Abstract
Background: To prioritize the development of antiviral compounds, it is necessary to compare their relative preclinical activity and clinical efficacy. Methods: We reviewed in vitro, animal model, and clinical studies of candidate anti-coronavirus compounds and placed extracted data in an online relational database. Results: As of August 2020, the Coronavirus Antiviral Research Database (CoV-RDB; covdb.stanford.edu) contained over 2800 cell culture, entry assay, and biochemical experiments, 259 animal model studies, and 73 clinical studies from over 400 published papers. SARS-CoV-2, SARS-CoV, and MERS-CoV account for 85% of the data. Approximately 75% of experiments involved compounds with known or likely mechanisms of action, including monoclonal antibodies and receptor binding inhibitors (21%), viral protease inhibitors (17%), miscellaneous host-acting inhibitors (10%), polymerase inhibitors (9%), interferons (7%), fusion inhibitors (5%), and host protease inhibitors (5%). Of 975 compounds with known or likely mechanism, 135 (14%) are licensed in the U.S. for other indications, 197 (20%) are licensed outside the U.S. or are in human trials, and 595 (61%) are pre-clinical investigational compounds. Conclusion: CoV-RDB facilitates comparisons between different candidate antiviral compounds, thereby helping scientists, clinical investigators, public health officials, and funding agencies prioritize the most promising compounds and repurposed drugs for further development.
1. Introduction
The Coronavirus Antiviral Research Database (CoV-RDB) is designed to promote uniform reporting of experimental results; to facilitate comparisons between different candidate antiviral compounds; and to help scientists, clinical investigators, public health officials, and funding agencies prioritize the most promising compounds and repurposed drugs for further development. By comprehensively reviewing published laboratory, animal model, and clinical data on potential coronavirus therapies, CoV-RDB also makes it unlikely that promising treatment approaches will be overlooked.
2. Methods/Results
CoV-RDB contains four main types of antiviral experimental data, six main lookup/explanation tables, and a registry of ongoing or planned clinical trials. The four main types of antiviral experimental data include (i) cell culture and entry assay experiments; (ii) biochemical experiments; (iii) animal model studies; and (iv) clinical studies. The six main lookup/explanation tables provide information on viruses, virus strains/isolates, tested compounds, compound targets, cell types, and animal models.
CoV-RDB data are stored in a PostgreSQL relational database, but there is not necessarily a one-to-one relationship for the tables displayed on the web and their underlying database structure. Indeed, several of the website tables contain information from more than one underlying database table. As of 14 August 2020, the CoV-RDB contains data from more than 1800 virus cell culture experiments, 465 entry assay experiments, 519 biochemical experiments, 259 animal model experiments, and 71 clinical studies from more than 310 peer-reviewed publications and 90 preprints.
Research articles are identified through incremental daily searches of PubMed and bioRxiv using the search term “coronavirus” and by citations identified through reading these papers. Publications containing experimental data are imported into a staging Zotero database and then annotated to extract the data described in the sections below. Preprints that are subsequently published in a peer-reviewed journal are identfied using a computer script that parses datasets downloaded from the Stephen B. Thacker CDC (Centers for Disease Control and Prevention) Library [1]. Clinical trial datasets are retrieved from clinicaltrials.gov and WHO (World Health Organization) ICTRP (International Clinical Trials Registry Platform). Clinical trials are linked to corresponding articles when they are published (Figure 1). The following sections describe the purpose and contents of the CoV-RDB tables displayed on the web.
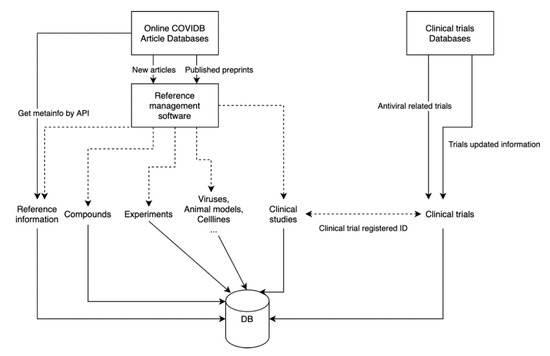
Figure 1.
Experiments and clinial trials are semi-automatically downloaded and processed, data are periodically updated.
2.1. Experimental Data Tables
2.1.1. Cell Culture and Entry Assay Experiments
The cell culture experiments table contains 13 fields, including four fields present in each of the experiment tables: reference, compound, virus category, and virus isolate/strain. The nine fields unique to the cell culture experiments table include six that describe experimental conditions and three that contain experimental results. The six experimental conditions include the (i) cells used for antiviral testing, (ii) multiplicity of infection (MOI; the virus titer divided by the number of cells), (iii) time between addition of drug and addition of virus, (iv) drug concentration(s), (v) duration of virus infection, and (vi) indicator of virus replication.
The three experiment results are the half-maximal effective concentration (EC50), percent inhibition, and the 50% cytotoxic concentration (CC50). The EC50 can only be determined using a series of compound dilutions. While the EC50 is usually reported as μM, inhibitory activity for interferons is also often reported as international units (IU)/mL and inhibitory activity for monoclonal antibodies is often reported as ng/mL. The EC50 is available for the vast majority of in vitro cell culture experiments. However, for a few experiments, the experimental setup involved a single compound concentration (rather than a dilution series). For these experiments, the percent virus inhibition with the single compound concentration is reported.
There are two tables for entry assay experiments—one for pseudovirus entry assays and another for cell–cell fusion assays. The pseudovirus assay table contains the following six unique fields: (i) pseudovirus vector, (ii) pseudovirus number, (iii) target cell type, (iv) time to addition of drug, (v) indicator of virus replication, and (vi) EC50. In the pseudovirus experiments, the virus strain is a virus construct composed of a virus that does not require a Biosafety Level 3 (BSL-3) laboratory, such as vesicular stomatitis virus (VSV) or human immunodeficiency virus type 1 (HIV-1), into which the coronavirus spike (S) gene has been cloned. This construct also has a reporter gene such as luciferase or GFP. The cell–cell fusion assay table contains the following seven unique fields: (i) effector cell type, (ii) effector cell number, (iii) target cell type, (iv) target cell number, (v) time to addition of drug, (vi) indicator of virus replication, and (vii) EC50.
2.1.2. Biochemical Experiments
The biochemical experiments table contains two unique fields: the biochemical target and the half maximal inhibitory concentration (IC50). The biochemical target is usually one of the virus enzymes including RNA-dependent RNA polymerase (RdRP), main protease (also called 3C-like protease; 3CLpro), papain-like protease (PLpro), and helicase. However, cell-free assays that test inhibitors of the spike (S) protein binding to angiotensin converting enzyme 2 (ACE2) are also included.
2.1.3. Animal Model Studies
The animal model experiments are characterized by comparisons between a group of animals receiving a treatment intervention either shortly before or after virus infection and a group of untreated virus-infected control animals. The animal model experiments table has two parts—experimental conditions and experimental results. The experimental conditions include the (i) animal model, (ii) size and route of virus challenge, (iii) treatment intervention, (iv) treatment dosage, (v) treatment timing in relation to the addition of virus, (vi) number of treated subjects, and (vii) number of control subjects. The experimental results, which often depend on the study, include endpoints such as mortality, weight loss, fever, respiratory rate, lung pathology, and virus load measurements. The reduction of endpoint severity is reported on an ordinal scale ranging from 0 to 3.
There are more than 59 references containing more than 259 animal model experiments, nearly all involving SARS-CoV-2, SARS-CoV, or MERS-CoV. Approximately 70% of the studies involve mice; 10% involve non-human primates (rhesus macaques, marmosets, and cynomolgus macaques); and 20% involve hamsters, ferrets, cats, dogs, or rabbits. The most commonly studied interventions have included monoclonal antibodies, fusion inhibitors, interferons, and the nucleoside analog polymerase inhibitors.
2.1.4. Clinical Studies
The clinical studies are represented using several enumerated and free-text fields. The enumerated fields include the reference, virus category, and type of study (e.g., observational, randomized trial, randomized placebo-controlled trial). The free-text fields include descriptions of the interventions and regimen details, the study population and methods, and the study findings. CoV-RDB does not provide an assessment of study quality such as validity and risk of bias as there are other research groups providing this type of assessment.
2.2. Lookup/Explanation Tables
2.2.1. Virus Categories
Antiviral data on coronaviruses other than SARS-CoV-2 provide insight into the robustness of an antiviral compound, in that, compounds that are active against multiple viral species will be more likely to inhibit future pandemic coronaviruses and will be less vulnerable to the development of drug-resistance mutations. Indeed, for many drug targets, such as the virus RdRP and 3CLpro enzymes, and for host processes upon which coronaviruses depend, inhibitory compounds will likely have broad-spectrum activity.
