RNAseq Analysis Reveals Virus Diversity within Hawaiian Apiary Insect Communities
Abstract
1. Introduction
2. Materials and Methods
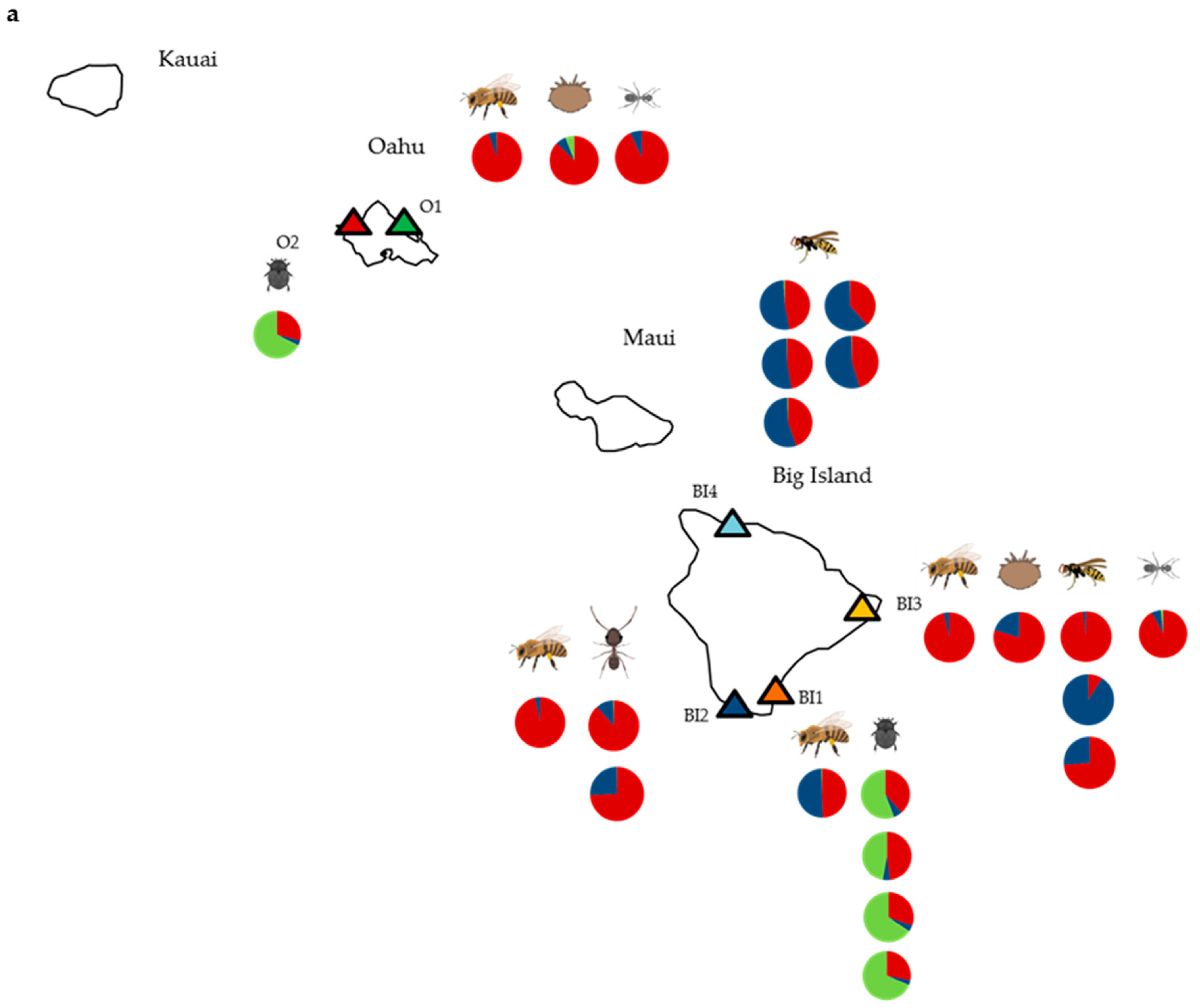
2.1. Site and Species Selection
2.2. RNA Extraction and Next Generation Sequencing
2.3. Bioinformatic Analysis
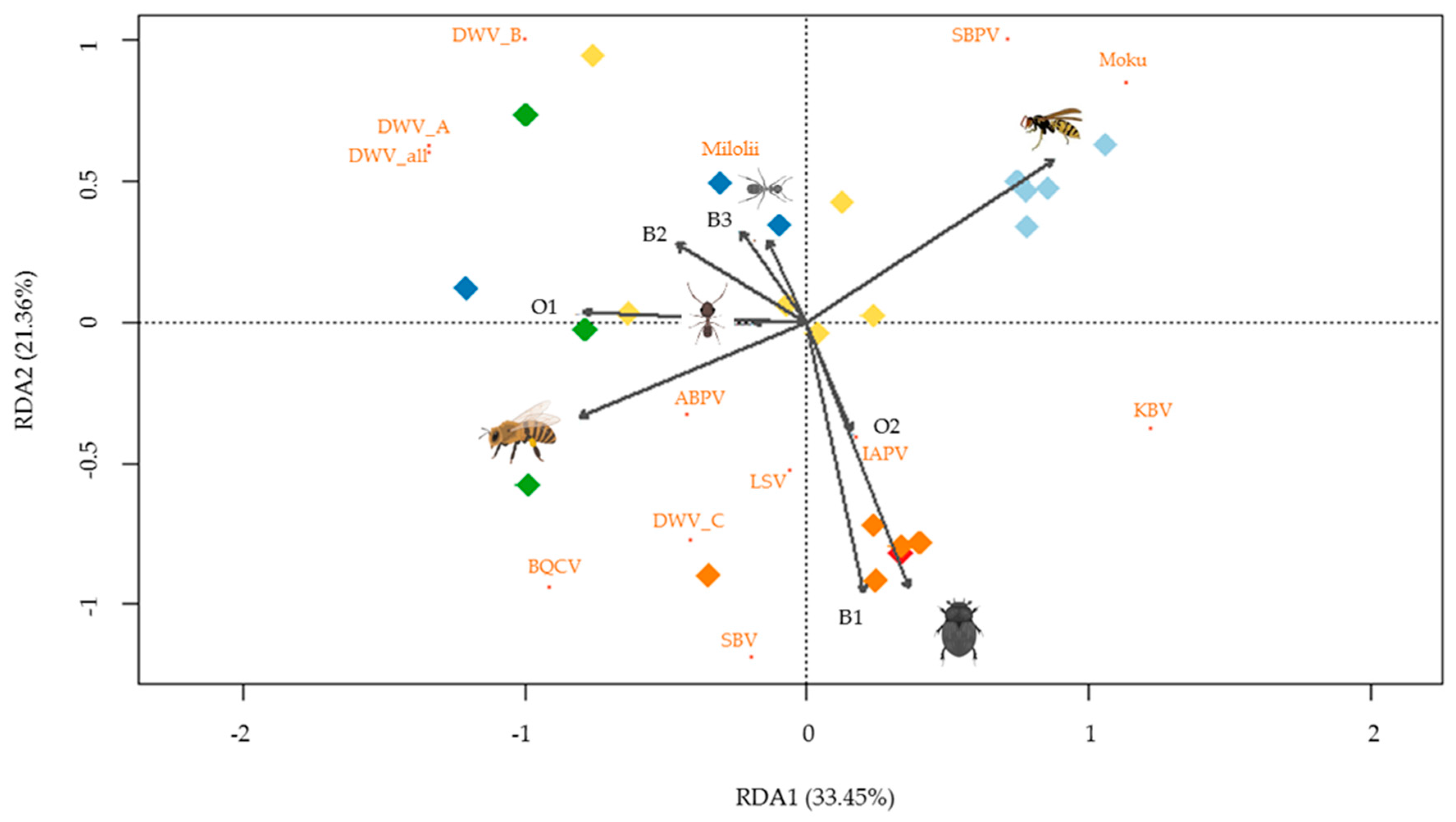
2.4. Statistical Analysis
3. Results
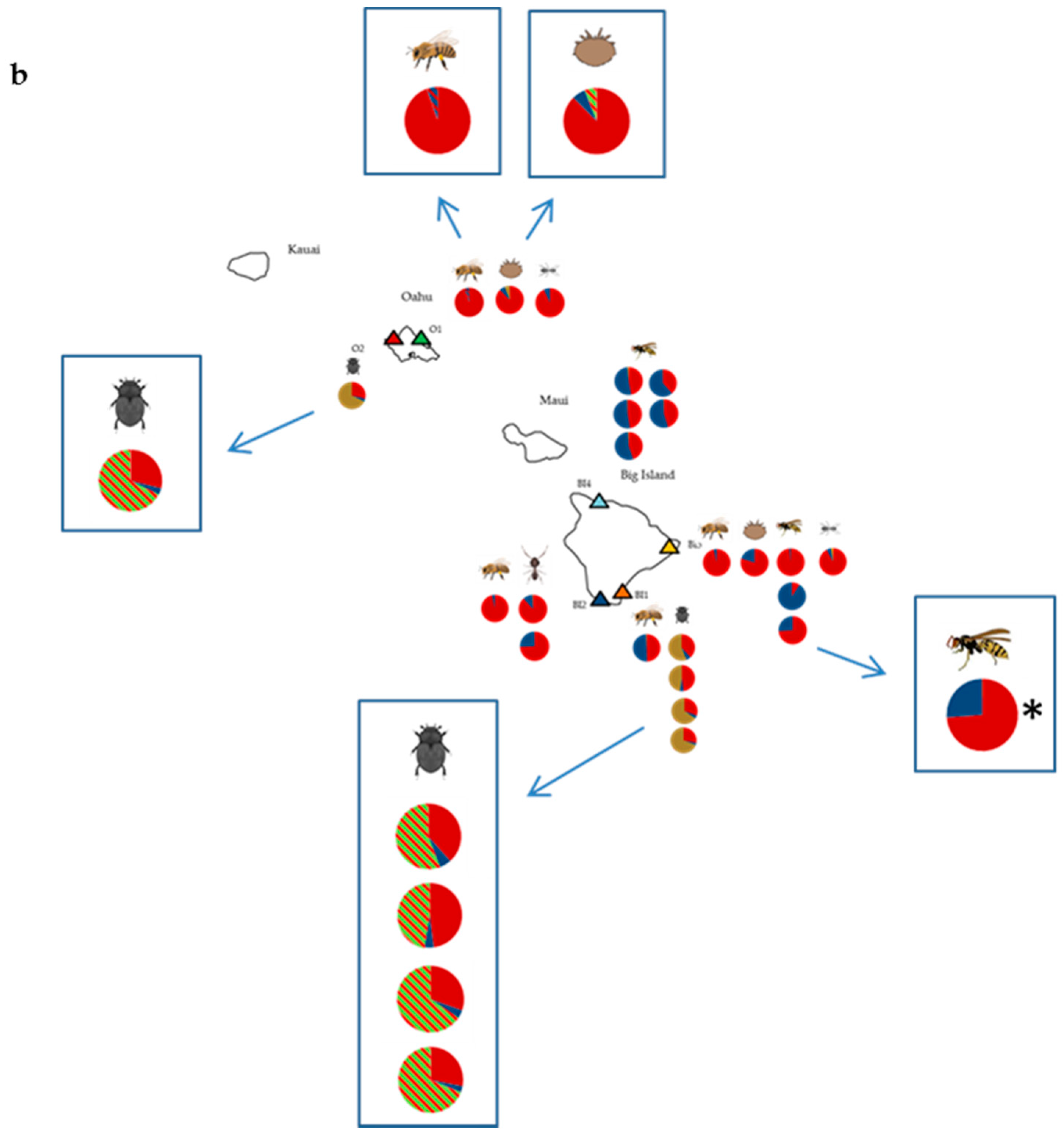
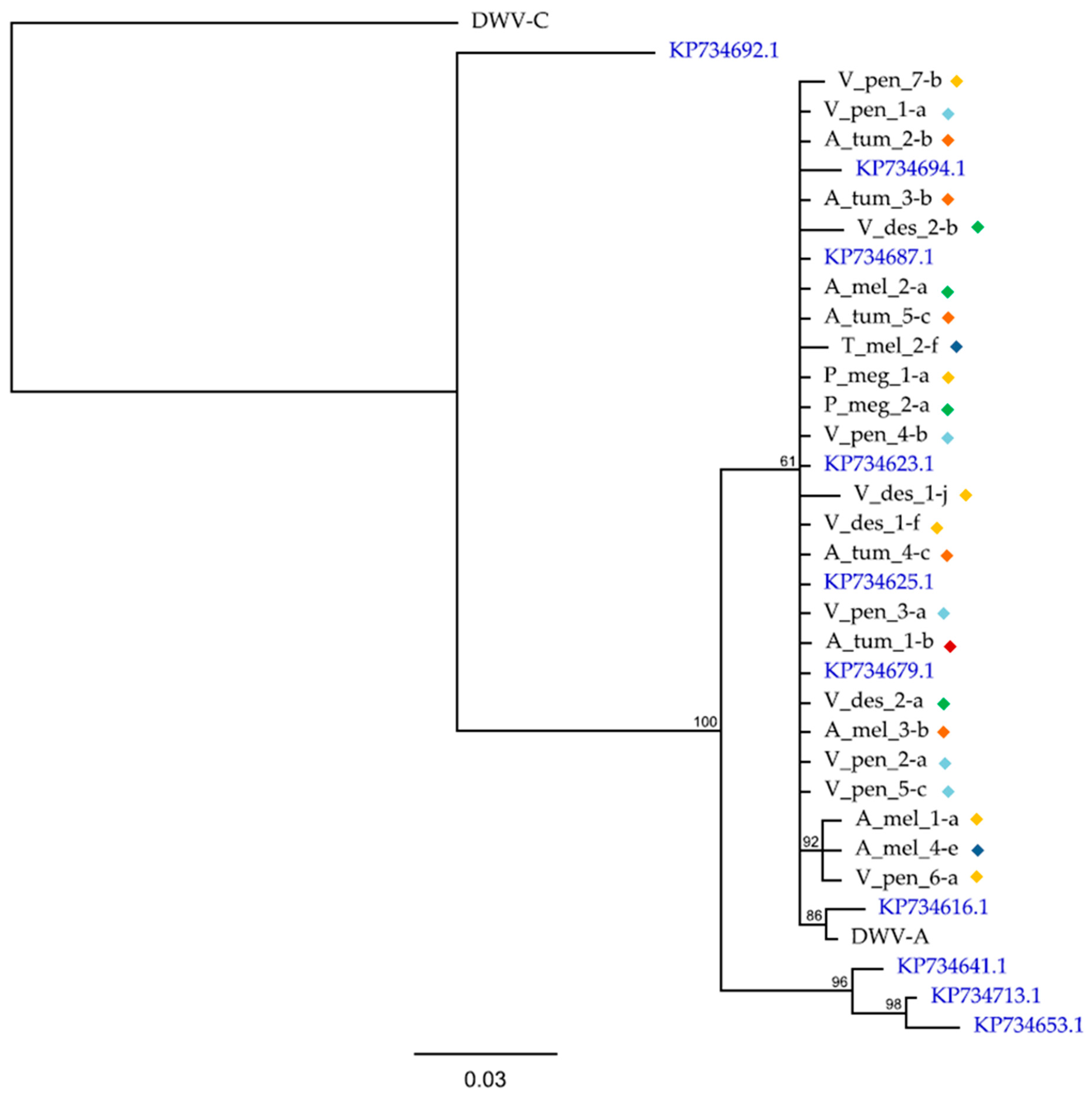
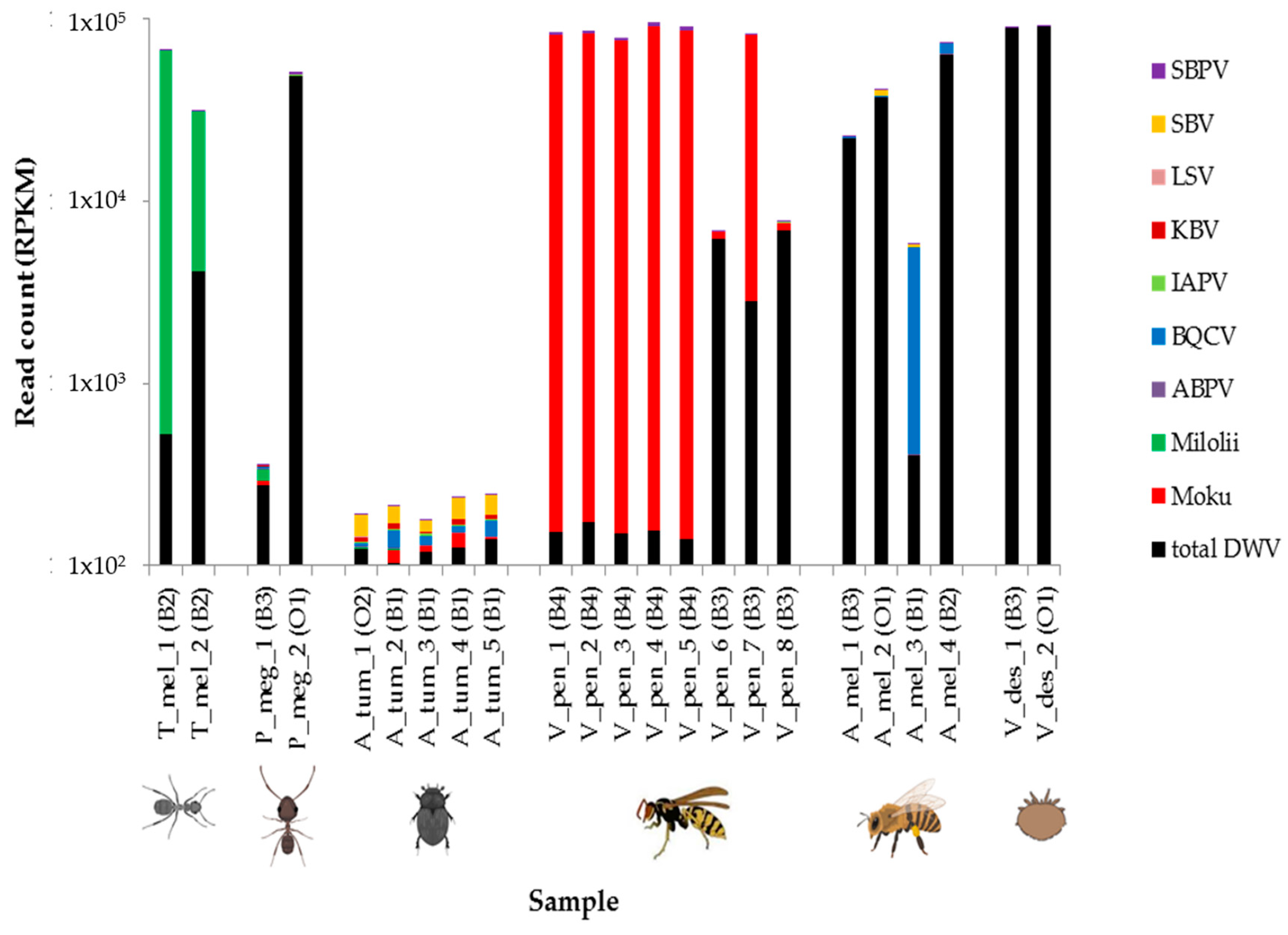
3.1. Deformed Wing Virus
3.2. Other Honey Bee-Associated Viruses
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Daszak, P.; Cunningham, A.A.; Hyatt, A.D. Emerging infectious diseases of wildlife--threats to biodiversity and human health. Science 2000, 287, 443. [Google Scholar] [CrossRef] [PubMed]
- Blaustein, A.R.; Johnson, P.T.J. Conservation biology: When an infection turns lethal. Nature 2010, 465, 881–882. [Google Scholar]
- Schroeder, D.C.; Martin, S.J. Deformed wing virus: The main suspect in unexplained honeybee deaths worldwide. Virulence 2012, 3, 589–591. [Google Scholar] [CrossRef] [PubMed]
- Wilfert, L.; Long, G.; Leggett, H.C.; Schmid-Hempel, P.; Butlin, R.; Martin, S.J.; Boots, M. Deformed wing virus is a recent global epidemic in honeybees driven by Varroa mites. Science 2016, 351, 594–597. [Google Scholar] [CrossRef]
