The Natural Flavonoid Compound Deguelin Inhibits HCMV Lytic Replication within Fibroblasts
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cells, Viruses, and Compounds
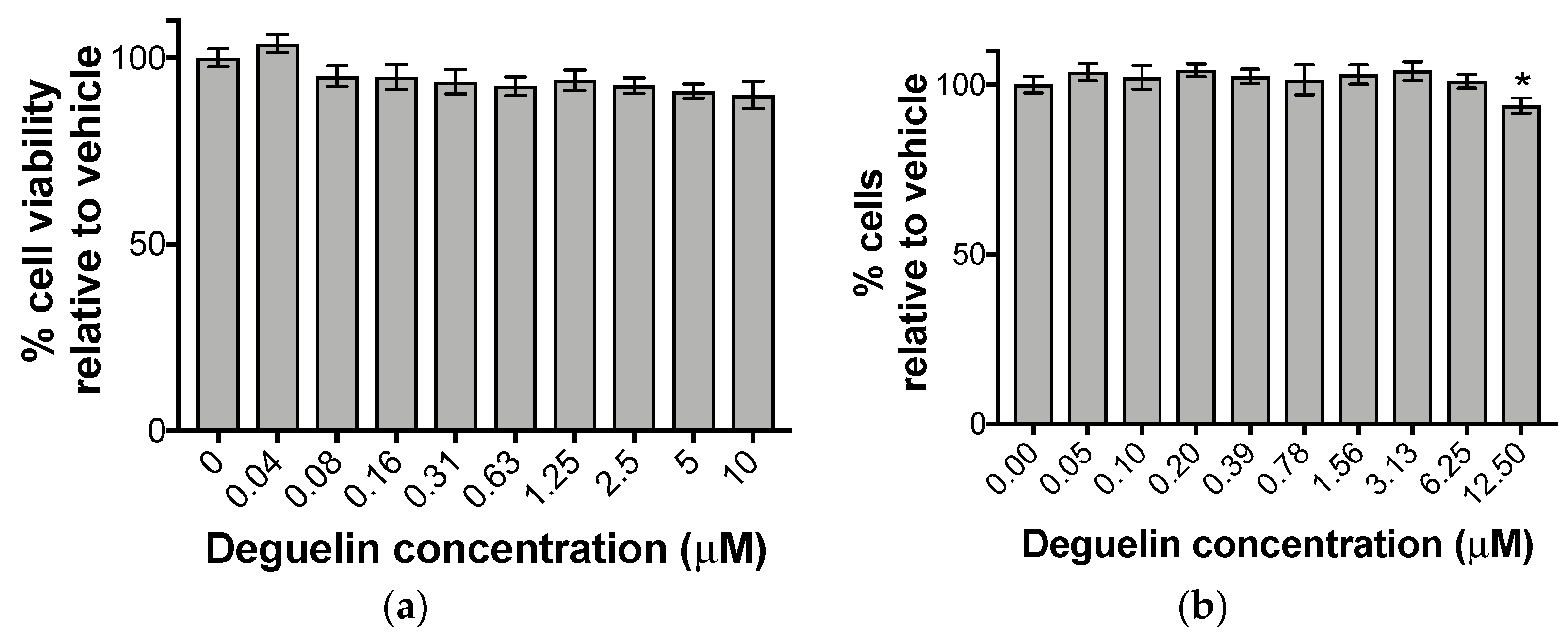
2.2. Cytotoxicity and Cytostaticity Assays
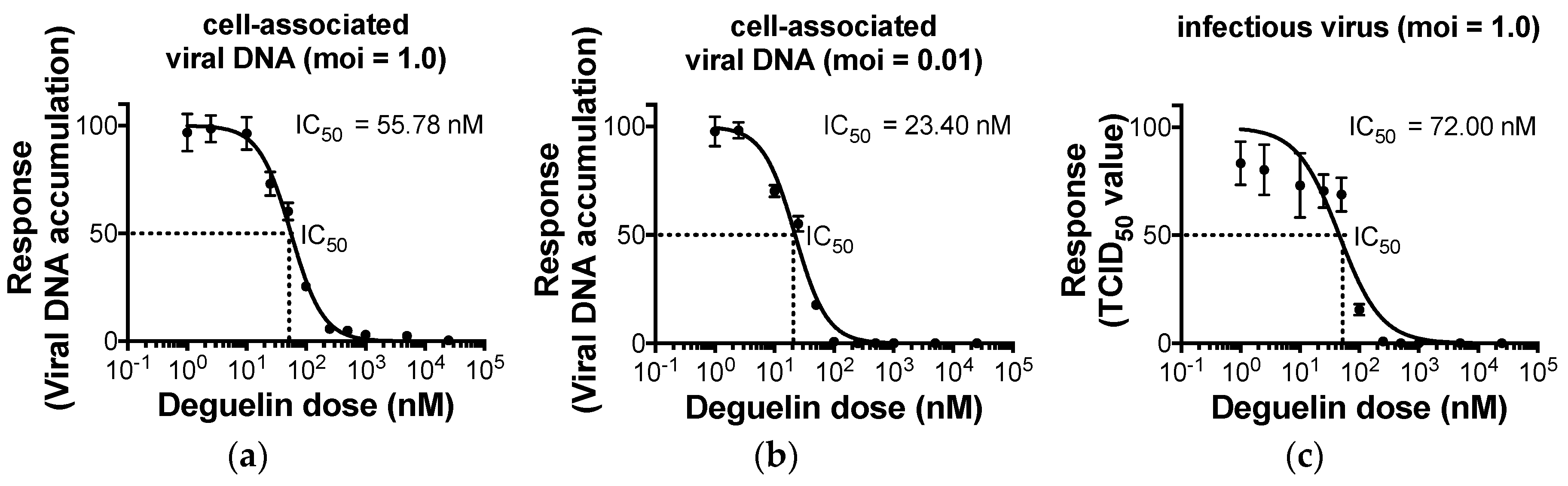
2.3. Half-Maximal Inhibitory Concentration (IC50) Measurements
2.4. Viral RNA, DNA, and Protein Analyses
3. Results
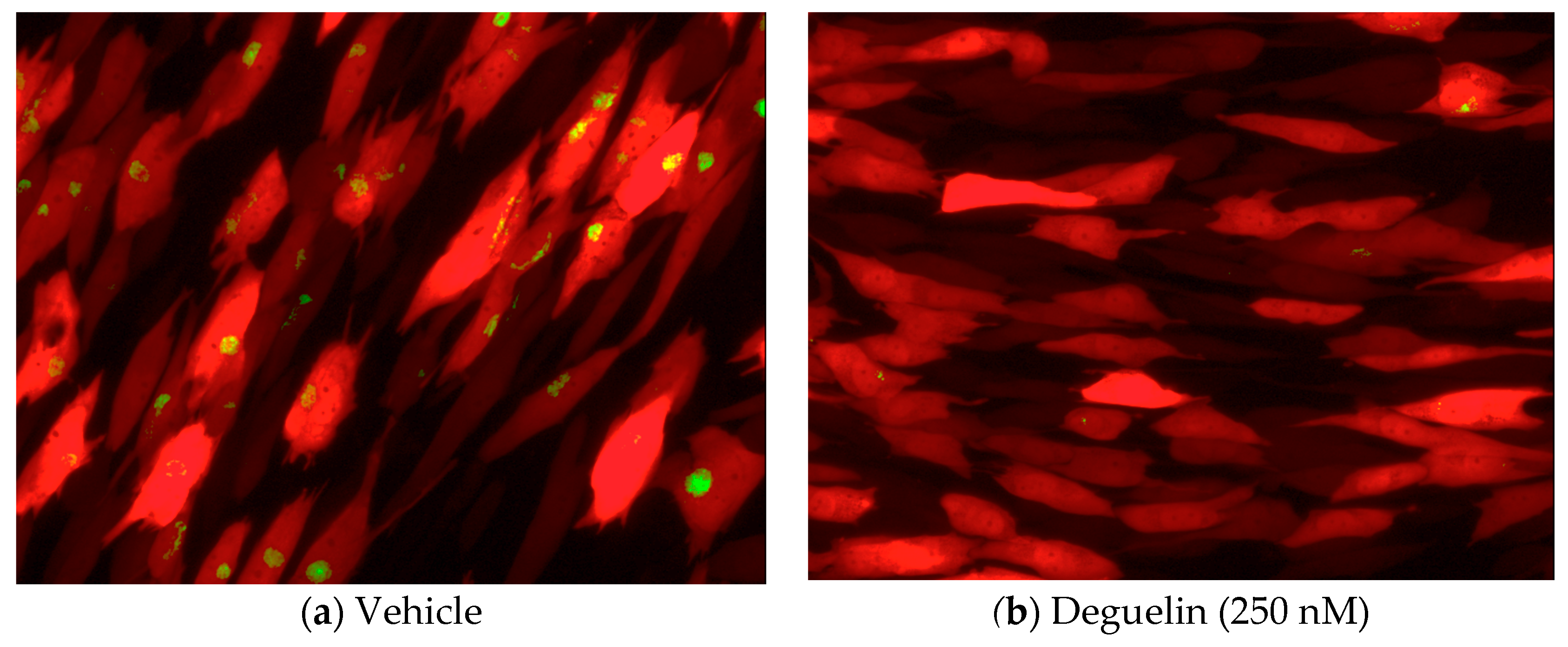
3.1. Deguelin Treatment Inhibits HCMV Lytic Replication
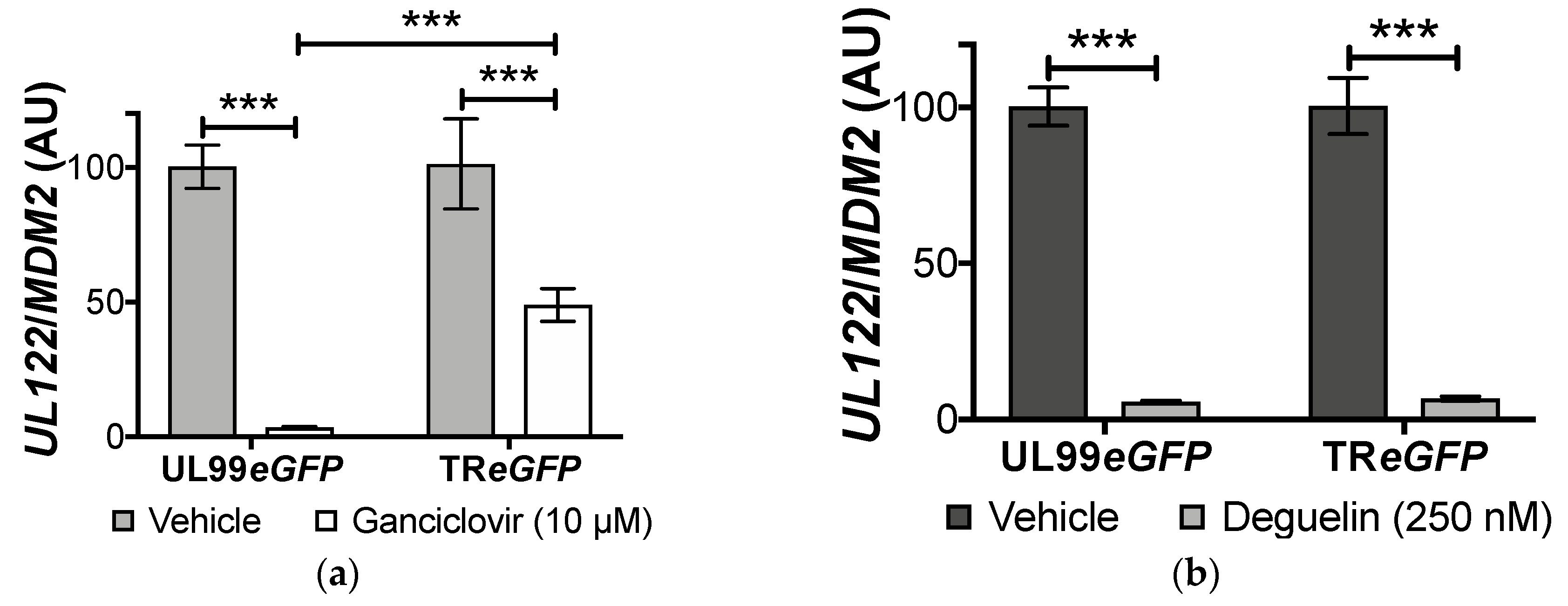
3.2. Deguelin Treatment Effectively Inhibits Ganciclovir-Resistant HCMV
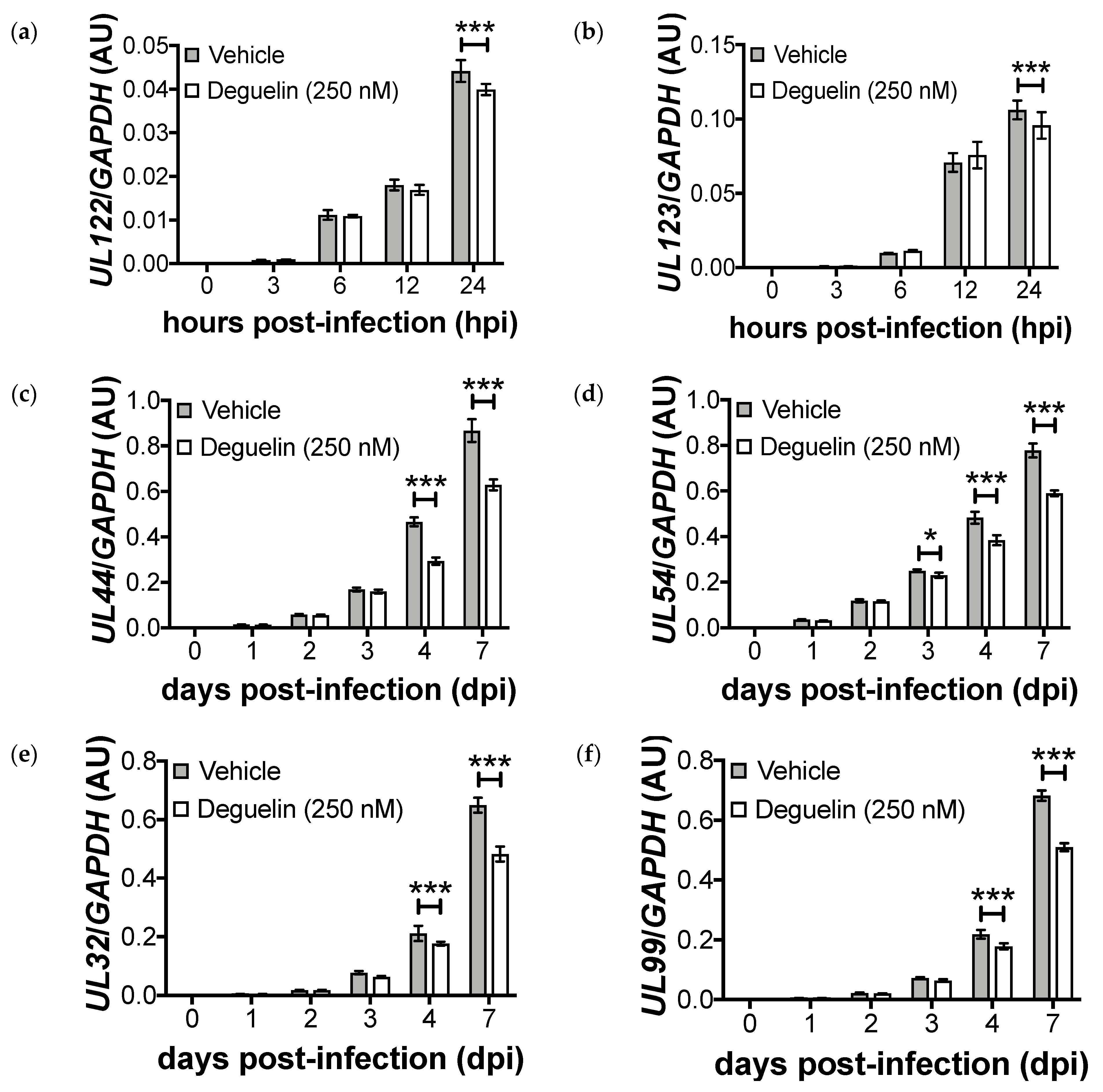
3.3. Lytic Gene Transcription Is Inhibited by Deguelin Treatment
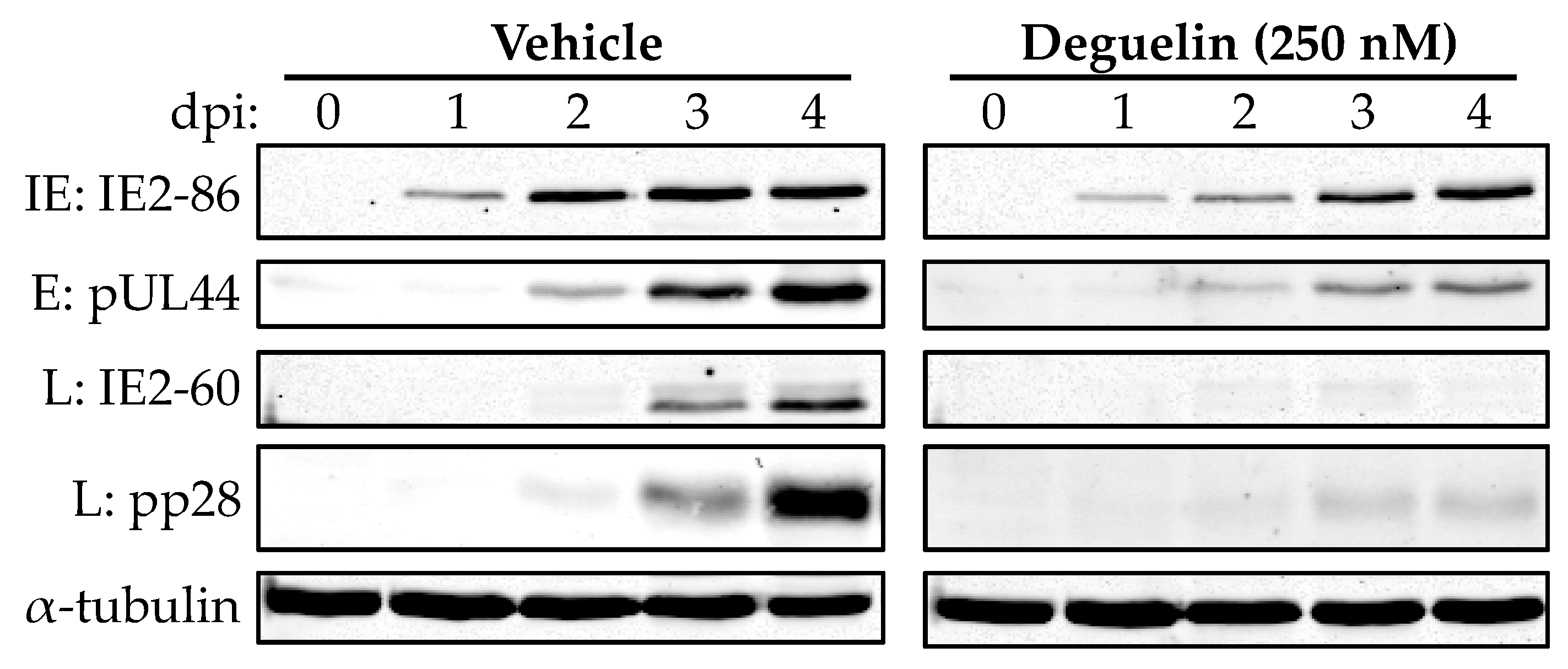
3.4. Deguelin Inhibits Viral Early Protein Expression
4. Discussion
5. Conclusions
Supplementary Materials
Supplementary File 1Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Poole, C.L.; James, S.H. Antiviral therapies for herpesviruses: Current agents and new directions. Clin. Ther. 2018, 40, 1282–1298. [Google Scholar] [CrossRef] [PubMed]
- Littler, E.; Stuart, A.D.; Chee, M.S. Human cytomegalovirus UL97 open reading frame encodes a protein that phosphorylates the antiviral nucleoside analogue ganciclovir. Nature 1992, 358, 160–162. [Google Scholar] [CrossRef] [PubMed]
- Jung, D.; Dorr, A. Single-dose pharmacokinetics of valganciclovir in HIV- and CMV-seropositive subjects. J. Clin. Pharmacol. 1999, 39, 800–804. [Google Scholar] [CrossRef] [PubMed]
- Biron, K.K.; Harvey, R.J.; Chamberlain, S.C.; Good, S.S.; Smith, A.A., 3rd; Davis, M.G.; Talarico, C.L.; Miller, W.H.; Ferris, R.; Dornsife, R.E.; et al. Potent and selective inhibition of human cytomegalovirus replication by 1263w94, a benzimidazole l-riboside with a unique mode of action. Antimicrob. Agents Chemother. 2002, 46, 2365–2372. [Google Scholar] [CrossRef] [PubMed]
- Ringden, O.; Wilczek, H.; Lonnqvist, B.; Gahrton, G.; Wahren, B.; Lernestedt, J.O. Foscarnet for cytomegalovirus infections. Lancet 1985, 1, 1503–1504. [Google Scholar] [CrossRef]
- Lalezari, J.P.; Drew, W.L.; Glutzer, E.; James, C.; Miner, D.; Flaherty, J.; Fisher, P.E.; Cundy, K.; Hannigan, J.; Martin, J.C.; et al. (S)-1-[3-hydroxy-2-(phosphonylmethoxy)propyl]cytosine (cidofovir): Results of a phase i/ii study of a novel antiviral nucleotide analogue. J. Infect. Dis. 1995, 171, 788–796. [Google Scholar] [CrossRef] [PubMed]