CoV-RDB contains antiviral data for six categories of coronaviruses: SARS-CoV-2, SARS-CoV, MERS-CoV, endemic human coronaviruses, bat coronaviruses, and non-bat mammalian coronaviruses. SARS-CoV-2 (37%), SARS-CoV (31%), and MERS-CoV (17%) account for 85% of the data. However, the proportion of data associated with SARS-CoV-2 is rapidly increasing. Figure 2 shows the distribution of study types for SARS-CoV-2, SARS-CoV, and MERS-CoV.
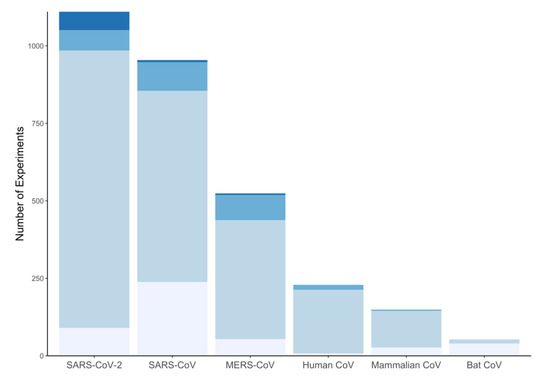
Figure 2.
The distribution of biochemical experiments (lightest), cell culture experiments (light), animal model experiments (dark), and clinical studies (darkest) for the six categories of virus in the Coronavirus Antiviral Research Database (CoV-RDB). The cell culture experiments also include entry assay experiments.
SARS-CoV and SARS-CoV-2 belong to the same betacoronavirus 2b (sarbecovirus) clade, and their amino acids are approximately 97% identical in the RdRP and 3CLpro enzymes and 84% identical in the spike protein. In contrast, MERS, a clade 2c betacoronavirus, is approximately 75% identical to SARS-CoV and SARS-CoV-2 in the RdRP gene, 60% identical in the 3CLpro gene, and 40% identical in the S gene. Within each of these three viruses, there is little diversity, with median pairwise distances ranging between 0% and 0.2%.
The four endemic human coronaviruses include two clade 2a betacoronaviruses and two alphacoronaviruses. Bat coronaviruses are distributed widely among different clades [2,3]. Indeed, 4 of the 9 betacoronavirus clades and 7 of 11 coronavirus clades are found only in bats. The mammalian coronaviruses include murine hepatitis virus (MHV), which is a longstanding experimental model for coronavirus infection, and several other coronaviruses that have been studied because they are important livestock diseases [4]. Although infectious bronchitis virus is an avian gammacoronavirus, we have included it in the non-bat mammalian coronavirus category.
2.2.2. Virus Isolates/Strains
CoV-RDB uses the terms isolate and strains to describe the different viruses used in antiviral studies, although “strains” is usually reserved for describing isolates that have distinct phenotypic properties [5]. Where possible, isolates are named according to the recommendations from the International Committee on Taxonomy of Viruses [6], i.e., virus/host/location/isolate/date.
Most SARS-CoV-2 isolates are nearly identical to one another with the upper limit for the pairwise amino acid distance being about 0.1%, although this number varies depending upon the gene [7]. Therefore, the biological significance of the isolate used for a particular study is not known. However, for some treatments such as monoclonal antibodies, changes in the sequence encoding the relevant epitope, specifically in the S protein receptor binding domain may prove to have biological and clinical significance [8,9]. Although SARS-CoV resulted from at least two zoonotic introductions from civet cats [4], and although MERS-CoV resulted from multiple zoonotic introductions from dromedary camels, these viruses also demonstrate little genetic variability.
Several of the most commonly used isolates have been cloned, either as intact viruses (e.g., by plaque purification or limiting dilution) or by constructing a cDNA copy representing a single sequence variant. Modification of these clones, such as selection of a resistant variant in vitro [10] or introduction of a reporter gene like GFP or luciferase, presumably retains the characteristics of the original parental virus isolate or strain [11]. Commonly used isolates that have been cloned and manipulated in the laboratory include MERS-CoV/human/Amsterdam/EMC/2012 [12], SARS-CoV/human/Hanoi/Urbani/2003 [13], SARS-CoV-2/human/USA/WA1/2020 [14], and SARS-CoV-2/human/Munich/929/2020 [15].
2.2.3. Cell Lines
The cell lines table provides descriptions for the cell lines used in cell culture and entry assay experiments. It contains four fields: (i) the cell line’s commonly used name, (ii) the source of the cell line, (iii) closely related cell lines, and (iv) a description of the cell line and one or more of the closely related cell lines. The most commonly used cell lines for SARS-CoV and SARS-CoV-2 include a variety of different Vero cell clones [16,17,18,19,20], Huh7 [16,21], Caco-2 [22], Calu-3 [23], and 293T/ACE2 cells [16,17,18] (Table 1). While each of these cell lines expresses ACE2, only Calu-3 cells were originally derived from lung epithelial cells. The 293T cells are typically used for cell–cell fusion and pseudovirus entry experiments. Several studies have also used human alveolar epithelial cells or a variety of different respiratory system or kidney organoids [24,25]. The cell lines used for MERS-CoV are similar, with the main exception that 293T/DPP4 (dipeptidyl peptidase 4) cells are used instead of 293T/ACE2 cells because DPP4 (aka CD26) is the MERS-CoV receptor [18].

Table 1.
Frequently used cells for culturing pandemic coronavirus antiviral research.
2.2.4. Animal Models
The over 10 different animal models used in experiments described in the CoV-RDB include three non-human primate models (rhesus macaques, cynomolgus macaque, and marmosets), multiple transgenic and non-transgenic mouse models, and several additional rodent models including hamsters and ferrets [26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43]. The transgenic mice have been modified in multiple ways, including to express hDPP4 so that they can be infected with MERS-CoV, to knock out the interferon (IFN)-α/β receptor to compromise innate immunity [44]; to knock out recombination activating gene 1 (RAG1) to compromise adaptive immunity [45]; to knock out carboxylesterase 1c, which causes poor plasma stability of remdesivir; and to express human rather than mouse ACE2 [39,40,41]. Table 2 describes the utility of the most common non-human primate and mammalian models for studies of the pandemic coronaviruses.

Table 2.
Animal models for pandemic coronavirus antiviral research.
2.2.5. Target and Compound Class
The target table has two main fields: name and description. The target classification organizes drugs, treatments, and compounds according to the virus or host process targeted by a compound including virus enzymes, virus entry into cells, host immunological responses, and other host processes. There are three virus enzyme inhibitor classes: RdRP, protease (including 3CLpro, PLpro), and helicase inhibitors. There are four inhibitor classes targeting virus entry: convalescent plasma and polyclonal antibody preparations, monoclonal antibodies, miscellaneous other compounds that inhibit virus receptor binding, and fusion inhibitors. There are two classes that target host immunological responses: interferons and other potential immunostimulatory compounds. Although there are potentially many mechanisms by which targeting a host processes may interfere with virus replication, we have divided these into two broad categories: host protease enzymes used by coronaviruses to cleave the spike protein, thus, priming it for fusion and other host proteins or pathways utilized by coronaviruses. The classification of host-acting compounds is likely to continue to evolve as mechanistic pathways become better defined. Table 3 describes each of the targets and compound classes described above. Figure 3 shows the distribution of experimental data types according to target.

Table 3.
Antiviral coronavirus therapy targets.
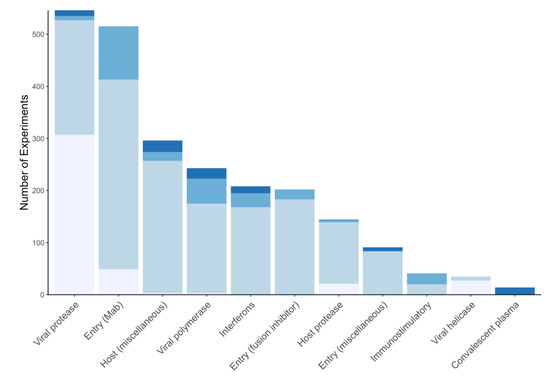
Figure 3.
The distribution of biochemical experiments (lightest), cell culture experiments (light), animal model studies (dark), and clinical studies (darkest) for the different targets of antiviral therapy in the Coronavirus Antiviral Research Database (CoV-RDB). The cell culture experiments also include entry assay experiments. The results for approximately 600 experiments involving compounds with an unknown or uncertain mechanism of action are not shown.