- Potts, S.G.; Vulliamy, B.; Dafni, A.; Ne’eman, G.; Willmer, P. Linking bees and flowers: How do floral communities structure pollinator communities? Ecology 2003, 84, 2628–2642. [Google Scholar]
- Steffan-Dewenter, I.; Tscharntke, T. Resource overlap and possible competition between honeybees and wild bees in central Europe. Oecologia 2000, 122, 288–296. [Google Scholar] [CrossRef]
- Steffan-Dewenter, I.; Tscharntke, T. Insect communities and biotic interactions on fragmented calcareous grasslands—a mini review. Biol. Cons. 2002, 104, 275–284. [Google Scholar]
- Thomson, D. Competitive interactions between the invasive European honeybee and native bumble bees. Ecology 2004, 85, 458–470. [Google Scholar] [CrossRef]
- Potts, S.G.; Biesmeijer, J.C.; Kremen, C.; Neumann, P.; Schweiger, O.; Kunin, W.E. Global pollinator declines: Trends, impacts and drivers. Trends Ecol. Evol. 2010, 25, 345–353. [Google Scholar] [CrossRef]
- Brown, M.J.; Dicks, L.V.; Paxton, R.J.; Baldock, K.C.; Barron, A.B.; Chauzat, M.P.; Freitas, B.M.; Goulson, D.; Jepsen, S.; Kremen, C.; et al. A horizon scan of future threats and opportunities for pollinators and pollination. PeerJ 2016, 4, e2249. [Google Scholar] [CrossRef] [PubMed]