- Jacobsen, T.; Sifontis, N. Drug interactions and toxicities associated with the antiviral management of cytomegalovirus infection. Am. J. Health Syst. Pharm. 2010, 67, 1417–1425. [Google Scholar] [CrossRef] [PubMed]
- Bonatti, H.; Sifri, C.D.; Larcher, C.; Schneeberger, S.; Kotton, C.; Geltner, C. Use of cidofovir for cytomegalovirus disease refractory to ganciclovir in solid organ recipients. Surg. Infect. 2017, 18, 128–136. [Google Scholar] [CrossRef] [PubMed]
- Marty, F.M.; Ljungman, P.; Chemaly, R.F.; Maertens, J.; Dadwal, S.S.; Duarte, R.F.; Haider, S.; Ullmann, A.J.; Katayama, Y.; Brown, J.; et al. Letermovir prophylaxis for cytomegalovirus in hematopoietic-cell transplantation. N. Engl. J. Med. 2017, 377, 2433–2444. [Google Scholar] [CrossRef] [PubMed]
- Chou, S. A third component of the human cytomegalovirus terminase complex is involved in letermovir resistance. Antiviral Res. 2017, 148, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Chou, S.; Satterwhite, L.E.; Ercolani, R.J. New locus of drug resistance in the human cytomegalovirus UL56 gene revealed by in vitro exposure to letermovir and ganciclovir. Antimicrob. Agents Chemother. 2018, 62. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Ma, W.; Zheng, W. Deguelin, a novel anti-tumorigenic agent targeting apoptosis, cell cycle arrest and anti-angiogenesis for cancer chemoprevention. Mol. Clin. Oncol. 2013, 1, 215–219. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, C.M.; Shenk, T. Human cytomegalovirus pus27 g protein-coupled receptor homologue is required for efficient spread by the extracellular route but not for direct cell-to-cell spread. J. Virol. 2011, 85, 3700–3707. [Google Scholar] [CrossRef] [PubMed]
- Murphy, E.; Yu, D.; Grimwood, J.; Schmutz, J.; Dickson, M.; Jarvis, M.A.; Hahn, G.; Nelson, J.A.; Myers, R.M.; Shenk, T.E. Coding potential of laboratory and clinical strains of human cytomegalovirus. Proc. Natl. Acad. Sci. USA 2003, 100, 14976–14981. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Connor, C.M.; Nukui, M.; Gurova, K.V.; Murphy, E.A. Inhibition of the fact complex reduces transcription from the human cytomegalovirus major immediate early promoter in models of lytic and latent replication. J. Virol. 2016, 90, 4249–4253. [Google Scholar] [CrossRef] [PubMed]
