.V.; da Silva Peixoto, P.; Kraemer, M.U.G.; et al. Resurgence of COVID-19 in Manaus, Brazil, despite high seroprevalence. Lancet 2021, 397, 452–455. [Google Scholar] [CrossRef]
- Taylor, L. Covid-19: Researchers find higher than expected reinfections with P.1 variant among the Brazilian Amazon. BMJ 2021, 373, n1353. [Google Scholar] [CrossRef]
- Creech, C.B.; Walker, S.C.; Samuels, R.J. SARS-CoV-2 Vaccines. JAMA 2021, 325, 1318–1320. [Google Scholar] [CrossRef]
- Tanriover, M.D.; Doganay, H.L.; Akova, M.; Guner, H.R.; Azap, A.; Akhan, S.; Kose, S.; Erdinc, F.S.; Akalin, E.H.; Tabak, O.F.; et al. Efficacy and safety of an inactivated whole-virion SARS-CoV-2 vaccine (CoronaVac): Interim results of a double-blind, randomised, placebo-controlled, phase 3 trial in Turkey. Lancet 2021, 398, 213–222. [Google Scholar] [CrossRef]
- Al Kaabi, N.; Zhang, Y.; Xia, S.; Yang, Y.; Al Qahtani, M.M.; Abdulrazzaq, N.; Al Nusair, M.; Hassany, M.; Jawad, J.S.; Abdalla, J.; et al. Effect of 2 Inactivated SARS-CoV-2 Vaccines on Symptomatic COVID-19 Infection in Adults: A Randomized Clinical Trial. JAMA 2021, 326, 35–45. [Google Scholar] [CrossRef]
- Logunov, D.Y.; Dolzhikova, I.V.; Zubkova, O.V.; Tukhvatulin, A.I.; Shcheblyakov, D.V.; Dzharullaeva, A.S.; Grousova, D.M.; Erokhova, A.S.; Kovyrshina, A.V.; Botikov, A.G.; et al. Safety and immunogenicity of an rAd26 and rAd5 vector-based heterologous prime-boost COVID-19 vaccine in two formulations: Two open, non-randomised phase 1/2 studies from Russia. Lancet 2020, 396, 887–897. [Google Scholar] [CrossRef]
- Sadoff, J.; Gray, G.; Vandebosch, A.; Cardenas, V.; Shukarev, G.; Grinsztejn, B.; Goepfert, P.A.; Truyers, C.; Fennema, H.; Spiessens, B.; et al. Safety and Efficacy of Single-Dose Ad26.COV2.S Vaccine against Covid-19. N. Engl. J. Med. 2021, 384, 2187–2201. [Google Scholar] [CrossRef]
- Voysey, M.; Clemens, S.A.C.; Madhi, S.A.; Weckx, L.Y.; Folegatti, P.M.; Aley, P.K.; Angus, B.; Baillie, V.L.; Barnabas, S.L.; Bhorat, Q.E.; et al. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: An interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet 2021, 397, 99–111. [Google Scholar] [CrossRef]
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Perez Marc, G.; Moreira, E.D.; Zerbini, C.; et al. Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N. Engl. J. Med. 2020, 383, 2603–2615. [Google Scholar] [CrossRef] [PubMed]
- Haas, E.J.; Angulo, F.J.; McLaughlin, J.M.; Anis, E.; Singer, S.R.; Khan, F.; Brooks, N.; Smaja, M.; Mircus, G.; Pan, K.; et al. Impact and effectiveness of mRNA BNT162b2 vaccine against SARS-CoV-2 infections and COVID-19 cases, hospitalisations, and deaths following a nationwide vaccination campaign in Israel: An observational study using national surveillance data. Lancet 2021, 397, 1819–1829. [Google Scholar] [CrossRef]
- Sahin, U.; Muik, A.; Derhovanessian, E.; Vogler, I.; Kranz, L.M.; Vormehr, M.; Baum, A.; Pascal, K.; Quandt, J.; Maurus, D.; et al. COVID-19 vaccine BNT162b1 elicits human antibody and TH1 T cell responses. Nature 2020, 586, 594–599. [Google Scholar] [CrossRef] [PubMed]
- Baden, L.R.; El Sahly, H.M.; Essink, B.; Kotloff, K.; Frey, S.; Novak, R.; Diemert, D.; Spector, S.A.; Rouphael, N.; Creech, C.B.; et al. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 2021, 384, 403–416. [Google Scholar] [CrossRef] [PubMed]
- Lombardi, A.; Bozzi, G.; Ungaro, R.; Villa, S.; Castelli, V.; Mangioni, D.; Muscatello, A.; Gori, A.; Bandera, A. Mini Review Immunological Consequences of Immunization With COVID-19 mRNA Vaccines: Preliminary Results. Front. Immunol. 2021, 12, 657711. [Google Scholar] [CrossRef]
- FDA. mRNA-1273 Sponsor Briefing Document Addendum. Vaccines and Related Biological Products Advisory Committee. Available online: https://www.fda.gov/media/144453/download (acessed on 17 December 2020).
