Reassessment of Viroid RNA Cytosine Methylation Status at the Single Nucleotide Level
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and RNA Preparations
2.2. Bisulfite Sequencing Protocols
2.3. RT-PCR, Cloning and Sequencing
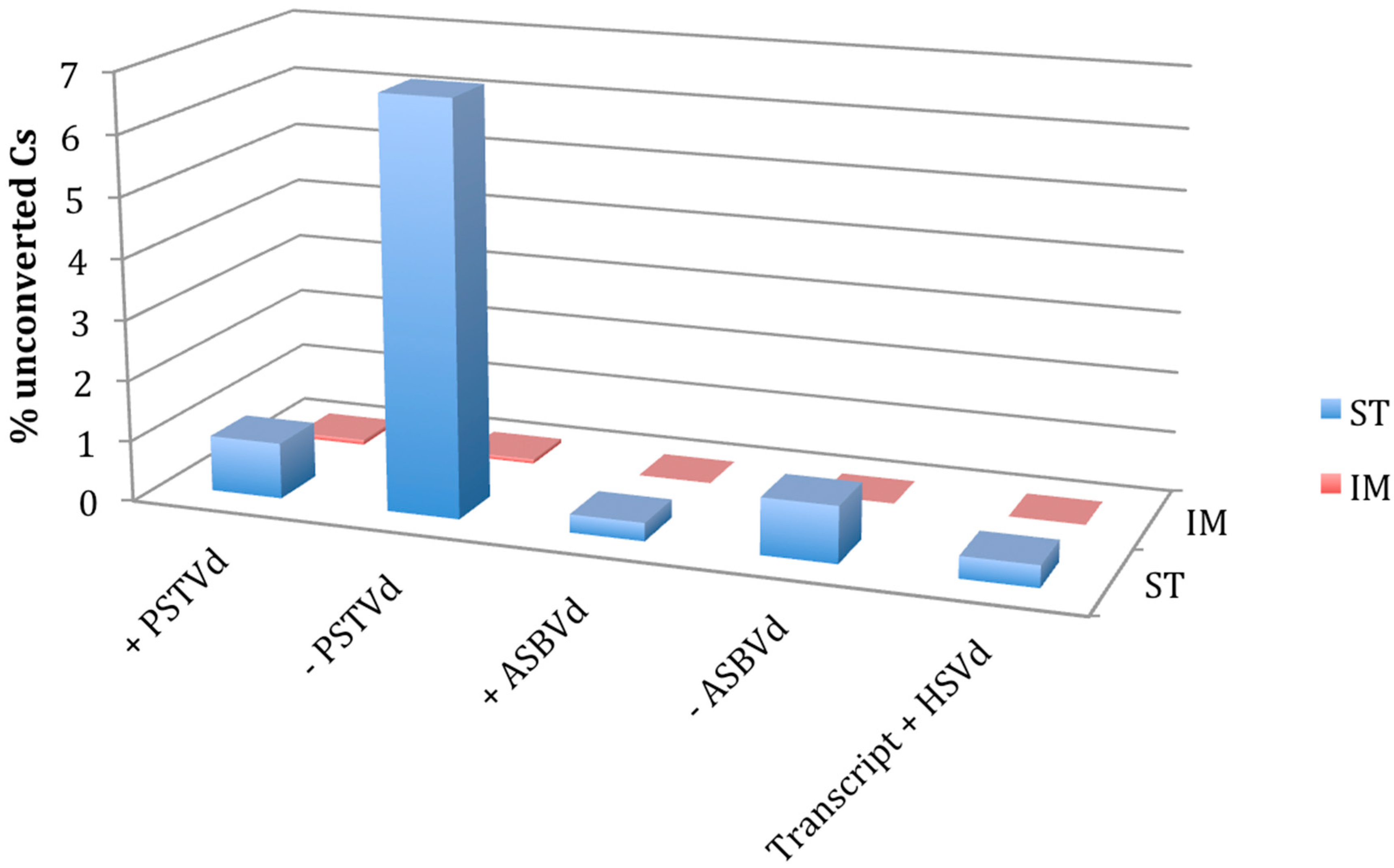
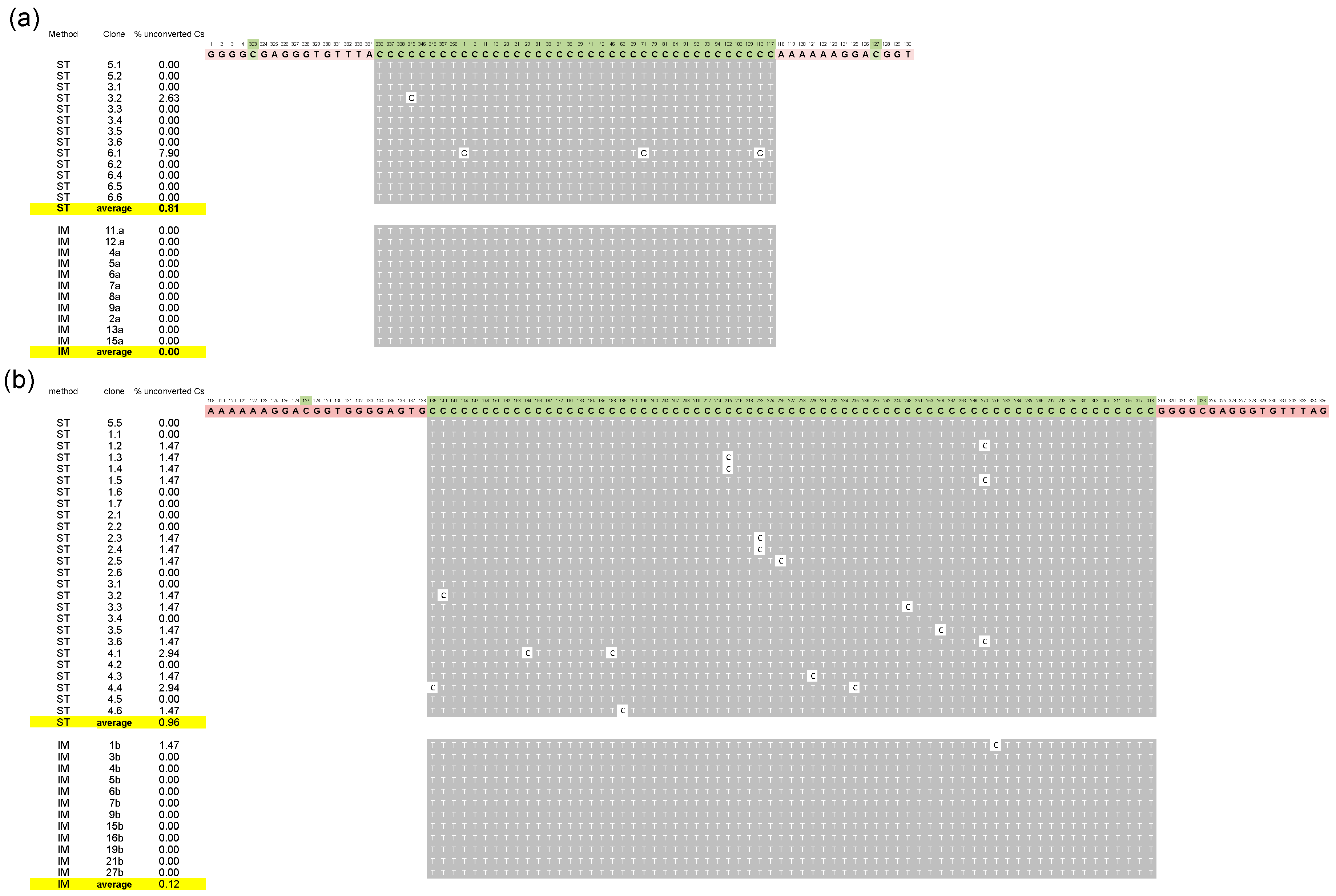
3. Results and Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Sharp, P.A. The centrality of RNA. Cell 2009, 136, 577–580. [Google Scholar] [CrossRef] [PubMed]
- Motorin, Y.; Lyko, F.; Helm, M. 5-methylcytosine in RNA: Detection, enzymatic formation and biological functions. Nucleic Acids Res. 2010, 38, 1415–1430. [Google Scholar] [CrossRef] [PubMed]
- Yi, C.; Pan, T. Cellular dynamics of RNA modification. Acc. Chem. Res. 2011, 44, 1380–1388. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Ma, P.; Liu, Y.; Li, W.; Shu, Y. Multiple functions of m(6)A RNA methylation in cancer. J. Hematol. Oncol. 2018, 11, 48. [Google Scholar] [CrossRef] [PubMed]
- Karikó, K.; Buckstein, M.; Ni, H.; Weissman, D. Suppression of RNA recognition by Toll-like receptors: The impact of nucleoside modification and the evolutionary origin of RNA. Immunity 2005, 23, 165–175. [Google Scholar] [CrossRef]
- Schwartz, S.; Agarwala, S.D.; Mumbach, M.R.; Jovanovic, M.; Mertins, P.; Shishkin, A.; Tabach, Y.; Mikkelsen, T.S.; Satija, R.; Ruvkun, G.; et al. High-resolution mapping reveals a conserved, widespread, dynamic mRNA methylation program in yeast meiosis. Cell 2013, 155, 1409–1421. [Google Scholar] [CrossRef] [PubMed]
- Pan, T. N6-methyl-adenosine modification in messenger and long non-coding RNA. Trends Biochem. Sci. 2013, 38, 204–209. [Google Scholar] [CrossRef]
- Grosjean, H. Modification and editing of RNA: Historical overview and important facts to remember. In Fine-Tuning of RNA Functions by Modification and Editing; Grosjean, H., Ed.; Springer: Berlin, Germany, 2005; pp. 1–22. [Google Scholar]
- Song, X.; Nazar, R.N. Modification of rRNA as a ‘quality control mechanism’ in ribosome biogenesis. FEBS Lett. 2002, 523, 182–186. [Google Scholar] [CrossRef]
- Agris, P.F. Decoding the genome: A modified view. Nucleic Acids Res. 2004, 32, 223–238. [Google Scholar] [CrossRef]