- Murphy, E.; Vanicek, J.; Robins, H.; Shenk, T.; Levine, A.J. Suppression of immediate-early viral gene expression by herpesvirus-coded micrornas: Implications for latency. Proc. Natl. Acad. Sci. USA 2008, 105, 5453–5458. [Google Scholar] [CrossRef] [PubMed]
- Warming, S.; Costantino, N.; Court, D.L.; Jenkins, N.A.; Copeland, N.G. Simple and highly efficient BAC recombineering using galk selection. Nucleic Acids Res. 2005, 33, e36. [Google Scholar] [CrossRef] [PubMed]
- Terhune, S.; Torigoi, E.; Moorman, N.; Silva, M.; Qian, Z.; Shenk, T.; Yu, D. Human cytomegalovirus ul38 protein blocks apoptosis. J. Virol. 2007, 81, 3109–3123. [Google Scholar] [CrossRef] [PubMed]
- Nevels, M.; Paulus, C.; Shenk, T. Human cytomegalovirus immediate-early 1 protein facilitates viral replication by antagonizing histone deacetylation. Proc. Natl. Acad. Sci. USA 2004, 101, 17234–17239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silva, M.C.; Yu, Q.C.; Enquist, L.; Shenk, T. Human cytomegalovirus UL99-encoded pp28 is required for the cytoplasmic envelopment of tegument-associated capsids. J. Virol. 2003, 77, 10594–10605. [Google Scholar] [CrossRef] [PubMed]
- Smith, I.L.; Taskintuna, I.; Rahhal, F.M.; Powell, H.C.; Ai, E.; Mueller, A.J.; Spector, S.A.; Freeman, W.R. Clinical failure of CMV retinitis with intravitreal cidofovir is associated with antiviral resistance. Arch. Ophthalmol. 1998, 116, 178–185. [Google Scholar] [CrossRef] [PubMed]
- Ji, B.C.; Yu, C.C.; Yang, S.T.; Hsia, T.C.; Yang, J.S.; Lai, K.C.; Ko, Y.C.; Lin, J.J.; Lai, T.Y.; Chung, J.G. Induction of DNA damage by deguelin is mediated through reducing DNA repair genes in human non-small cell lung cancer NCI-h460 cells. Oncol. Rep. 2012, 27, 959–964. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Gao, F.; Ma, X.; Wang, R.; Dong, X.; Wang, W. Deguelin inhibits non-small cell lung cancer via down-regulating hexokinases II-mediated glycolysis. Oncotarget 2017, 8, 32586–32599. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Yu, X.; Ma, X.; Xie, L.; Xia, Z.; Liu, L.; Yu, X.; Wang, J.; Zhou, H.; Zhou, X.; et al. Deguelin attenuates non-small cell lung cancer cell metastasis through inhibiting the ctsz/fak signaling pathway. Cell. Signal. 2018, 50, 131–141. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Yu, X.; Xia, Z.; Yu, X.; Xie, L.; Ma, X.; Zhou, H.; Liu, L.; Wang, J.; Yang, Y.; et al. Repression of noxa by BMI1 contributes to deguelin-induced apoptosis in non-small cell lung cancer cells. J. Cell. Mol. Med. 2018. [Google Scholar] [CrossRef] [PubMed]