- Callaway, E.; Mallapaty, S. Novavax offers first evidence that COVID vaccines protect people against variants. Nature 2021, 590, 17. [Google Scholar] [CrossRef]
- Shinde, V.; Bhikha, S.; Hoosain, Z.; Archary, M.; Bhorat, Q.; Fairlie, L.; Lalloo, U.; Masilela, M.S.L.; Moodley, D.; Hanley, S.; et al. Efficacy of NVX-CoV2373 Covid-19 Vaccine against the B.1.351 Variant. N. Engl. J. Med. 2021, 384, 1899–1909. [Google Scholar] [CrossRef]
- Nasreen, S.; He, S.; Chung, H.; Brown, K.A.; Gubbay, J.B.; Buchan, S.A.; Wilson, S.E.; Sundaram, M.E.; Fell, D.B.; Chen, B. Effectiveness of COVID-19 Vaccines against Variants of Concern, Canada. medRxiv 2021. [Google Scholar] [CrossRef]
- Hall, V.J.; Foulkes, S.; Saei, A.; Andrews, N.; Oguti, B.; Charlett, A.; Wellington, E.; Stowe, J.; Gillson, N.; Atti, A.; et al. COVID-19 vaccine coverage in health-care workers in England and effectiveness of BNT162b2 mRNA vaccine against infection (SIREN): A prospective, multicentre, cohort study. Lancet 2021, 397, 1725–1735. [Google Scholar] [CrossRef]
- Abu-Raddad, L.J.; Chemaitelly, H.; Butt, A.A.; National Study Group for COVID-19 Vaccination. Effectiveness of the BNT162b2 Covid-19 Vaccine against the B.1.1.7 and B.1.351 Variants. N. Engl. J. Med. 2021, 385, 187–189. [Google Scholar] [CrossRef] [PubMed]
- Bernal, J.L.; Andrews, N.; Gower, C.; Gallagher, E.; Simmons, R.; Thelwall, S.; Tessier, E.; Groves, N.; Dabrera, G.; Myers, R. Effectiveness of COVID-19 vaccines against the B. 1.617. 2 variant. medRxiv 2021. [Google Scholar]
- Iacobucci, G. Covid-19: Two doses of Pfizer vaccine are “highly effective” against infection, hospital admission, and death, study finds. BMJ 2021, 373, n1164. [Google Scholar] [CrossRef] [PubMed]
- Hitchings, M.D.T.; Ranzani, O.T.; Scaramuzzini Torres, M.S.; de Oliveira, S.B.; Almiron, M.; Said, R.; Borg, R.; Schulz, W.L.; de Oliveira, R.D.; da Silva, P.V.; et al. Effectiveness of CoronaVac among healthcare workers in the setting of high SARS-CoV-2 Gamma variant transmission in Manaus, Brazil: A test-negative case-control study. medRxiv 2021. [Google Scholar] [CrossRef]
- Mizrahi, B.; Lotan, R.; Kalkstein, N.; Peretz, A.; Perez, G.; Ben-Tov, A.; Chodick, G.; Gazit, S.; Patalon, T. Correlation of SARS-CoV-2 Breakthrough Infections to Time-from-vaccine; Preliminary Study. medRxiv 2021. [Google Scholar] [CrossRef]
- Bar-On, Y.M.; Goldberg, Y.; Mandel, M.; Bodenheimer, O.; Freedman, L.; Kalkstein, N.; Mizrahi, B.; Alroy-Preis, S.; Ash, N.; Milo, R.; et al. BNT162b2 vaccine booster dose protection: A nationwide study from Israel. medRxiv 2021. [Google Scholar] [CrossRef]
- Tauzin, A.; Nayrac, M.; Benlarbi, M.; Gong, S.Y.; Gasser, R.; Beaudoin-Bussieres, G.; Brassard, N.; Laumaea, A.; Vezina, D.; Prevost, J.; et al. A single dose of the SARS-CoV-2 vaccine BNT162b2 elicits Fc-mediated antibody effector functions and T cell responses. Cell Host Microbe 2021, 29, 1137–1150. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Schmidt, F.; Weisblum, Y.; Muecksch, F.; Barnes, C.O.; Finkin, S.; Schaefer-Babajew, D.; Cipolla, M.; Gaebler, C.; Lieberman, J.A.; et al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature 2021, 59, 616–622. [Google Scholar] [CrossRef]