2.2.6. Compounds
The database contains experiments involving approximately 1650 compounds. More than 1240 of these compounds appear in the online compounds tables, which contain the following fields: (i) name, (ii) synonyms including abbreviations, (iii) closely related compounds, (iv) drug availability, (v) drug class, (vi) target, and (vii) description. For interferons, the drug class is the type of interferon (α, β, γ, or λ). For monoclonal antibodies, additional data are stored and displayed, including the antibody source and information on sequence and structure availability.
The closely related compounds are subjectively defined as those that we intend to be returned by a query even if that compound was not entered by the user. There are five broad categories of closely related compounds: (i) monoclonal antibodies described in the same publication, (ii) interferons belonging to the same type (i.e., α, β, γ, or λ), (iii) a series of compounds derived from the same lead compound, (iv) prodrugs such as those for GS-441524 (i.e., remdesivir) and β-N-hydroxycytidine (i.e, EIDD-28014), (v) drug combinations such as lopinavir and ritonavir-boosted lopinavir (lopinavir/r), and (vi) compounds presumed to act by a highly similar mechanism of action (e.g., hydroxychloroquine and chloroquine).
The drug availability category indicates whether the compound has been licensed in the U.S. or another country or has been studied in humans. Of 975 compounds with a known or likely mechanism of action, 135 (14%) are U.S. FDA (U.S. Food and Drug Administration) -approved drugs (for indications other than COVID-19), 197 (20%) have been or are currently being evaluated in human clinical trials or are approved outside the U.S., and 595 (61%) are preclinical investigational compounds.
Figure 4 displays EC50 values for many of the directly acting antiviral compounds currently in clinical trials for the treatment of COVID-19 including six polymerase inhibitors (remdesivir, EIDD-2801, favipiravir, ribavirin, galidesivir, and sofosbuvir), three HIV-1 protease inhibitors (lopinavir, atazanavir, and darunavir), and three entry inhibitors (receptor binding monoclonal antibodies, soluble recombinant human ACE2, and umifenovir). Figure 5 displays EC50 values for many of the repurposed compounds that target host processes required for virus replication including two host PIs that target the transmembrane serine protease 2 (TMPRSS2) enzyme (camostat and nafamostat), three chloroquine analogs that interfere with endosomal acidification (chloroquine, hydroxychloroquine, and mefloquine), three other compounds believed to interfere with membrane trafficking (niclosamide, imatinib, and chlorpromazine), and four compounds acting by a variety of different cellular mechanisms (ivermectin, nitazoxanide, ciclesonide, and cyclosporin).
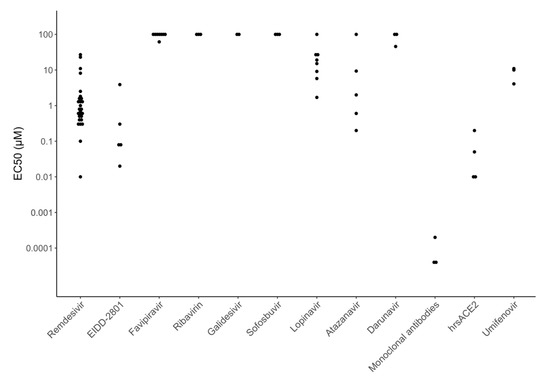
Figure 4.
Half maximal effective concentration (EC50) values for many of the directly acting antiviral compounds in clinical trials including six polymerase inhibitors (remdesivir, EIDD-2801, favipiravir, ribavirin, galidesivir, and sofosbuvir), three protease inhibitors (lopinavir, atazanavir, and darunavir), and three entry inhibitors (receptor binding monoclonal antibodies, soluble recombinant human angiotensin converting enzyme 2 (ACE2), and umifenovir). EC50 values above 100 μM are plotted at 100 μM.
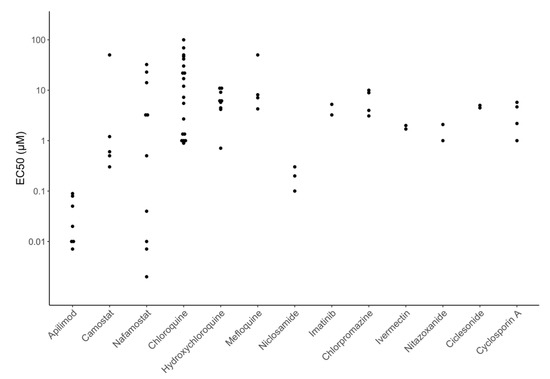
Figure 5.
EC50 values for many of the repurposed host-acting compounds currently in clinical trials including the host protease inhibitors (camostat and nafamostat), six possible endosomal trafficking inhibitors (chloroquine, hydroxychloroquine, mefloquine, niclosamide, imatinib, chlorpromazine) and four inhibitors acting by a variety of different mechanisms (ivermectin, nitazoxanide, ciclesonide, and cyclosporin).
Figure 4 and Figure 5 show that the potency of currently studied compounds extends over several orders of magnitude with monoclonal antibodies having EC50s in the high picomolar to low nanomolar range and some compounds displaying no activity at concentrations above 100 μM. However, there is also marked heterogeneity in the EC50 values for the same compound in different experiments. For several drugs, the heterogeneity can be explained by the type of cells used, inoculum size, drug timing, and culture duration. For example, the host TMPRSS2 inhibitors camostat and nafamostat are practically inactive against SARS-CoV-2 in Vero cells but have EC50s consistently below 1 μM in Caco-2 and Calu-3 cells, because these cells require TMPRSS2 for virus replication whereas Vero cells do not [22,51,52,53,54,55,56].
Table 4 describes a set of the most promising compounds for the treatment of SARS-CoV-2 based on the following criteria: (i) act by a validated direct or indirect antiviral mechanism, (ii) display sub-micromolar activity in vitro and/or inhibitory activity in an animal model, and (iii) have a record of safety and favorable pharmacokinetics in human subjects. The majority of these compounds are being studied in clinical trials, although the numbers of these trials are far fewer than those for less promising compounds.

Table 4.
Promising SARS-CoV-2 antiviral compounds.
2.3. Clinical Trials Registry
The Clinical Trials Registry table is a regularly updated, annotated list of ongoing, planned, or completed clinical trials obtained from the ClinicalTrials.gov, WHO ICTRP, and Chinese Clinical Trial websites. It contains trials of compounds with potential antiviral activity but not studies of non-antiviral interventions, such as those designed to optimize intensive-care management or reduce the inflammatory response associated with severe COVID-19. The Clinical Trials Registry classifies trials according to the compound target, the type of trial (e.g., observational or randomized controlled study), the status of the trial (pending, active, or completed), and the population studied. As of 6 August 2020, it contains more than 700 trials of which about 73% are listed on ClinicalTrials.gov and 27% are listed only on the WHO International Clinical Trials Platform.
Figure 6A displays the distribution of planned, ongoing, and published studies according to the compound targets of the drugs studied. Figure 6B displays the same distribution for those drugs in three or more studies. It is notable that many of the most commonly studied compounds have either little or no activity against SARS-CoV-2, including several drugs used for non-coronavirus infections such as the HIV protease inhibitors lopinavir and darunavir and the influenza inhibitors favipiravir, oseltamivir, and umifenovir. The chloroquine analogs, chloroquine and hydroxychloroquine, have weak in vitro activity but have failed to show clinical efficacy in multiple clinical trials [104,105,106,107,108,109,110].
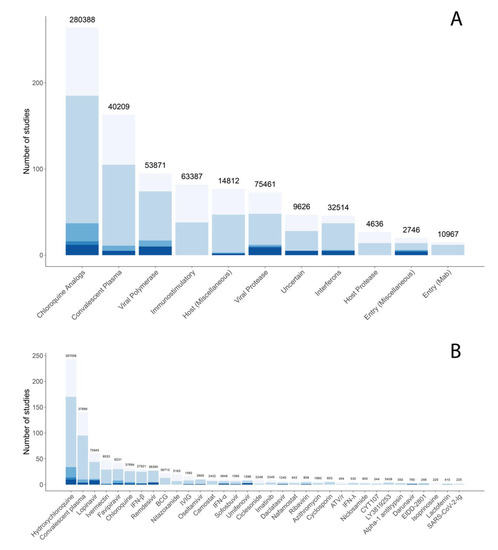
Figure 6.
Distribution of targets (A) and the most commonly studied compounds (B) for published (bottom), ongoing (middle), and planned (top) antiviral clinical trials through August 6. Although chloroquine analogs are considered to act primarily through the inhibition of virus endosomal trafficking, they are separated out from other endosomal trafficking inhibitors in (A). (B) shows compounds included in three or more trials.