- Mehta, R.; Katta, H.; Alimirah, F.; Patel, R.; Murillo, G.; Peng, X.; Muzzio, M.; Mehta, R.G. Deguelin action involves C-MET and EGFR signaling pathways in triple negative breast cancer cells. PLoS ONE 2013, 8, e65113. [Google Scholar] [CrossRef] [PubMed]
- Suh, Y.A.; Kim, J.H.; Sung, M.A.; Boo, H.J.; Yun, H.J.; Lee, S.H.; Lee, H.J.; Min, H.Y.; Suh, Y.G.; Kim, K.W.; et al. A novel antitumor activity of deguelin targeting the insulin-like growth factor (IGF) receptor pathway via up-regulation of IGF-binding protein-3 expression in breast cancer. Cancer Lett. 2013, 332, 102–109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thamilselvan, V.; Menon, M.; Thamilselvan, S. Anticancer efficacy of deguelin in human prostate cancer cells targeting glycogen synthase kinase-3 beta/beta-catenin pathway. Int. J. Cancer 2011, 129, 2916–2927. [Google Scholar] [CrossRef] [PubMed]
- Kang, W.; Zheng, X.; Wang, P.; Guo, S. Deguelin exerts anticancer activity of human gastric cancer MGC-803 and MKN-45 cells in vitro. Int. J. Mol. Med. 2018, 41, 3157–3166. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Lee, J.H.; Jung, K.H.; Hong, S.S. Deguelin promotes apoptosis and inhibits angiogenesis of gastric cancer. Oncol. Rep. 2010, 24, 957–963. [Google Scholar] [PubMed]
- Li, Z.; Wu, C.; Wu, J.; Ji, M.; Shi, L.; Jiang, J.; Xu, B.; Yuan, J. Synergistic antitumor effects of combined deguelin and cisplatin treatment in gastric cancer cells. Oncol. Lett. 2014, 8, 1603–1607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.H.; Lee, D.H.; Lee, H.S.; Choi, J.S.; Kim, K.W.; Hong, S.S. Deguelin inhibits human hepatocellular carcinoma by antiangiogenesis and apoptosis. Oncol. Rep. 2008, 20, 129–134. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Yu, X.; Li, W.; Liu, T.; Deng, G.; Liu, W.; Liu, H.; Gao, F. Deguelin suppresses angiogenesis in human hepatocellular carcinoma by targeting HGF-C-MET pathway. Oncotarget 2018, 9, 152–166. [Google Scholar] [CrossRef] [PubMed]