- Vanbergen, A.J. Threats to an ecosystem service: Pressures on pollinators. Front. Ecol. Environ. 2013, 11, 251–259. [Google Scholar] [CrossRef]
- Cornman, R.S.; Tarpy, D.R.; Chen, Y.; Jeffreys, L.; Lopez, D.; Pettis, J.S.; Evans, J.D. Pathogen webs in collapsing honeybee colonies. PLoS ONE 2012. [Google Scholar] [CrossRef]
- Collison, E.; Hird, H.; Cresswell, J.; Tyler, C. Interactive effects of pesticide exposure and pathogen infection on bee health—A critical analysis. Biol. Rev. Camb. Philos. Soc. 2016, 91, 1006–1019. [Google Scholar] [CrossRef]
- Parrish, C.R.; Holmes, E.C.; Morens, D.M.; Park, E.C.; Burke, D.S.; Calisher, C.H.; Laughlin, C.A.; Saif, L.J.; Daszak, P. Cross-species virus transmission and the emergence of new epidemic diseases. Microbiol. Mol. Biol. Rev. 2008, 72, 457–470. [Google Scholar] [CrossRef] [PubMed]
- Domingo, E.J.; Holland, J.J. RNA virus mutations and fitness for survival. Annu. Rev. Microbiol. 1997, 51, 151–178. [Google Scholar] [CrossRef] [PubMed]
- Ryabov, E.V.; Wood, G.R.; Fannon, J.M.; Moore, J.D.; Bull, J.C.; Chandler, D.; Mead, A.; Burroughs, N.; Evans, D.J. A virulent strain of Deformed wing virus (DWV) of honeybees (Apis mellifera) prevails after Varroa destructor-mediated, or in vitro, transmission. PLoS Pathog. 