- Lustig, Y.; Nemet, I.; Kliker, L.; Zuckerman, N.; Yishai, R.; Alroy-Preis, S.; Mendelson, E.; Mandelboim, M. Neutralizing Response against Variants after SARS-CoV-2 Infection and One Dose of BNT162b2. N. Engl. J. Med. 2021, 384, 2453–2454. [Google Scholar] [CrossRef] [PubMed]
- Kardani, K.; Bolhassani, A.; Shahbazi, S. Prime-boost vaccine strategy against viral infections: Mechanisms and benefits. Vaccine 2016, 34, 413–423. [Google Scholar] [CrossRef]
- Vaine, M.; Wang, S.; Hackett, A.; Arthos, J.; Lu, S. Antibody responses elicited through homologous or heterologous prime-boost DNA and protein vaccinations differ in functional activity and avidity. Vaccine 2010, 28, 2999–3007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Parker, C.; Taaffe, J.; Solorzano, A.; Garcia-Sastre, A.; Lu, S. Heterologous HA DNA vaccine prime--inactivated influenza vaccine boost is more effective than using DNA or inactivated vaccine alone in eliciting antibody responses against H1 or H3 serotype influenza viruses. Vaccine 2008, 26, 3626–3633. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levine, M.Z.; Holiday, C.; Jefferson, S.; Gross, F.L.; Liu, F.; Li, S.; Friel, D.; Boutet, P.; Innis, B.L.; Mallett, C.P.; et al. Heterologous prime-boost with A(H5N1) pandemic influenza vaccines induces broader cross-clade antibody responses than homologous prime-boost. NPJ Vaccines 2019, 4, 22. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Shaw, R.H.; Stuart, A.S.V.; Greenland, M.; Aley, P.K.; Andrews, N.J.; Cameron, J.C.; Charlton, S.; Clutterbuck, E.A.; Collins, A.M.; et al. Safety and immunogenicity of heterologous versus homologous prime-boost schedules with an adenoviral vectored and mRNA COVID-19 vaccine (Com-COV): A single-blind, randomised, non-inferiority trial. Lancet 2021, 398, 856–869. [Google Scholar] [CrossRef]
- Mahase, E. Covid-19: Moderna and Novavax vaccines to be tested in mixing vaccines trial. BMJ 2021, 373, n971. [Google Scholar] [CrossRef]
- Borobia, A.M.; Carcas, A.J.; Perez-Olmeda, M.; Castano, L.; Bertran, M.J.; Garcia-Perez, J.; Campins, M.; Portoles, A.; Gonzalez-Perez, M.; Garcia Morales, M.T.; et al. Immunogenicity and reactogenicity of BNT162b2 booster in ChAdOx1-S-primed participants (CombiVacS): A multicentre, open-label, randomised, controlled, phase 2 trial. Lancet 2021, 398, 121–130. [Google Scholar] [CrossRef]
- Hillus, D.; Schwarz, T.; Tober-Lau, P.; Vanshylla, K.; Hastor, H.; Thibeault, C.; Jentzsch, S.; Helbig, E.T.; Lippert, L.J.; Tscheak, P.; et al. Safety, reactogenicity, and immunogenicity of homologous and heterologous prime-boost immunisation with ChAdOx1 nCoV-19 and BNT162b2: A prospective cohort study. Lancet Respir. Med. 2021. [Google Scholar] [CrossRef]
- Wu, K.; Choi, A.; Koch, M.; Ma, L.; Hill, A.; Nunna, N.; Huang, W.; Oestreicher, J.; Colpitts, T.; Bennett, H.; et al. Preliminary Analysis of Safety and Immunogenicity of a SARS-CoV-2 Variant Vaccine Booster. medRxiv 2021. [Google Scholar] [CrossRef]