- Baba, Y.; Kato, Y. Deguelin, a novel anti-tumorigenic agent in human esophageal squamous cell carcinoma. EBioMedicine 2017, 26, 10. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Liang, Q.; Liu, W.; Zhou, L.; Li, W.; Liu, H. Deguelin, an aurora B kinase inhibitor, exhibits potent anti-tumor effect in human esophageal squamous cell carcinoma. EBioMedicine 2017, 26, 100–111. [Google Scholar] [CrossRef] [PubMed]
- Bortul, R.; Tazzari, P.L.; Billi, A.M.; Tabellini, G.; Mantovani, I.; Cappellini, A.; Grafone, T.; Martinelli, G.; Conte, R.; Martelli, A.M. Deguelin, a PI3K/AKT inhibitor, enhances chemosensitivity of leukaemia cells with an active pi3k/akt pathway. Br. J. Haematol. 2005, 129, 677–686. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.P.; Yi, S.; Wen, L.; Zhang, B.P.; Zhao, Z.; Hu, J.Y.; Zhao, F.; He, J.; Fang, J.; Zhang, C.; et al. Nontoxic-dose of deguelin induce NPMC+ aml cell differentiation by selectively targeting MT NPM1/SIRT1 instead of HDAC1/3. Curr. Cancer Drug Targets 2014, 14, 685–699. [Google Scholar] [CrossRef] [PubMed]
- Yi, S.; Wen, L.; He, J.; Wang, Y.; Zhao, F.; Zhao, J.; Zhao, Z.; Cui, G.; Chen, Y. Deguelin, a selective silencer of the NPM1 mutant, potentiates apoptosis and induces differentiation in aml cells carrying the npm1 mutation. Ann. Hematol. 2015, 94, 201–210. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhao, Z.; Yi, S.; Wen, L.; He, J.; Hu, J.; Ruan, J.; Fang, J.; Chen, Y. Deguelin induced differentiation of mutated npm1 acute myeloid leukemia in vivo and in vitro. Anticancer Drugs 2017, 28, 723–738. [Google Scholar] [CrossRef] [PubMed]
- Boreddy, S.R.; Srivastava, S.K. Deguelin suppresses pancreatic tumor growth and metastasis by inhibiting epithelial-to-mesenchymal transition in an orthotopic model. Oncogene 2013, 32, 3980–3991. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.D.; Zhao, Y.; Zhang, M.; He, R.Z.; Shi, X.H.; Guo, X.J.; Shi, C.J.; Peng, F.; Wang, M.; Shen, M.; et al. Inhibition of autophagy by deguelin sensitizes pancreatic cancer cells to doxorubicin. Int. J. Mol. Sci. 2017, 18, 370. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Lu, S.; Cai, H.; Kang, M.; Qin, W.; Li, C.; Wu, Y. Deguelin inhibits proliferation and migration of human pancreatic cancer cells in vitro targeting hedgehog pathway. Oncol. Lett. 2016, 12, 2761–2765. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baba, Y.; Fujii, M.; Maeda, T.; Suzuki, A.; Yuzawa, S.; Kato, Y. Deguelin induces apoptosis by targeting both EGFR-AKT and IGF1R-AKT pathways in head and neck squamous cell cancer cell lines. Biomed. Res. Int. 2015, 2015, 657179. [Google Scholar] [CrossRef] [PubMed]