2.4. Search Functions
The search function allows users to specify one or more of the following options from four drop-down lists: (i) compound target, (ii) compound, (iii) virus category, and (iv) study type. If the user selects “Any” for one of these and leaves the others in their default position, the search function returns the database’s complete set of cell culture experiments, biochemical experiments, entry assay experiments, animal model studies, and published clinical studies. By selecting one or more of the above options, the search function restricts the data returned to those meeting the search criteria. By using the “Copy to clipboard” link, users can import the results of any query into a spreadsheet where they can further sort and filter query results. The search function also provides a link to the trials in the Clinical Trials registry for selected compounds and compound targets.
The compound drop-down list displays 60 of the most well recognized compounds. Selecting a compound returns the data for that compound as well as for an additional 363 closely related compounds (as described in the compound table Section 2.2.6). If the user selects the compound target from the dropdown menu, then the compound menu will list all compounds designed to inhibit the selected target. The compounds entry on the compounds page also links to all the data on that compound in the database.
3. Discussion
To prioritize licensed drugs and investigational compounds for the treatment of COVID-19, it is necessary to compare their relative antiviral activities. Compounds that are not active in vitro will almost certainly not be useful clinically. Therefore, pre-clinical data are necessary to prioritize animal model and clinical studies. Compounds that are active in vitro, however, may also not be clinically useful if their associated in vitro data do not reflect physiologic conditions or if standard dosing with these compounds does not result in sufficient inhibitory concentrations at sites of infection.
The creation of the CoV-RDB was primarily motivated by the observation that many of the drugs being evaluated in CoVID-19 clinical trials demonstrate little or no in vitro anti-coronavirus activity. For example, as recently as July 2020, four of the most commonly studied drugs—chloroquine analogs, azithromycin, lopinavir/r, and favipiravir—demonstrated little if any in vitro activity.
The creation of the CoV-RDB was secondarily motivated by the observation that results for the same compound often vary across different laboratories as a result of experimental design such as cell line, inoculum size, drug-addition timing, duration of culture, and method for measuring virus replication. Given sufficient data, a database makes it possible to eventually identify the experimental features responsible for the heterogeneity in published results, thus improving the ability to compare the antiviral activity of different compounds. Indeed, we have already noted in this manuscript several compound classes for which viral inhibition is influenced by the cells used for virus culture.
The CoV-RDB is also designed to be educational as it provides multiple lookup tables for the viruses, drugs, cell lines, and animal models used in reported experiments. These tables contain descriptions of viruses, virus isolate/strains, cell lines, animal models, and more than 300 licensed and investigational compounds. Work is underway to also add comprehensive, yet detailed, summaries of SARS-CoV-2 monoclonal antibodies and of pharmacokinetic data for those drugs with well-documented in vitro inhibitory activity.
There are several additional web resources devoted to coronavirus drug development including sites devoted to high-throughput drug screening [111], the genetics of monoclonal antibodies [112], and meta-analyses of published clinical trials [113,114]. The National Institutes of Health (NIH) recognizes the importance of such resources and has recently announced a Notice of Special Interest: National Institute of Allergy and Infectious Diseases (NIAID) Priorities for Biomedical Knowledgebases and Repositories (NOT-AI-20-044). The CoV-RDB database, user interface, and underlying computer code represent a framework for organizing a vast amount of data and for facilitating data curation. However, the value of this resource depends upon ongoing manual data curation and annotation.
In conclusion, the CoV-RDB provides a uniquely integrated interdisciplinary synthesis of in vitro, animal model, and clinical studies of compounds with proven or possible anti-coronavirus activity. It helps researchers place their findings in the context of previously published data and it facilitates comparisons between different candidate antiviral compounds, thereby helping scientists, clinical investigators, public health officials, and funding agencies to prioritize the most promising compounds and repurposed drugs for further development.
Author Contributions
Writing and concept, R.W.S.; database and website, K.T. and P.L.T.; data curation and annotation, J.N., S.-Y.R., R.W.S., B.D.H., S.P., and N.P. All authors have read and agreed to the published version of the manuscript.
Funding
R.W.S., P.L.T, K.T., and S.-Y.R. were supported, in part, by grants from the National Institutes of Health (NIH/NIAID R24AI136618).
Conflicts of Interest
The authors declare no conflict of interest.
References
- U.S. Centers for Disease Control and Prevention (CDC) The Stepehn B. Thacker CDC Library. COVID-19 Research Articles Downloadable Database. Available online: https://www.cdc.gov/library/researchguides/2019novelcoronavirus/researcharticles.html (accessed on 1 September 2020).
- Anthony, S.J.; Johnson, C.K.; Greig, D.J.; Kramer, S.; Che, X.; Wells, H.; Hicks, A.L.; Joly, D.O.; Wolfe, N.D.; Daszak, P.; et al. Global patterns in coronavirus diversity. Virus Evol. 2017, 3, vex012. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Li, F.; Shi, Z.-L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019, 17, 181–192. [Google Scholar] [CrossRef] [PubMed]
- Bolles, M.; Donaldson, E.; Baric, R. SARS-CoV and emergent coronaviruses: Viral determinants of interspecies transmission. Curr. Opin. Virol. 2011, 1, 624–634. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, J.H.; Bao, Y.; Bavari, S.; Becker, S.; Bradfute, S.; Brister, J.R.; Bukreyev, A.A.; Chandran, K.; Davey, R.A.; Dolnik, O.; et al. Virus nomenclature below the species level: A standardized nomenclature for natural variants of viruses assigned to the family Filoviridae. Arch. Virol. 2013, 158, 301–311. [Google Scholar] [CrossRef] [PubMed]
- Coronaviridae Study Group of the International Committee on Taxonomy of Viruses. The species Severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020. [Google Scholar] [CrossRef]