2014, 10, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Martin, S.J.; Highfield, A.C.; Brettell, L.; Villalobos, E.M.; Budge, G.E.; Powell, M.; Nikaido, S.; Schroeder, D.C. Global honeybee viral landscape altered by a parasitic mite. Science 2012, 336, 1304–1306. [Google Scholar] [CrossRef]
- Mondet, F.; de Miranda, J.R.; Kretzschmar, A.; Le Conte, Y.; Mercer, A.R. On the front line: Quantitative virus dynamics in honeybee (Apis mellifera L.) colonies along a new expansion front of the parasite Varroa destructor. PLoS Pathog. 2014, 10, e1004323. [Google Scholar] [CrossRef]
- Martin, S.J.; Brettell, L.E. Deformed wing virus in Honeybees and Other Insects. Annu. Rev. Virol. 2019, 6. [Google Scholar] [CrossRef]
- Eyer, M.; Chen, Y.P.; Schäfer, M.O.; Pettis, J.; Neumann, P. Small hive beetle, Aethina tumida, as a potential biological vector of honeybee viruses. Apidologie 2009, 40, 419–428. [Google Scholar] [CrossRef]
- Fürst, M.A.; McMahon, D.P.; Osborne, J.L.; Paxton, R.J.; Brown, M.J.F. Disease associations between honeybees and bumblebees as a threat to wild pollinators. Nature 2014, 506, 364–366. [Google Scholar] [CrossRef]