- Alexandrov, A.; Chernyakov, I.; Gu, W.; Hiley, S.L.; Hughes, T.R.; Grayhack, E.J.; Phizicky, E.M. Rapid tRNA decay can result from lack of nonessential modifications. Mol. Cell 2006, 21, 87–96. [Google Scholar] [CrossRef]
- Schaefer, M.; Lyko, F. Solving the Dnmt2 enigma. Chromosoma 2010, 119, 35–40. [Google Scholar] [CrossRef]
- Dominissini, D.; Moshitch-Moshkovitzm, S.; Schwartz, S.; Salmon-Divon, M.; Ungar, L.; Osenberg, S.; Cesarkas, K.; Jacob-Hirsch, J.; Amariglio, N.; Kupiec, M.; et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012, 485, 201–206. [Google Scholar] [CrossRef]
- Meyer, K.D.; Saletore, Y.; Zumbo, P.; Elemento, O.; Mason, C.E.; Jaffrey, S.R. Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell 2012, 149, 1635–1646. [Google Scholar] [CrossRef]
- Cui, X.; Liang, Z.; Shen, L.; Zhang, Q.; Bao, S.; Geng, Y.; Zhang, B.; Leo, V.; Vardy, L.A.; Lu, T.; et al. 5-Methylcytosine RNA methylation in Arabidopsis Thaliana. Mol. Plant 2017, 10, 1387–1399. [Google Scholar] [CrossRef]
- Edelheit, S.; Schwartz, S.; Mumbach, M.R.; Wurtzel, O.; Sorek, R. Transcriptome-Wide Mapping of 5-methylcytidine RNA Modifications in Bacteria, Archaea, and Yeast Reveals m5C within Archaeal mRNAs. PLoS Genet. 2013, 9, e1003602. [Google Scholar] [CrossRef]
- Schaefer, M.; Pollex, T.; Hanna, K.; Lyko, F. RNA cytosine methylation analysis by bisulfite sequencing. Nucleic Acids Res. 2009, 37, e12. [Google Scholar] [CrossRef] [PubMed]
- Pollex, T.; Hanna, K.; Schaefer, M. Detection of cytosine methylation in RNA using bisulfite sequencing. Cold Spring Harb. Protoc. 2010, 10, pdb.prot5505. [Google Scholar] [CrossRef] [PubMed]
- Squires, J.E.; Patel, H.R.; Nousch, M.; Sibbritt, T.; Humphreys, D.T.; Parker, B.J.; Suter, C.M.; Preiss, T. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 2012, 40, 5023–5033. [Google Scholar] [CrossRef] [PubMed]
- Amort, T.; Soulière, M.F.; Wille, A.; Jia, X.Y.; Fiegl, H.; Wörle, H.; Micura, R.; Lusser, A. Long non-coding RNAs as targets for cytosine methylation. RNA Biol. 2013, 10, 1003–1008. [Google Scholar] [CrossRef] [PubMed]
- Motorin, Y.; Helm, M. RNA nucleotide methylation. Wiley Interdiscip. Rev. RNA 2011, 2, 611–631. [Google Scholar] [CrossRef] [PubMed]
- Ding, B. The biology of viroid-host interactions. Annu. Rev. Phytopathol. 2009, 47, 105–131. [Google Scholar] [CrossRef]