- Wang, C.; Liu, Z.; Chen, Z.; Huang, X.; Xu, M.; He, T.; Zhang, Z. The establishment of reference sequence for SARS-CoV-2 and variation analysis. J. Med. Virol. 2020. [Google Scholar] [CrossRef]
- Baum, A.; Fulton, B.O.; Wloga, E.; Copin, R.; Pascal, K.E.; Russo, V.; Giordano, S.; Lanza, K.; Negron, N.; Ni, M.; et al. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science 2020. [Google Scholar] [CrossRef]
- Weisblum, Y.; Schmidt, F.; Zhang, F.; DaSilva, J.; Poston, D.; Lorenzi, J.C.C.; Muecksch, F.; Rutkowska, M.; Hoffmann, H.-H.; Michailidis, E.; et al. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. bioRxiv 2020. [Google Scholar] [CrossRef]
- Agostini, M.L.; Andres, E.L.; Sims, A.C.; Graham, R.L.; Sheahan, T.P.; Lu, X.; Smith, E.C.; Case, J.B.; Feng, J.Y.; Jordan, R.; et al. Coronavirus Susceptibility to the Antiviral Remdesivir (GS-5734) Is Mediated by the Viral Polymerase and the Proofreading Exoribonuclease. mBio 2018, 9. [Google Scholar] [CrossRef]
- Almazán, F.; Sola, I.; Zuñiga, S.; Marquez-Jurado, S.; Morales, L.; Becares, M.; Enjuanes, L. Coronavirus reverse genetic systems: Infectious clones and replicons. Virus Res. 2014, 189, 262–270. [Google Scholar] [CrossRef]
- Scobey, T.; Yount, B.L.; Sims, A.C.; Donaldson, E.F.; Agnihothram, S.S.; Menachery, V.D.; Graham, R.L.; Swanstrom, J.; Bove, P.F.; Kim, J.D.; et al. Reverse genetics with a full-length infectious cDNA of the Middle East respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. USA 2013, 110, 16157–16162. [Google Scholar] [CrossRef] [PubMed]
- Yount, B.; Curtis, K.M.; Fritz, E.A.; Hensley, L.E.; Jahrling, P.B.; Prentice, E.; Denison, M.R.; Geisbert, T.W.; Baric, R.S. Reverse genetics with a full-length infectious cDNA of severe acute respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. USA 2003, 100, 12995–13000. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Muruato, A.; Lokugamage, K.G.; Narayanan, K.; Zhang, X.; Zou, J.; Liu, J.; Schindewolf, C.; Bopp, N.E.; Aguilar, P.V.; et al. An Infectious cDNA Clone of SARS-CoV-2. Cell Host Microbe 2020. [Google Scholar] [CrossRef] [PubMed]
- Thao, T.T.N.; Labroussaa, F.; Ebert, N.; V’kovski, P.; Stalder, H.; Portmann, J.; Kelly, J.; Steiner, S.; Holwerda, M.; Kratzel, A.; et al. Rapid reconstruction of SARS-CoV-2 using a synthetic genomics platform. Nature 2020, 1–8. [Google Scholar] [CrossRef]
- Ogando, N.; Dalebout, T.; Zevenhoven-Dobbe, J.C.; Limpens, R.W.A.L.; van der Meer, Y.; Caly, L.; Druce, J.; de Vries, J.; Kikkert, M.; Barcena, M.; et al. SARS-coronavirus-2 replication in Vero E6 cells: Replication kinetics, rapid adaptation and cytopathology. bioRxiv 2020. [Google Scholar] [CrossRef]
- Kaye, M. SARS-associated coronavirus replication in cell lines. Emerg. Infect. Dis. 2006, 12, 128–133. [Google Scholar] [CrossRef]
- Chan, J.F.-W.; Chan, K.-H.; Choi, G.K.-Y.; To, K.K.-W.; Tse, H.; Cai, J.-P.; Yeung, M.L.; Cheng, V.C.-C.; Chen, H.; Che, X.-Y.; et al. Differential cell line susceptibility to the emerging novel human betacoronavirus 2c EMC/2012: Implications for disease pathogenesis and clinical manifestation. J. Infect. Dis. 2013, 207, 1743–1752. [Google Scholar] [CrossRef]
- Emeny, J.M.; Morgan, M.J. Regulation of the Interferon System: Evidence that Vero Cells have a Genetic Defect in Interferon Production. J. Gen. Virol. 1979, 43, 247–252. [Google Scholar] [CrossRef]
- Matsuyama, S.; Nao, N.; Shirato, K.; Kawase, M.; Saito, S.; Takayama, I.; Nagata, N.; Sekizuka, T.; Katoh, H.; Kato, F.; et al. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc. Natl. Acad. Sci. USA 2020, 117, 7001–7003. [Google Scholar] [CrossRef]
- Mirabelli, C.; Wotring, J.W.; Zhang, C.J.; McCarty, S.M.; Fursmidt, R.; Kadambi, N.S.; Amin, A.T.; O’Meara, T.R.; Pretto-Kernahan, C.D.; Spence, J.R.; et al. Morphological Cell Profiling of SARS-CoV-2 Infection Identifies Drug Repurposing Candidates for COVID-19. bioRxiv 2020. [Google Scholar] [CrossRef]
- Ellinger, B. Identification of inhibitors of SARS-CoV-2 in-vitro cellular toxicity in human (Caco-2) cells using a large scale drug repurposing collection. Drug Discov. Des. Dev. 2020. [Google Scholar] [CrossRef]
- Ko, M.; Jeon, S.; Ryu, W.-S.; Kim, S. Comparative analysis of antiviral efficacy of FDA-approved drugs against SARS-CoV-2 in human lung cells: Nafamostat is the most potent antiviral drug candidate. bioRxiv 2020. [Google Scholar] [CrossRef]
- Pizzorno, A.; Padey, B.; Julien, T.; Trouillet-Assant, S.; Traversier, A.; Errazuriz-Cerda, E.; Fouret, J.; Dubois, J.; Gaymard, A.; Lescure, X.; et al. Characterization and treatment of SARS-CoV-2 in nasal and bronchial human airway epithelia. bioRxiv 2020. [Google Scholar] [CrossRef]
- Milewska, A.; Kula-Pacurar, A.; Wadas, J.; Suder, A.; Szczepanski, A.; Dabrowska, A.; Owczarek, K.; Marcello, A.; Ochman, M.; Stacel, T.; et al. Replication of SARS-CoV-2 in human respiratory epithelium. J. Virol. 2020. [Google Scholar] [CrossRef] [PubMed]
- McAuliffe, J.; Vogel, L.; Roberts, A.; Fahle, G.; Fischer, S.; Shieh, W.-J.; Butler, E.; Zaki, S.; St Claire, M.; Murphy, B.; et al. Replication of SARS coronavirus administered into the respiratory tract of African Green, rhesus and cynomolgus monkeys. Virology 2004, 330, 8–15. [Google Scholar] [CrossRef]
- Greenough, T.C.; Carville, A.; Coderre, J.; Somasundaran, M.; Sullivan, J.L.; Luzuriaga, K.; Mansfield, K. Pneumonitis and Multi-Organ System Disease in Common Marmosets (Callithrix jacchus) Infected with the Severe Acute Respiratory Syndrome-Associated Coronavirus. Am. J. Pathol. 2005, 167, 455–463. [Google Scholar] [CrossRef]
- Roberts, A.; Subbarao, K. Animal models for SARS. Adv. Exp. Med. Biol. 2006, 581, 463–471. [Google Scholar] [CrossRef]
- Gretebeck, L.M.; Subbarao, K. Animal models for SARS and MERS coronaviruses. Curr. Opin. Virol. 2015, 13, 123–129. [Google Scholar] [CrossRef]
- Falzarano, D.; de Wit, E.; Rasmussen, A.L.; Feldmann, F.; Okumura, A.; Scott, D.P.; Brining, D.; Bushmaker, T.; Martellaro, C.; Baseler, L.; et al. Treatment with interferon-α2b and ribavirin improves outcome in MERS-CoV-infected rhesus macaques. Nat. Med. 2013, 19, 1313–1317. [Google Scholar] [CrossRef]
- Lawler, J.V.; Endy, T.P.; Hensley, L.E.; Garrison, A.; Fritz, E.A.; Lesar, M.; Baric, R.S.; Kulesh, D.A.; Norwood, D.A.; Wasieloski, L.P.; et al. Cynomolgus macaque as an animal model for severe acute respiratory syndrome. PLoS Med. 2006, 3, e149. [Google Scholar] [CrossRef]
- Munster, V.J.; Feldmann, F.; Williamson, B.N.; van Doremalen, N.; Pérez-Pérez, L.; Schulz, J.; Meade-White, K.; Okumura, A.; Callison, J.; Brumbaugh, B.; et al. Respiratory disease in rhesus macaques inoculated with SARS-CoV-2. Nature 2020. [Google Scholar] [CrossRef] [PubMed]
- Rockx, B.; Kuiken, T.; Herfst, S.; Bestebroer, T.; Lamers, M.M.; Oude Munnink, B.B.; de Meulder, D.; van Amerongen, G.; van den Brand, J.; Okba, N.M.A.; et al. Comparative pathogenesis of COVID-19, MERS, and SARS in a nonhuman primate model. Science 2020. [Google Scholar] [CrossRef] [PubMed]