- Baba, Y.; Maeda, T.; Suzuki, A.; Takada, S.; Fujii, M.; Kato, Y. Deguelin potentiates apoptotic activity of an egfr tyrosine kinase inhibitor (AG1478) in PIK3CA-mutated head and neck squamous cell carcinoma. Int. J. Mol. Sci. 2017, 18, 262. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.L.; Ji, C.; Bi, Z.G.; Lu, C.C.; Wang, R.; Gu, B.; Cheng, L. Deguelin induces both apoptosis and autophagy in cultured head and neck squamous cell carcinoma cells. PLoS ONE 2013, 8, e54736. [Google Scholar] [CrossRef] [PubMed]
- Yan, B.; Zhao, D.; Yao, Y.; Bao, Z.; Lu, G.; Zhou, J. Deguelin induces the apoptosis of lung squamous cell carcinoma cells through regulating the expression of galectin-1. Int. J. Biol. Sci. 2016, 12, 850–860. [Google Scholar] [CrossRef] [PubMed]
- Robles, A.J.; Cai, S.X.; Cichewicz, R.H.; Mooberry, S.L. Selective activity of deguelin identifies therapeutic targets for androgen receptor-positive breast cancer. Breast Cancer Res. Treat. 2016, 157, 475–488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oh, S.H.; Woo, J.K.; Jin, Q.; Kang, H.J.; Jeong, J.W.; Kim, K.W.; Hong, W.K.; Lee, H.Y. Identification of novel antiangiogenic anticancer activities of deguelin targeting hypoxia-inducible factor-1 alpha. Int. J. Cancer 2008, 122, 5–14. [Google Scholar] [CrossRef] [PubMed]
- Vincent, H.A.; Ziehr, B.; Moorman, N.J. Human cytomegalovirus strategies to maintain and promote mRNA translation. Viruses 2016, 8, 97. [Google Scholar] [CrossRef] [PubMed]
- Chun, K.H.; Kosmeder, J.W., 2nd; Sun, S.; Pezzuto, J.M.; Lotan, R.; Hong, W.K.; Lee, H.Y. Effects of deguelin on the phosphatidylinositol 3-kinase/AKT pathway and apoptosis in premalignant human bronchial epithelial cells. J. Natl. Cancer Inst. 2003, 95, 291–302. [Google Scholar] [CrossRef] [PubMed]
- Botto, S.; Streblow, D.N.; DeFilippis, V.; White, L.; Kreklywich, C.N.; Smith, P.P.; Caposio, P. Il-6 in human cytomegalovirus secretome promotes angiogenesis and survival of endothelial cells through the stimulation of survivin. Blood 2011, 117, 352–361. [Google Scholar] [CrossRef] [PubMed]