- McMahon, D.P.; Fürst, M.A.; Caspar, J.; Theodorou, P.; Brown, M.J.F.; Paxton, R.J. A sting in the spit: Widespread cross-infection of multiple RNA viruses across wild and managed bees. J. Anim. Ecol. 2015, 84, 615–624. [Google Scholar] [CrossRef]
- Santamaria, J.; Villalobos, E.M.; Brettell, L.E.; Nikaido, S.; Graham, J.R.; Martin, S. Evidence of Varroa-mediated deformed wing virus spillover in Hawaii. J. Invertebr. Pathol. 2018, 151, 126–130. [Google Scholar] [CrossRef]
- Loope, K.J.; Baty, J.W.; Lester, P.J.; Wilson Rankin, E.E. Pathogen shifts in a honeybee predator following the arrival of the Varroa mite. Proc. R. Soc. B 2019, 286. [Google Scholar] [CrossRef]
- Mordecai, G.J.; Brettell, L.E.; Martin, S.J.; Dixon, D.; Jones, I.M.; Schroeder, D.C. Superinfection exclusion and the long-term survival of honeybees in Varroa-infested colonies. Isme J. 2016, 10, 1182–1191. [Google Scholar] [CrossRef]
- Brettell, L.E.; University of Salford, Manchester, UK. Results of screening for Deformed wing virus in common apiary pest insects using RT-PCR. 2012. [Google Scholar]
- Yang, X.; Charlebois, P.; Gnerre, S.; Coole, M.G.; Lennon, N.J.; Levin, J.Z.; Qu, J.; Ryan, E.M.; Zody, M.C.; Henn, M.R. De novo assembly of highly diverse viral populations. BMC Genomics 2012, 13, 475. [Google Scholar] [CrossRef]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef]