- Rogers, T.F.; Zhao, F.; Huang, D.; Beutler, N.; Abbott, R.K.; Callaghan, S.; Garcia, E.; He, W.; Hurtado, J.; Limbo, O.; et al. Rapid isolation of potent SARS-CoV-2 neutralizing antibodies and protection in a small animal model. bioRxiv 2020. [Google Scholar] [CrossRef]
- Shi, J.; Wen, Z.; Zhong, G.; Yang, H.; Wang, C.; Huang, B.; Liu, R.; He, X.; Shuai, L.; Sun, Z.; et al. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science 2020, 368, 1016–1020. [Google Scholar] [CrossRef]
- Park, S.-J.; Yu, K.-M.; Kim, Y.-I.; Kim, S.-M.; Kim, E.-H.; Kim, S.-G.; Kim, E.J.; Casel, M.A.B.; Rollon, R.; Jang, S.-G.; et al. Antiviral Efficacies of FDA-Approved Drugs against SARS-CoV-2 Infection in Ferrets. mBio 2020, 11. [Google Scholar] [CrossRef]
- Kim, Y.-I.; Kim, S.-G.; Kim, S.-M.; Kim, E.-H.; Park, S.-J.; Yu, K.-M.; Chang, J.-H.; Kim, E.J.; Lee, S.; Casel, M.A.B.; et al. Infection and Rapid Transmission of SARS-CoV-2 in Ferrets. Cell Host Microbe 2020, 27, 704–709.e2. [Google Scholar] [CrossRef]
- Sia, S.F.; Yan, L.-M.; Chin, A.W.H.; Fung, K.; Choy, K.-T.; Wong, A.Y.L.; Kaewpreedee, P.; Perera, R.A.P.M.; Poon, L.L.M.; Nicholls, J.M.; et al. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature 2020. [Google Scholar] [CrossRef]
- Bao, L.; Deng, W.; Huang, B.; Gao, H.; Liu, J.; Ren, L.; Wei, Q.; Yu, P.; Xu, Y.; Qi, F.; et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature 2020. [Google Scholar] [CrossRef]
- Jiang, R.-D.; Liu, M.-Q.; Chen, Y.; Shan, C.; Zhou, Y.-W.; Shen, X.-R.; Li, Q.; Zhang, L.; Zhu, Y.; Si, H.-R.; et al. Pathogenesis of SARS-CoV-2 in Transgenic Mice Expressing Human Angiotensin-Converting Enzyme 2. Cell 2020. [Google Scholar] [CrossRef]
- Lutz, C.; Maher, L.; Lee, C.; Kang, W. COVID-19 preclinical models: Human angiotensin-converting enzyme 2 transgenic mice. Hum. Genomics 2020, 14, 20. [Google Scholar] [CrossRef]
- Shan, C.; Yao, Y.-F.; Yang, X.-L.; Zhou, Y.-W.; Gao, G.; Peng, Y.; Yang, L.; Hu, X.; Xiong, J.; Jiang, R.-D.; et al. Infection with novel coronavirus (SARS-CoV-2) causes pneumonia in Rhesus macaques. Cell Res. 2020, 1–8. [Google Scholar] [CrossRef]
- Imai, M.; Iwatsuki-Horimoto, K.; Hatta, M.; Loeber, S.; Halfmann, P.J.; Nakajima, N.; Watanabe, T.; Ujie, M.; Takahashi, K.; Ito, M.; et al. Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development. Proc. Natl. Acad. Sci. USA 2020. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Perera, R.A.P.M.; Kayali, G.; Meyerholz, D.; Perlman, S.; Peiris, M. Passive immunotherapy with dromedary immune serum in an experimental animal model for Middle East respiratory syndrome coronavirus infection. J. Virol. 2015, 89, 6117–6120. [Google Scholar] [CrossRef] [PubMed]
- Channappanavar, R.; Lu, L.; Xia, S.; Du, L.; Meyerholz, D.K.; Perlman, S.; Jiang, S. Protective Effect of Intranasal Regimens Containing Peptidic Middle East Respiratory Syndrome Coronavirus Fusion Inhibitor Against MERS-CoV Infection. J. Infect. Dis. 2015, 212, 1894–1903. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.F.-W.; Yao, Y.; Yeung, M.-L.; Deng, W.; Bao, L.; Jia, L.; Li, F.; Xiao, C.; Gao, H.; Yu, P.; et al. Treatment With Lopinavir/Ritonavir or Interferon-β1b Improves Outcome of MERS-CoV Infection in a Nonhuman Primate Model of Common Marmoset. J. Infect. Dis. 2015, 212, 1904–1913. [Google Scholar] [CrossRef]
- Park, A.; Iwasaki, A. Type I and Type III Interferons—Induction, Signaling, Evasion, and Application to Combat COVID-19. Cell Host Microbe 2020. [Google Scholar] [CrossRef]
- Lokugamage, K.G.; Schindewolf, C.; Menachery, V.D. SARS-CoV-2 sensitive to type I interferon pretreatment. bioRxiv 2020. [Google Scholar] [CrossRef]
- Tang, T.; Bidon, M.; Jaimes, J.A.; Whittaker, G.R.; Daniel, S. Coronavirus membrane fusion mechanism offers a potential target for antiviral development. Antiviral Res. 2020, 178, 104792. [Google Scholar] [CrossRef]
- Zumla, A.; Chan, J.F.W.; Azhar, E.I.; Hui, D.S.C.; Yuen, K.-Y. Coronaviruses - drug discovery and therapeutic options. Nat. Rev. Drug Discov. 2016, 15, 327–347. [Google Scholar] [CrossRef]
- Yamamoto, M.; Kiso, M.; Sakai-Tagawa, Y.; Iwatsuki-Horimoto, K.; Imai, M.; Takeda, M.; Kinoshita, N.; Ohmagari, N.; Gohda, J.; Semba, K.; et al. The Anticoagulant Nafamostat Potently Inhibits SARS-CoV-2 S Protein-Mediated Fusion in a Cell Fusion Assay System and Viral Infection In Vitro in a Cell-Type-Dependent Manner. Viruses 2020, 12, 629. [Google Scholar] [CrossRef]
- Jeon, S.; Ko, M.; Lee, J.; Choi, I.; Byun, S.Y.; Park, S.; Shum, D.; Kim, S. Identification of antiviral drug candidates against SARS-CoV-2 from FDA-approved drugs. Antimicrob. Agents Chemother. 2020. [Google Scholar] [CrossRef] [PubMed]
- Bojkova, D.; McGreig, J.E.; McLaughlin, K.-M.; Masterson, S.G.; Widera, M.; Kraehling, V.; Ciesek, S.; Wass, M.N.; Michaelis, M.; Cinatl, J.N. SARS-CoV-2 and SARS-CoV differ in their cell tropism and drug sensitivity profiles. bioRxiv 2020. [Google Scholar] [CrossRef]
- Ko, M.; Jeon, S.; Ryu, W.-S.; Kim, S. Comparative analysis of antiviral efficacy of FDA-approved drugs against SARS-CoV-2 in human lung cells. J. Med. Virol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Schroeder, S.; Kleine-Weber, H.; Müller, M.A.; Drosten, C.; Pöhlmann, S. Nafamostat mesylate blocks activation of SARS-CoV-2: New treatment option for COVID-19. Antimicrob. Agents Chemother. 2020. [Google Scholar] [CrossRef] [PubMed]
- Riva, L.; Yuan, S.; Yin, X.; Martin-Sancho, L.; Matsunaga, N.; Burgstaller, S.; Pache, L.; Jesus, P.D.; Hull, M.V.; Chang, M.; et al. A Large-scale Drug Repositioning Survey for SARS-CoV-2 Antivirals. bioRxiv 2020. [Google Scholar] [CrossRef]
- Pruijssers, A.J.; George, A.S.; Schäfer, A.; Leist, S.R.; Gralinksi, L.E.; Dinnon, K.H.; Yount, B.L.; Agostini, M.L.; Stevens, L.J.; Chappell, J.D.; et al. Remdesivir Inhibits SARS-CoV-2 in Human Lung Cells and Chimeric SARS-CoV Expressing the SARS-CoV-2 RNA Polymerase in Mice. Cell Rep. 2020, 107940. [Google Scholar] [CrossRef]
- Dittmar, M.; Lee, J.S.; Whig, K.; Segrist, E.; Li, M.; Jurado, K.; Samby, K.; Ramage, H.; Schultz, D.; Cherry, S. Drug repurposing screens reveal FDA approved drugs active against SARS-Cov-2. bioRxiv 2020. [Google Scholar] [CrossRef]
- Williamson, B.N.; Feldmann, F.; Schwarz, B.; Meade-White, K.; Porter, D.P.; Schulz, J.; van Doremalen, N.; Leighton, I.; Yinda, C.K.; Pérez-Pérez, L.; et al. Clinical benefit of remdesivir in rhesus macaques infected with SARS-CoV-2. Nature 2020. [Google Scholar] [CrossRef]
- Beigel, J.H.; Tomashek, K.M.; Dodd, L.E.; Mehta, A.K.; Zingman, B.S.; Kalil, A.C.; Hohmann, E.; Chu, H.Y.; Luetkemeyer, A.; Kline, S.; et al. Remdesivir for the Treatment of Covid-19 - Preliminary Report. N. Engl. J. Med. 2020. [Google Scholar] [CrossRef]
- Sheahan, T.P.; Sims, A.C.; Zhou, S.; Graham, R.L.; Pruijssers, A.J.; Agostini, M.L.; Leist, S.R.; Schäfer, A.; Dinnon, K.H.; Stevens, L.J.; et al. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci. Transl. Med. 2020. [Google Scholar] [CrossRef] [PubMed]