- Tsagris, E.M.; Martínez de Alba, A.E.; Gozmanova, M.; Kalantidis, K. Viroids. Cell Microbiol. 2008, 10, 2168–2179. [Google Scholar] [CrossRef] [PubMed]
- Flores, R.; Minoia, S.; Carbonell, A.; Gisel, A.; Delgado, S.; López-Carrasco, A.; Navarro, B.; Di Serio, F. Viroids, the simplest RNA replicons: How they manipulate their hosts for being propagated and how their hosts react for containing the infection. Virus Res. 2015, 209, 136–145. [Google Scholar] [CrossRef]
- Navarro, B.; Gisel, A.; Rodio, M.E.; Delgado, S.; Flores, R.; Di Serio, F. Viroids: How to infect a host and cause disease without encoding proteins. Biochimie 2012, 94, 1474–1480. [Google Scholar] [CrossRef]
- Flores, R.; Serra, P.; Minoia, S.; Di Serio, F.; Navarro, B. Viroids: From genotype to phenotype just relying on RNA sequence and structural motifs. Front. Microbiol. 2012, 3, 217. [Google Scholar] [CrossRef] [PubMed]
- Ding, B. Viroids: Self-replicating, mobile, and fast-evolving noncoding regulatory RNAs. Wiley Interdiscip. Rev. RNA 2010, 1, 362–375. [Google Scholar] [CrossRef]
- Di Serio, F.; Flores, R.; Verhoeven, J.T.; Li, S.F.; Pallás, V.; Randles, J.W.; Sano, T.; Vidalakis, G.; Owens, R.A. Current status of viroid taxonomy. Arch. Virol. 2014, 159, 3467–3478. [Google Scholar] [CrossRef]
- Di Serio, F.; Li, S.F.; Matoušek, J.; Owens, R.A.; Pallás, V.; Randles, J.W.; Sano, T.; Verhoeven, J.T.J.; Vidalakis, G.; Flores, R. ICTV Virus Taxonomy Profile: Avsunviroidae. J. Gen. Virol. 2018, 99, 611–612. [Google Scholar] [CrossRef]
- Domdey, H.; Jank, P.; Sänger, L.; Gross, H.J. Studies on the primary and secondary structure of potato spindle tuber viroid: Products of digestion with ribonuclease A and ribonuclease T1, and modification with bisulfite. Nucleic Acids Res. 1978, 5, 1221–1236. [Google Scholar] [CrossRef]
- Goddard, J.P.; Schulman, L.H. Conversion of exposed cytidine residues to uridine residues in Escherichia coli formylmethionine transfer ribonucleic acid. J. Biol. Chem. 1972, 247, 3864–3867. [Google Scholar]
- Goddard, J.P.; Maden, B.E. Reaction of HeLa cell methyl-labelled 28S ribosomal RNA with sodium bisulfite: A conformational probe for methylated sequences. Nucleic Acids Res. 1976, 3, 431–440. [Google Scholar] [CrossRef]