- Mordecai, G.J.; Brettell, L.E.; Pachori, P.; Villalobos, E.M.; Martin, S.J.; Jones, I.M.; Schroeder, D.C. Moku virus; a new Iflavirus found in wasps, honeybees and Varroa. Sci. Rep. 2016, 6, 34983. [Google Scholar] [CrossRef]
- Brettell, L.E.; Mordecai, G.; Pachori, P.; Martin, S. Novel RNA virus genome discovered in Ghost ants (Tapinoma melanocephalum) from Hawaii. Genome Announc. 2017, 5, e00669-17. [Google Scholar] [CrossRef]
- Villalobos, E.M. The mite that jumped, the bee that traveled, the disease that followed. Science 2016, 351, 554–556. [Google Scholar] [CrossRef]
- Brooks, E.M.; Sheflin, L.G.; Spaulding, S.W. Secondary structure in the 3’UTR of EGF and the choice of reverse transcriptases affect the detection of message diversity by RT-PCR. Biotechniques 1995, 19, 806–812. [Google Scholar]
- Lanzi, G.; de Miranda, J.R.; Boniotti, M.B.; Cameron, C.E.; Lavazza, A.; Capucci, L.; Carmazine, S.M.; Rossi, C. Molecular and Biological Characterization of Deformed Wing Virus of Honeybees (Apis mellifera L.). J. Virol. 2006, 80, 4998–5009. [Google Scholar] [CrossRef]