- Hansen, J.; Baum, A.; Pascal, K.E.; Russo, V.; Giordano, S.; Wloga, E.; Fulton, B.O.; Yan, Y.; Koon, K.; Patel, K.; et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science 2020. [Google Scholar] [CrossRef] [PubMed]
- Baum, A.; Copin, R.; Ajithdoss, D.; Zhou, A.; Lanza, K.; Negron, N.; Ni, M.; Wei, Y.; Atwal, G.S.; Oyejide, A.; et al. REGN-COV2 antibody cocktail prevents and treats SARS-CoV-2 infection in rhesus macaques and hamsters. bioRxiv 2020. [Google Scholar] [CrossRef]
- Shi, R.; Shan, C.; Duan, X.; Chen, Z.; Liu, P.; Song, J.; Song, T.; Bi, X.; Han, C.; Wu, L.; et al. A human neutralizing antibody targets the receptor binding site of SARS-CoV-2. Nature 2020. [Google Scholar] [CrossRef]
- Ju, B.; Zhang, Q.; Ge, J.; Wang, R.; Sun, J.; Ge, X.; Yu, J.; Shan, S.; Zhou, B.; Song, S.; et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature 2020, 1–8. [Google Scholar] [CrossRef]
- Pinto, D.; Park, Y.-J.; Beltramello, M.; Walls, A.C.; Tortorici, M.A.; Bianchi, S.; Jaconi, S.; Culap, K.; Zatta, F.; De Marco, A.; et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature 2020. [Google Scholar] [CrossRef]
- Robbiani, D.F.; Gaebler, C.; Muecksch, F.; Lorenzi, J.C.C.; Wang, Z.; Cho, A.; Agudelo, M.; Barnes, C.O.; Gazumyan, A.; Finkin, S.; et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature 2020. [Google Scholar] [CrossRef]
- Rogers, T.F.; Zhao, F.; Huang, D.; Beutler, N.; Burns, A.; He, W.-T.; Limbo, O.; Smith, C.; Song, G.; Woehl, J.; et al. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science 2020. [Google Scholar] [CrossRef]
- Brouwer, P.J.M.; Caniels, T.G.; van der Straten, K.; Snitselaar, J.L.; Aldon, Y.; Bangaru, S.; Torres, J.L.; Okba, N.M.A.; Claireaux, M.; Kerster, G.; et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science 2020. [Google Scholar] [CrossRef]
- Cao, Y.; Su, B.; Guo, X.; Sun, W.; Deng, Y.; Bao, L.; Zhu, Q.; Zhang, X.; Zheng, Y.; Geng, C.; et al. Potent Neutralizing Antibodies against SARS-CoV-2 Identified by High-Throughput Single-Cell Sequencing of Convalescent Patients’ B Cells. Cell 2020. [Google Scholar] [CrossRef]
- Chi, X.; Yan, R.; Zhang, J.; Zhang, G.; Zhang, Y.; Hao, M.; Zhang, Z.; Fan, P.; Dong, Y.; Yang, Y.; et al. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science 2020. [Google Scholar] [CrossRef] [PubMed]
- Huo, J.; Zhao, Y.; Ren, J.; Zhou, D.; Duyvesteyn, H.M.E.; Ginn, H.M.; Carrique, L.; Malinauskas, T.; Ruza, R.R.; Shah, P.N.M.; et al. Neutralization of SARS-CoV-2 by Destruction of the Prefusion Spike. Cell Host Microbe 2020. [Google Scholar] [CrossRef] [PubMed]
- Huo, J.; Le Bas, A.; Ruza, R.R.; Duyvesteyn, H.M.E.; Mikolajek, H.; Malinauskas, T.; Tan, T.K.; Rijal, P.; Dumoux, M.; Ward, P.N.; et al. Neutralizing nanobodies bind SARS-CoV-2 spike RBD and block interaction with ACE2. Nat. Struct. Mol. Biol. 2020, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Zost, S.J.; Gilchuk, P.; Case, J.B.; Binshtein, E.; Chen, R.E.; Nkolola, J.P.; Schäfer, A.; Reidy, J.X.; Trivette, A.; Nargi, R.S.; et al. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature 2020, 1–7. [Google Scholar] [CrossRef]
- Ejemel, M.; Li, Q.; Hou, S.; Schiller, Z.A.; Tree, J.A.; Wallace, A.; Amcheslavsky, A.; Kurt Yilmaz, N.; Buttigieg, K.R.; Elmore, M.J.; et al. A cross-reactive human IgA monoclonal antibody blocks SARS-CoV-2 spike-ACE2 interaction. Nat. Commun. 2020, 11, 4198. [Google Scholar] [CrossRef]
- Wu, Y.; Wang, F.; Shen, C.; Peng, W.; Li, D.; Zhao, C.; Li, Z.; Li, S.; Bi, Y.; Yang, Y.; et al. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science 2020, 368, 1274–1278. [Google Scholar] [CrossRef]
- Wec, A.Z.; Wrapp, D.; Herbert, A.S.; Maurer, D.P.; Haslwanter, D.; Sakharkar, M.; Jangra, R.K.; Dieterle, M.E.; Lilov, A.; Huang, D.; et al. Broad neutralization of SARS-related viruses by human monoclonal antibodies. Science 2020. [Google Scholar] [CrossRef]
- Alsoussi, W.B.; Turner, J.S.; Case, J.B.; Zhao, H.; Schmitz, A.J.; Zhou, J.Q.; Chen, R.E.; Lei, T.; Rizk, A.A.; McIntire, K.M.; et al. A Potently Neutralizing Antibody Protects Mice against SARS-CoV-2 Infection. J. Immunol. Baltim. Md 1950 2020. [Google Scholar] [CrossRef]
- Seydoux, E.; Homad, L.J.; MacCamy, A.J.; Parks, K.R.; Hurlburt, N.K.; Jennewein, M.F.; Akins, N.R.; Stuart, A.B.; Wan, Y.-H.; Feng, J.; et al. Analysis of a SARS-CoV-2-Infected Individual Reveals Development of Potent Neutralizing Antibodies with Limited Somatic Mutation. Immunity 2020, 53, 98–105.e5. [Google Scholar] [CrossRef]
- Felgenhauer, U.; Schoen, A.; Gad, H.H.; Hartmann, R.; Schaubmar, A.R.; Failing, K.; Drosten, C.; Weber, F. Inhibition of SARS-CoV-2 by type I and type III interferons. J. Biol. Chem. 2020. [Google Scholar] [CrossRef]
- Mantlo, E.; Bukreyeva, N.; Maruyama, J.; Paessler, S.; Huang, C. Antiviral activities of type I interferons to SARS-CoV-2 infection. Antiviral Res. 2020, 179, 104811. [Google Scholar] [CrossRef] [PubMed]
- Vanderheiden, A.; Ralfs, P.; Chirkova, T.; Upadhyay, A.A.; Zimmerman, M.G.; Bedoya, S.; Aoued, H.; Tharp, G.M.; Pellegrini, K.L.; Manfredi, C.; et al. Type I and Type III IFN Restrict SARS-CoV-2 Infection of Human Airway Epithelial Cultures. J. Virol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Zheng, F.; Zhou, Y.; Zhou, Z.; Ye, F.; Huang, B.; Huang, Y.; Ma, J.; Zuo, Q.; Tan, X.; Xie, J.; et al. SARS-CoV-2 Clearance in COVID-19 Patients with Novaferon Treatment: A Randomized, Open-label, Parallel Group Trial. Int. J. Infect. Dis. IJID Off. Publ. Int. Soc. Infect. Dis. 2020. [Google Scholar] [CrossRef] [PubMed]
- Hung, I.F.-N.; Lung, K.-C.; Tso, E.Y.-K.; Liu, R.; Chung, T.W.-H.; Chu, M.-Y.; Ng, Y.-Y.; Lo, J.; Chan, J.; Tam, A.R.; et al. Triple combination of interferon beta-1b, lopinavir–ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID-19: An open-label, randomised, phase 2 trial. Lancet 2020, 395, 1695–1704. [Google Scholar] [CrossRef]
- Wettstein, L.; Conzelmann, C.; Müller, J.A.; Weil, T.; Groß, R.; Hirschenberger, M.; Seidel, A.; Klute, S.; Zech, F.; Bozzo, C.P.; et al. Alpha-1 antitrypsin inhibits SARS-CoV-2 infection. bioRxiv 2020. [Google Scholar] [CrossRef]
- Hempel, T.; Raich, L.; Olsson, S.; Azouz, N.P.; Klingler, A.M.; Rothenberg, M.E.; Noe, F. Molecular mechanism of SARS-CoV-2 cell entry inhibition via TMPRSS2 by Camostat and Nafamostat mesylate. bioRxiv 2020. [Google Scholar] [CrossRef]
- Shrimp, J.H.; Kales, S.C.; Sanderson, P.E.; Simeonov, A.; Shen, M.; Hall, M.D. An Enzymatic TMPRSS2 Assay for Assessment of Clinical Candidates and Discovery of Inhibitors as Potential Treatment of COVID-19. bioRxiv 2020. [Google Scholar] [CrossRef]
- Riva, L.; Yuan, S.; Yin, X.; Martin-Sancho, L.; Matsunaga, N.; Pache, L.; Burgstaller-Muehlbacher, S.; De Jesus, P.D.; Teriete, P.; Hull, M.V.; et al. Discovery of SARS-CoV-2 antiviral drugs through large-scale compound repurposing. Nature 2020. [Google Scholar] [CrossRef]