- Pallas, V.; Navarro, A.; Flores, R. Isolation of a viroid-like RNA from hop different from hop stunt viroid. J. Gen. Virol. 1987, 68, 3201–3205. [Google Scholar] [CrossRef]
- Goll, M.G.; Kirpekar, F.; Maggert, K.A.; Yoder, J.A.; Hsieh, C.L.; Zhang, X.; Golic, K.G.; Jacobsen, S.E.; Bestor, T.H. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science 2006, 311, 395–398. [Google Scholar] [CrossRef] [PubMed]
- Ohno, T.; Takamatsu, N.; Meshi, T.; Okada, Y. Hop stunt viroid: Molecular cloning and nucleotide sequence of the complete cDNA copy. Nucleic Acids Res. 1983, 11, 6185–6197. [Google Scholar] [CrossRef] [PubMed]
- Duran-Vila, N.; Elena, S.F.; Daròs, J.A.; Flores, R. Structure and Evolution of Viroids. In Origin and Evolution of Viruses, 2nd ed.; Domingo, E., Parrish, C.R., Holland, J.J., Eds.; Academic Press: Cambridge, MA, USA, 2008; Chapter 2; pp. 43–64. ISBN 978-0-12-374153-0. [Google Scholar]
- Trixl, L.; Lusser, A. The dynamic RNA modification 5-methylcytosine and its emerging role as an epitranscriptomic mark. WIREs RNA 2019, 10, e1510. [Google Scholar] [CrossRef] [PubMed]
- García-Vílchez, R.; Sevilla, A.; Blanco, S. Post-transcriptional regulation by cytosine-5 methylation of RNA. Biochim. Biophys. Acta Gene Regul. Mech. 2019, 1862, 240–252. [Google Scholar] [CrossRef] [PubMed]
- David, R.; Burgess, A.; Parker, B.; Li, J.; Pulsford, K.; Sibbritt, T.; Preiss, T.; Searle, I.R. Transcriptome-wide mapping of RNA 5-methylcytosine in Arabidopsis mRNAs and noncoding RNAs. Plant Cell 2017, 29, 445–460. [Google Scholar] [CrossRef] [PubMed]
- Aguilo, F.; Li, S.; Balasubramaniyan, N.; Sancho, A.; Benko, S.; Zhang, F.; Vashisht, A.; Rengasamy, M.; Andino, B.; Chen, C.H.; et al. Deposition of 5-Methylcytosine on Enhancer RNAs Enables the Coactivator Function of PGC-1α. Cell Rep. 2016, 14, 479–492. [Google Scholar] [CrossRef]
- López-Carrasco, A.; Flores, R. The predominant circular form of avocado sunblotch accumulates in planta as a free RNA adopting a rod-shaped secondary structure unprotected by tightly bound host proteins. J. Gen. Virol. 2017, 98, 1913–1922. [Google Scholar] [CrossRef]
- López-Carrasco, A.; Flores, R. Dissecting the secondary structure of the circular RNA of a nuclear viroid in vivo: A “naked” rod-like conformation similar but not identical to that observed in vitro. RNA Biol. 2017, 14, 1046–1054. [Google Scholar] [CrossRef]