- Wood, G.R.; Burroughs, N.J.; Evans, D.J.; Ryabov, E.V. Error correction and diversity analysis of population mixtures determined by NGS. PeerJ 2014, 2, e645. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kevill, J.L.; de Souza, F.; Sharples, C.; Schroeder, D.; Martin, S.J. DWV-A lethal to honey bees (Apis mellifera): A colony level survey of DWV variants (A, B & C) in England, Wales and 32 states across the US. Viruses 2019. (submitted). [Google Scholar]
- Villalobos, E.M.; (University of Hawaii, Hawaii, USA). Personal communication, 2017.
- De Souza, F.S.; Kevill, J.L.; Correia-Oliveira, M.E.; de Carvalho, C.A.; Martin, S.J. Occurrence of deformed wing virus variants in the stingless bee Melipona subnitida and honey bee Apis mellifera populations in Brazil. J. Gen. Virol. 2019, 100, 289–294. [Google Scholar] [CrossRef]
- Boncristiani, H.F.; Di Prisco, G.; Pettis, J.S.; Hamilton, M.; Chen, Y.P. Molecular approaches to the analysis of deformed wing virus replication and pathogenesis in the honey bee, Apis mellifera. Virol. J. 2009, 6, 221. [Google Scholar] [CrossRef]
- Fung, E.; Hill, K.; Hogendoorn, K.; Glatz, R.V.; Napier, K.R.; Bellgard, M.I.; Barrero, R.A. De novo assembly of honey bee RNA viral genomes by tapping into the innate insect antiviral response pathway. J. Invertebr. Pathol. 2018, 152, 38–47. [Google Scholar] [CrossRef]