- Vaira, V.; Lee, C.W.; Goel, H.L.; Bosari, S.; Languino, L.R.; Altieri, D.C. Regulation of survivin expression by IGF-1/MTOR signaling. Oncogene 2007, 26, 2678–2684. [Google Scholar] [CrossRef] [PubMed]
- DeMeritt, I.B.; Milford, L.E.; Yurochko, A.D. Activation of the NF-κb pathway in human cytomegalovirus-infected cells is necessary for efficient transactivation of the major immediate-early promoter. J. Virol. 2004, 78, 4498–4507. [Google Scholar] [CrossRef] [PubMed]
- Nair, A.S.; Shishodia, S.; Ahn, K.S.; Kunnumakkara, A.B.; Sethi, G.; Aggarwal, B.B. Deguelin, an akt inhibitor, suppresses ikappabalpha kinase activation leading to suppression of NF-κb-regulated gene expression, potentiation of apoptosis, and inhibition of cellular invasion. J. Immunol. 2006, 177, 5612–5622. [Google Scholar] [CrossRef] [PubMed]
- Murillo, G.; Peng, X.; Torres, K.E.; Mehta, R.G. Deguelin inhibits growth of breast cancer cells by modulating the expression of key members of the Wnt signaling pathway. Cancer Prev. Res. 2009, 2, 942–950. [Google Scholar] [CrossRef] [PubMed]
- De Wit, R.H.; Mujic-Delic, A.; van Senten, J.R.; Fraile-Ramos, A.; Siderius, M.; Smit, M.J. Human cytomegalovirus encoded chemokine receptor us28 activates the HIF-1α/PKM2 axis in glioblastoma cells. Oncotarget 2016, 7, 67966–67985. [Google Scholar] [CrossRef] [PubMed]
- Heiske, A.; Roettger, Y.; Bacher, M. Cytomegalovirus upregulates vascular endothelial growth factor and its second cellular kinase domain receptor in human fibroblasts. Viral Immunol. 2012, 25, 360–367. [Google Scholar] [CrossRef] [PubMed]
- Langemeijer, E.V.; Slinger, E.; de Munnik, S.; Schreiber, A.; Maussang, D.; Vischer, H.; Verkaar, F.; Leurs, R.; Siderius, M.; Smit, M.J. Constitutive β-catenin signaling by the viral chemokine receptor US28. PLoS ONE 2012, 7, e48935. [Google Scholar] [CrossRef] [PubMed]
- Maussang, D.; Langemeijer, E.; Fitzsimons, C.P.; Stigter-van Walsum, M.; Dijkman, R.; Borg, M.K.; Slinger, E.; Schreiber, A.; Michel, D.; Tensen, C.P.; et al. The human cytomegalovirus-encoded chemokine receptor us28 promotes angiogenesis and tumor formation via cyclooxygenase-2. Cancer Res. 2009, 69, 2861–2869. [Google Scholar] [CrossRef] [PubMed]