- Rana, A.K.; Ankri, S. Reviving the RNA World: An Insight into the Appearance of RNA Methyltransferases. Front Genet. 2016, 7, 99. [Google Scholar] [CrossRef] [PubMed]
- Di Serio, F.; Navarro, B.; Flores, F. Origin and evolution of viroids. In Viroids and Satellites; Hadidi, A., Flores, R., Randles, J.W., Palukaitis, P., Eds.; Academic Press: Cambridge, MA, USA, 2017; Chapter 12; pp. 125–134. ISBN 978-0-12-801498-1. [Google Scholar]
| Name | seq (5′ to 3′) * | Position # |
|---|---|---|
| PSTVd_met_1F_plus | GGGGCGAGGGTGTTTAG | 319–335 |
| PSTVd_met_2R_plus | CACTCCCCACCRTCCTTTTTT | 138–118 |
| PSTVd_met_3F_plus | AAAAAAGGAYGGTGGGGAGTG | 118–138 |
| PSTVd_met_4R_plus | CTAAACACCCTCRCCCC | 335–319 |
| PSTVd_met_5F_minus | GAAGAAAGGAAGGGTGAAAA | 196–177 |
| PSTVd_met_6R_minus | ACCACCCCTCRCCCCCTT | 222–239 |
| ASBVd_met_1F_plus | GTGGTGAAYTTTTATTAAAAAAATTAG | 106–132 |
| ASBVd_met_2R_plus | CCACRACTCCTCCTTCTCTCACAA | 109–86 |
| ASBVd_met_3F_minus | GAGTGAAYTAATTTTTTTAATAAAAGTT | 139–112 |
| ASBVd_met_4R_minus | TCTTCAATCTCTTRATCACTTC | 141–162 |
| HSVd_met_1F_plus | GAGAGGYGTGGAGAGAGGG | 106–125 |
| HSVd_met_2R_plus | CCTCCCTRCCTTATTTTTTCTTT | 56–34 |
| tRNA-Asp_1F | GTCGTTGTAGTATAGTGG | |
| tRNA-Asp_2R | ATCGTTCCCAGGTCAGGG |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Di Serio, F.; Torchetti, E.M.; Daròs, J.-A.; Navarro, B. Reassessment of Viroid RNA Cytosine Methylation Status at the Single Nucleotide Level. Viruses 2019, 11, 357. https://doi.org/10.3390/v11040357
Di Serio F, Torchetti EM, Daròs J-A, Navarro B. Reassessment of Viroid RNA Cytosine Methylation Status at the Single Nucleotide Level. Viruses. 2019; 11(4):357. https://doi.org/10.3390/v11040357
Chicago/Turabian StyleDi Serio, Francesco, Enza Maria Torchetti, José-Antonio Daròs, and Beatriz Navarro. 2019. "Reassessment of Viroid RNA Cytosine Methylation Status at the Single Nucleotide Level" Viruses 11, no. 4: 357. https://doi.org/10.3390/v11040357
APA StyleDi Serio, F., Torchetti, E. M., Daròs, J.-A., & Navarro, B. (2019). Reassessment of Viroid RNA Cytosine Methylation Status at the Single Nucleotide Level. Viruses, 11(4), 357. https://doi.org/10.3390/v11040357