- Singh, R.; Levitt, A.L.; Rajotte, E.G.; Holmes, E.C.; Ostiguy, N.; Lipkin, W.I.; Toth, A.L.; Cox-Foster, D.L. RNA viruses in Hymenopteran pollinators: Evidence of inter-taxa virus transmission via pollen and potential impact on non-Apis hymenopteran species. PLoS ONE 2010, 5, e14357. [Google Scholar] [CrossRef]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brettell, L.E.; Schroeder, D.C.; Martin, S.J. RNAseq Analysis Reveals Virus Diversity within Hawaiian Apiary Insect Communities. Viruses 2019, 11, 397. https://doi.org/10.3390/v11050397
Brettell LE, Schroeder DC, Martin SJ. RNAseq Analysis Reveals Virus Diversity within Hawaiian Apiary Insect Communities. Viruses. 2019; 11(5):397. https://doi.org/10.3390/v11050397
Chicago/Turabian StyleBrettell, Laura E., Declan C. Schroeder, and Stephen J. Martin. 2019. "RNAseq Analysis Reveals Virus Diversity within Hawaiian Apiary Insect Communities" Viruses 11, no. 5: 397. https://doi.org/10.3390/v11050397
APA StyleBrettell, L. E., Schroeder, D. C., & Martin, S. J. (2019). RNAseq Analysis Reveals Virus Diversity within Hawaiian Apiary Insect Communities. Viruses, 11(5), 397. https://doi.org/10.3390/v11050397