- Wu, K.; Choi, A.; Koch, M.; Elbashir, S.; Ma, L.; Lee, D.; Woods, A.; Henry, C.; Palandjian, C.; Hill, A.; et al. Variant SARS-CoV-2 mRNA vaccines confer broad neutralization as primary or booster series in mice. bioRxiv 2021. [Google Scholar] [CrossRef]
- He, Q.; Mao, Q.; An, C.; Zhang, J.; Gao, F.; Bian, L.; Li, C.; Liang, Z.; Xu, M.; Wang, J. Heterologous prime-boost: Breaking the protective immune response bottleneck of COVID-19 vaccine candidates. Emerg. Microbes. Infect. 2021, 10, 629–637. [Google Scholar] [CrossRef]
| WHO Label | Variants | GSAID Lineage | Nextstrain Clade | Date of Designation | Estimated Date of Emergence | Location of Emergence | Mutations (https://covdb.stanford.edu/page/mutation-viewer/#sec_alpha; accessed on 1 July 2021) |
|---|---|---|---|---|---|---|---|
| Alpha | 20I/501Y.V1, VOC 202012/01, or B.1.1.7 | GRY (formerly GR/501Y.V1) | 20I (V1) | December 2020 | September 2020 | United Kingdom | ORF1a: T183I (PLpro), A890D (PLpro), I1412T (PLpro), Δ106–108 (nsp6), P323L (RdRP) Spike: Δ69–70, Δ144, N501Y, D614G, P681H, T716I, S982A, D1118H ORF8: insQ27, R52I, Y73C N: D3L, 235F |
| Beta | 20H/501Y.V2 or B.1.351 | GH/501Y.V2 | 20H(V2) | December 2020 | May 2020 | South Africa | ORF1: T85I (nsp2), K837N (PLpro), H26Y(nsp4), S137L (nsp4), K90R (3CL), Δ106–108 (nsp6), D135Y (RdRP), P323L (RdRP), T588I (nsp3) Spike: L18F, D80A, D215G, Δ242–244, R246I, K417N, E484K, N501Y, D614G, A701V ORF3a: Q57H, 171L E: P71L, N: T205I |
| Gamma | P.1 | GR/501Y.V3 | 20J (V3) | January 2021 | November 2020 | Brazil | ORF1: S70L (PLpro), K977Q (PLpro), Δ106–108 (nsp6), P323L (PdRP), E341D (nsp13) Spike: L18F, T20N, P26S, D138Y, R190S, K417T, E484K, N501Y, D614G, H655Y, T1027I, V1176F ORF3a: S253P ORF8: 92K N: P80R, R203K, G204R |
| Delta | B.1.617.2 | G/478K.V1 | 21A | May 2021(VOI: Apr 2021) | October 2020 | India | ORF1: A488S (PLpro), P1228 (PLpro), P1469S (PLpro), V167L (nsp4), T492I (nsp4), T77A (nsp6), P323L (RdRP), G671S (RdRP), P77L (nsp13), A394V (nsp14) Spike: T19R, Δ157–158, L452R, T478K, D614G, P681R, D950N ORF3: S26L M: I82T ORF7a: T120I, V82a ORF7b: T40I ORF8: Δ119–120 N: D63G, R203M, N377Y |
| VOC | Transmissibility | Immune Evasiveness |
|---|---|---|
| Alpha | 75% (95% CI: 70–80%) more transmissible than the pre-existing variants from October 2020–November 2020 [57]. Estimated to be 50% more transmissible in Switzerland (report, https://ispmbern.github.io/covid-19/variants/ accessed on 18 June 2021) An estimated 29% increase in transmissibility [58]. 43–90% more transmissible than previous circulating strains. Increased transmissibility of 40–42% across the US [59]. | Enhanced innate immune evasion [60]. May be due to the mutations in nsp14 [61]. Mutations in NTD and N501 are associated with virus escape from neutralising antibodies [62,63]. Reduced potency of neutralising antibodies reported. Escape from mainly NTD-specific antibodies [64,65] No significant difference to 4.5-fold reduction in neutralisation of alpha variant and the ancestral strain by convalescent plasma [66,67,68,69,70] No significant difference to a 2.6-fold reduction in neutralisation of alpha variant and the ancestral strain by post vaccination (BNT162b2) sera [66,67,69,71,72]. No significant difference to a 1.8-fold reduction in neutralisation of alpha variant and the ancestral strain by post vaccination (mRNA-1273) sera [66,70,73]. Reduced (8.9 fold) ability of post-vaccination (ChAdOx1) sera to neutralise [74]. |