- Bouhaddou, M.; Memon, D.; Meyer, B.; White, K.M.; Rezelj, V.V.; Correa Marrero, M.; Polacco, B.J.; Melnyk, J.E.; Ulferts, S.; Kaake, R.M.; et al. The Global Phosphorylation Landscape of SARS-CoV-2 Infection. Cell 2020. [Google Scholar] [CrossRef]
- Chen, C.Z.; Shinn, P.; Itkin, Z.; Eastman, R.; Bostwick, R.; Rasmussen, L.; Huang, R.; Shen, M.; Hu, X.; Wilson, K.M.; et al. Drug Repurposing Screen for Compounds Inhibiting the Cytopathic Effect of SARS-CoV-2. bioRxiv 2020. [Google Scholar] [CrossRef]
- Bakowski, M.A.; Beutler, N.; Chen, E.; Nguyen, T.-T.H.; Kirkpatrick, M.G.; Parren, M.; Yang, L.; Ricketts, J.; Gupta, A.K.; Hull, M.V.; et al. Oral drug repositioning candidates and synergistic remdesivir combinations for the prophylaxis and treatment of COVID-19. bioRxiv 2020. [Google Scholar] [CrossRef]
- Ou, X.; Liu, Y.; Lei, X.; Li, P.; Mi, D.; Ren, L.; Guo, L.; Guo, R.; Chen, T.; Hu, J.; et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020, 11, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.-L.; Chou, Y.; Rothlauf, P.W.; Liu, Z.; Soh, T.K.; Cureton, D.; Case, J.B.; Chen, R.E.; Diamond, M.S.; Whelan, S.P.J.; et al. Inhibition of PIKfyve kinase prevents infection by Zaire ebolavirus and SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2020. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Weetall, M.; Trotta, C.; Cintron, K.; Ma, J.; Kim, M.J.; Furia, B.; Romfo, C.; Graci, J.D.; Li, W.; et al. Targeting of Hematologic Malignancies with PTC299, A Novel Potent Inhibitor of Dihydroorotate Dehydrogenase with Favorable Pharmaceutical Properties. Mol. Cancer Ther. 2019, 18, 3–16. [Google Scholar] [CrossRef]
- Cheung, N.N.; Lai, K.K.; Dai, J.; Kok, K.H.; Chen, H.; Chan, K.-H.; Yuen, K.-Y.; Kao, R.Y.T. Broad-spectrum inhibition of common respiratory RNA viruses by a pyrimidine synthesis inhibitor with involvement of the host antiviral response. J. Gen. Virol. 2017, 98, 946–954. [Google Scholar] [CrossRef]
- Liu, Q.; Gupta, A.; Okesli-Armlovich, A.; Qiao, W.; Fischer, C.R.; Smith, M.; Carette, J.E.; Bassik, M.C.; Khosla, C. Enhancing the Antiviral Efficacy of RNA-Dependent RNA Polymerase Inhibition by Combination with Modulators of Pyrimidine Metabolism. Cell Chem. Biol. 2020, 27, 668–677.e9. [Google Scholar] [CrossRef]
- Luban, J.; Sattler, R.; Muhlberger, E.; Graci, J.D.; Cao, L.; Weetall, M.; Trotta, C.; Colacino, J.M.; Bavari, S.; Strambio-De-Castillia, C.; et al. The DHODH Inhibitor PTC299 Arrests SARS-CoV-2 Replication and Suppresses Induction of Inflammatory Cytokines. bioRxiv 2020. [Google Scholar] [CrossRef]
- Wang, Q.; Guo, H.; Li, Y.; Jian, X.; Hou, X.; Zhong, N.; Fei, J.; Su, D.; Bian, Z.; Zhang, Y.; et al. Efficacy and Safety of Leflunomide for Refractory COVID-19: An Open-label Controlled Study. medRxiv 2020. [Google Scholar] [CrossRef]
- Imai, Y.; Kuba, K.; Rao, S.; Huan, Y.; Guo, F.; Guan, B.; Yang, P.; Sarao, R.; Wada, T.; Leong-Poi, H.; et al. Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature 2005, 436, 112–116. [Google Scholar] [CrossRef]
- Khan, A.; Benthin, C.; Zeno, B.; Albertson, T.E.; Boyd, J.; Christie, J.D.; Hall, R.; Poirier, G.; Ronco, J.J.; Tidswell, M.; et al. A pilot clinical trial of recombinant human angiotensin-converting enzyme 2 in acute respiratory distress syndrome. Crit. Care Lond. Engl. 2017, 21, 234. [Google Scholar] [CrossRef]
- Monteil, V.; Kwon, H.; Prado, P.; Hagelkrüys, A.; Wimmer, R.A.; Stahl, M.; Leopoldi, A.; Garreta, E.; Hurtado Del Pozo, C.; Prosper, F.; et al. Inhibition of SARS-CoV-2 Infections in Engineered Human Tissues Using Clinical-Grade Soluble Human ACE2. Cell 2020. [Google Scholar] [CrossRef]
- Lei, C.; Qian, K.; Li, T.; Zhang, S.; Fu, W.; Ding, M.; Hu, S. Neutralization of SARS-CoV-2 spike pseudotyped virus by recombinant ACE2-Ig. Nat. Commun. 2020, 11, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Horby, P.; Mafham, M.; Linsell, L.; Bell, J.L.; Staplin, N.; Emberson, J.R.; Wiselka, M.; Ustianowski, A.; Elmahi, E.; Prudon, B.; et al. Effect of Hydroxychloroquine in Hospitalized Patients with COVID-19: Preliminary results from a multi-centre, randomized, controlled trial. medRxiv 2020. [Google Scholar] [CrossRef]
- Skipper, C.P.; Pastick, K.A.; Engen, N.W.; Bangdiwala, A.S.; Abassi, M.; Lofgren, S.M.; Williams, D.A.; Okafor, E.C.; Pullen, M.F.; Nicol, M.R.; et al. Hydroxychloroquine in Nonhospitalized Adults With Early COVID-19: A Randomized Trial. Ann. Intern. Med. 2020. [Google Scholar] [CrossRef] [PubMed]
- Cavalcanti, A.B.; Zampieri, F.G.; Rosa, R.G.; Azevedo, L.C.P.; Veiga, V.C.; Avezum, A.; Damiani, L.P.; Marcadenti, A.; Kawano-Dourado, L.; Lisboa, T.; et al. Hydroxychloroquine with or without Azithromycin in Mild-to-Moderate Covid-19. N. Engl. J. Med. 2020. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Cao, Z.; Han, M.; Wang, Z.; Chen, J.; Sun, W.; Wu, Y.; Xiao, W.; Liu, S.; Chen, E.; et al. Hydroxychloroquine in patients with mainly mild to moderate coronavirus disease 2019: Open label, randomised controlled trial. BMJ 2020, 369. [Google Scholar] [CrossRef]
- Mitja, O.; Ubals, M.; Corbacho, M.; Alemany, A.; Suner, C.; Tebe, C.; Tobias, A.; Penafiel, J.; Ballana, E.; Perez, C.A.; et al. A Cluster-Randomized Trial of Hydroxychloroquine as Prevention of Covid-19 Transmission and Disease. medRxiv 2020. [Google Scholar] [CrossRef]
- Mitjà, O.; Corbacho-Monné, M.; Ubals, M.; Tebe, C.; Peñafiel, J.; Tobias, A.; Ballana, E.; Alemany, A.; Riera-Martí, N.; Pérez, C.A.; et al. Hydroxychloroquine for Early Treatment of Adults with Mild Covid-19: A Randomized-Controlled Trial. Clin. Infect. Dis. 2020, ciaa1009. [Google Scholar] [CrossRef]
- Boulware, D.R.; Pullen, M.F.; Bangdiwala, A.S.; Pastick, K.A.; Lofgren, S.M.; Okafor, E.C.; Skipper, C.P.; Nascene, A.A.; Nicol, M.R.; Abassi, M.; et al. A Randomized Trial of Hydroxychloroquine as Postexposure Prophylaxis for Covid-19. N. Engl. J. Med. 2020, 383, 517–525. [Google Scholar] [CrossRef]
- Brimacombe, K.R.; Zhao, T.; Eastman, R.T.; Hu, X.; Wang, K.; Backus, M.; Baljinnyam, B.; Chen, C.Z.; Chen, L.; Eicher, T.; et al. An OpenData portal to share COVID-19 drug repurposing data in real time. bioRxiv 2020. [Google Scholar] [CrossRef]
- Raybould, M.I.J.; Kovaltsuk, A.; Marks, C.; Deane, C.M. CoV-AbDab: The Coronavirus Antibody Database. bioRxiv 2020. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Shang, Y.-X.; Tian, Z.-Y.; Xiong, M.; Lu, C.-L.; Jiang, Y.; Zhang, Y.; Zhang, Y.-Y.; Jin, X.-Y.; Jin, Q.-B.; et al. Characteristics of registered studies for Coronavirus disease 2019 (COVID-19): A systematic review. Integr. Med. Res. 2020, 9, 100426. [Google Scholar] [CrossRef] [PubMed]
- Lythgoe, M.P.; Middleton, P. Ongoing Clinical Trials for the Management of the COVID-19 Pandemic. Trends Pharmacol. Sci. 2020. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).