- Maussang, D.; Verzijl, D.; van Walsum, M.; Leurs, R.; Holl, J.; Pleskoff, O.; Michel, D.; van Dongen, G.A.; Smit, M.J. Human cytomegalovirus-encoded chemokine receptor us28 promotes tumorigenesis. Proc. Natl. Acad. Sci. USA 2006, 103, 13068–13073. [Google Scholar] [CrossRef] [PubMed]
| Primer Use | Sequence | Primer Name |
|---|---|---|
| galK insertion 1 | CAACGTCCACCCACCCCCGGGACAAAAAAGCCCGCCGCCCCCTTGTCCTTTCCTGTTGACAATTAATCATCGGCA | UL99galK 5′ |
| galK insertion 1 | GTGTCCCATTCCCGACTCGCGAATCGTACGCGAGACCTGAAAGTTTATGAGTCAGCACTGTCCTGCTCCTT | UL99galK 3′ |
| Reversion primer | CAACGTCCACCCACCCCCGGGACAAAAAAGCCCGCCGCCCCCTTGTCCTTTGTGAGCAAGGGCGAGGAGCTGTTCACCG | UL99eGFP 5′ |
| Reversion primer | GTGTCCCATTCCCGACTCGCGAATCGTACGCGAGACCTGAAAGTTTATGAGTTACTTGTACAGCTCGTCCATGCCGAGAGT | UL99eGFP 3′ |
| qPCR/RTqPCR | ATGGTTTTGCAGGCTTTGATG | UL122 FOR |
| qPCR/RTqPCR | ACCTGCCCTTCACGATTCC | UL122 REV |
| RTqPCR | GCCTTCCCTAAGACCACCAAT | UL123 FOR |
| RTqPCR | ATTTTCTGGGCATAAGCCATAATC | UL123 REV |
| RTqPCR | GGC GTG AAA AAC ATG CGT ATC AAC | UL44 FOR |
| RTqPCR | TAC AAC AGC GTG TCG TGC TCC G | UL44 REV |
| RTqPCR | CCCTCGGCTTCTCACAACAAT | UL54 FOR |
| RTqPCR | CGAGTTAGTCTTGGCCATGCAT | UL54 REV |
| RTqPCR | CACACAACACCGTCGTCCGATTAC | UL32 FOR |
| RTqPCR | GGTTTCTGGCTCGTGGATGTCG | UL32 REV |
| RTqPCR | GTGTCCCATTCCCGACTCG | UL99 FOR |
| RTqPCR | TTCACAACGTCCACCCACC | UL99 REV |
| RTqPCR | ACCCACTCCTCCACCTTTGAC | GAPDH 2 FOR |
| RTqPCR | CTGTTGCTGTAGCCAAATTCGT | GAPDH REV |
| qPCR | CCCCTTCCATCACATTGCA | MDM2 3 FOR |
| qPCR | AGTTTGGCTTTCTCAGAGATTTCC | MDM2 REV |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nukui, M.; O’Connor, C.M.; Murphy, E.A. The Natural Flavonoid Compound Deguelin Inhibits HCMV Lytic Replication within Fibroblasts. Viruses 2018, 10, 614. https://doi.org/10.3390/v10110614
Nukui M, O’Connor CM, Murphy EA. The Natural Flavonoid Compound Deguelin Inhibits HCMV Lytic Replication within Fibroblasts. Viruses. 2018; 10(11):614. https://doi.org/10.3390/v10110614
Chicago/Turabian StyleNukui, Masatoshi, Christine M. O’Connor, and Eain A. Murphy. 2018. "The Natural Flavonoid Compound Deguelin Inhibits HCMV Lytic Replication within Fibroblasts" Viruses 10, no. 11: 614. https://doi.org/10.3390/v10110614
APA StyleNukui, M., O’Connor, C. M., & Murphy, E. A. (2018). The Natural Flavonoid Compound Deguelin Inhibits HCMV Lytic Replication within Fibroblasts. Viruses, 10(11), 614. https://doi.org/10.3390/v10110614