| Beta | Estimated to be 1.5 times more transmissible than the previous variants [75]. An estimated 25% increase in transmissibility [58]. | Resistance to antibody mainly due to RBD mutations (K417N, E484K, N501Y) [76]. Reduced (7.9 to 13.3-fold) neutralisation of beta variant compared with the ancestral strains by convalescent plasma [65,67,68,77,78,79]. Reduced ability (4.9 to 16-fold) of post-vaccination (BNT162b2) sera to neutralise [64,65,67,68,80]. Reduced ability (12.4-fold) of post-vaccination (mRNA-1273) sera to neutralise [65,73]. Reduced (9 fold to undetectable) ability of post-vaccination (ChAdOx1) sera to neutralise [64,68,79,81]. Reduced ability (9.7- and 14.5-fold reduction) of post-vaccination (mRNA-1273 or NVX-CoV2373 respectively) sera to neutralise [82]. |
| Gamma | An estimated 38% increase in transmissibility [58]. Between 1.7 to 2.4 times more transmissible than the earlier circulating variants [83]. Could be more transmissible due to the higher viral load in the upper respiratory tract compared to prior circulating variants [84]. | Resistance to antibody mainly due to RBD mutations (K417N, E484K, N501Y) [76] Reduced (3.1 to 8.2-fold) neutralisation of gamma variant compared with the ancestral strains by convalescent plasma [68,85,86]. Reduced ability (2.6 to 5.1-fold) of post-vaccination (BNT162b2) sera to neutralise [68,85,86,87]. Reduced ability (4.8-fold) of post-vaccination (mRNA-1273) sera to neutralise [73,86]. Reduced (2.9-fold to undetectable) ability of post-vaccination (ChAdOx1) sera to neutralise [68,87]. |
| Delta | An estimated 97% increase in transmissibility compared to non-VOCs [58]. | Escape due to the combination of RBD mutations (L452R, T478K) and NTD mutations [64,76] Reduced (2.4 to 6-fold) neutralisation of delta variant compared with the ancestral strains by convalescent plasma [64,68,88]. Reduced ability (2.5 to 11.3-fold) of post-vaccination (BNT162b2) sera to neutralise [64,68,72,80,88]. Reduced ability (3-fold) of post-vaccination (mRNA-1273) sera to neutralise [88]. Reduced (4.3 to 5-fold) ability of post-vaccination (ChAdOx1) sera to neutralise [64,68]. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Torbati, E.; Krause, K.L.; Ussher, J.E. The Immune Response to SARS-CoV-2 and Variants of Concern. Viruses 2021, 13, 1911. https://doi.org/10.3390/v13101911
Torbati E, Krause KL, Ussher JE. The Immune Response to SARS-CoV-2 and Variants of Concern. Viruses. 2021; 13(10):1911. https://doi.org/10.3390/v13101911
Chicago/Turabian StyleTorbati, Elham, Kurt L. Krause, and James E. Ussher. 2021. "The Immune Response to SARS-CoV-2 and Variants of Concern" Viruses 13, no. 10: 1911. https://doi.org/10.3390/v13101911
APA StyleTorbati, E., Krause, K. L., & Ussher, J. E. (2021). The Immune Response to SARS-CoV-2 and Variants of Concern. Viruses, 13(10), 1911. https://doi.org/10.3390/v13101911






