Comparative Analysis of MicroRNA Expression in Three Paulownia Species with Phytoplasma Infection
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Construction and Sequencing of Small RNA and Degradome Libraries
2.3. Bioinformatics Analysis of the sRNA Sequencing Data
2.4. Identification of miRNAs Related to PaWB
2.5. Analysis of Target Genes of the Differentially Expressed miRNA
2.6. Verification of DEMs and Their Targets by qRT-PCR Analysis
3. Results
3.1. sRNAs Sequencing and miRNA Identification
3.2. Differentially Expressed miRNAs in the Three Paulownia Species
3.3. Identification of Target Genes of the DEMs in the Three Paulownia Species
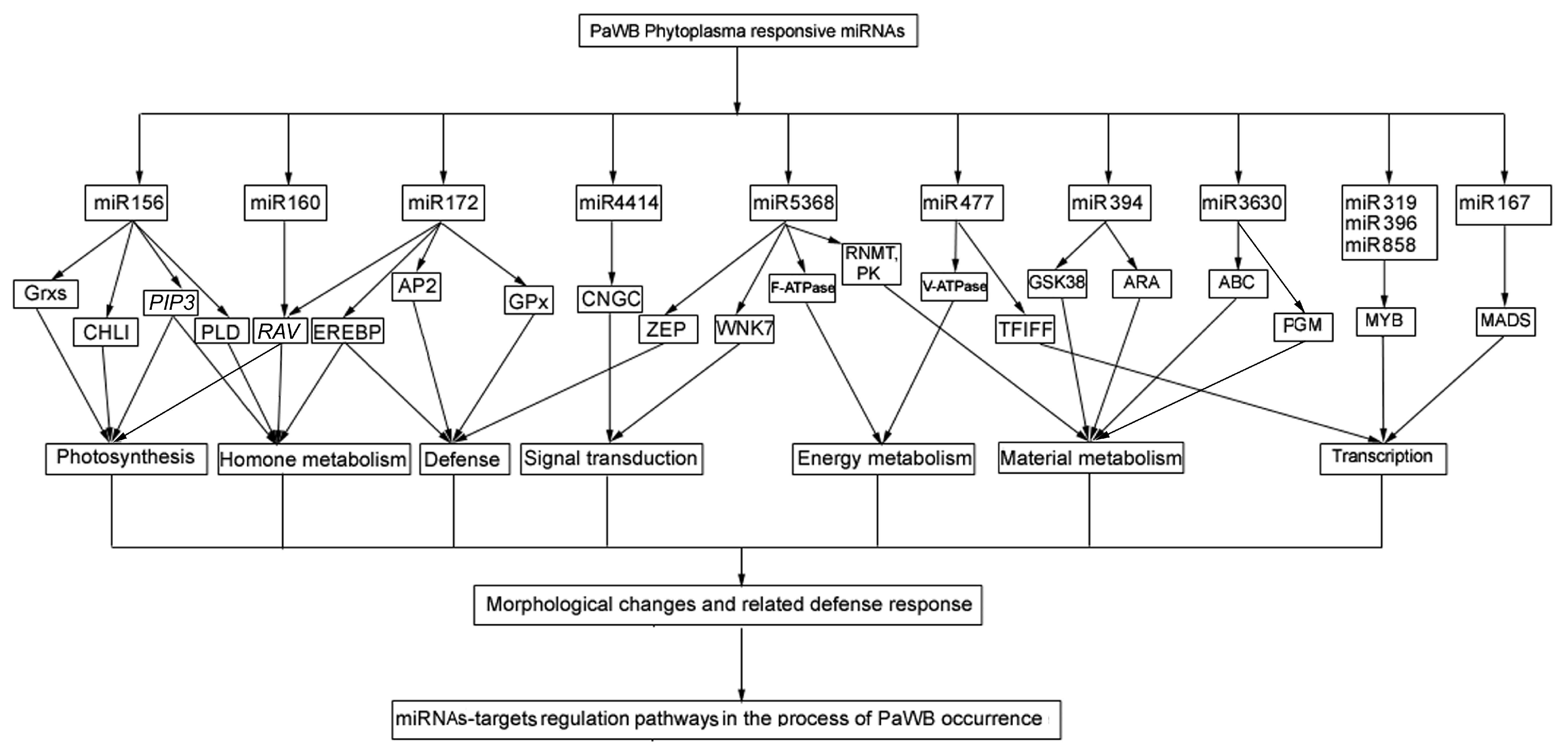
3.4. Functional Analysis of the Target Genes of the DEMs
3.4.1. Genes Associated with Photosynthesis
3.4.2. Genes Associated with Plant Hormones
3.4.3. Genes Involved in Plant Defense and Related Signal Pathways
3.4.4. Genes Involved in Energy Metabolism Pathway
3.4.5. Genes Involved in Material Metabolic Pathways
3.5. Expression Patterns of the DEMs and Their Target Genes by qRT-PCR
4. Discussion
4.1. MiR156 Is the Key Regulatory Factor Related to Witches’ Broom
4.2. A miR156–miR160–miR5368 Cluster Regulates Leaf Yellowing in Phytoplasma-Infected Paulownia
4.3. A miR160–miR172–miR397 Cluster Regulates Leaf Dwarf Morphology in Phytoplasma-Infected Paulownia
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Namba, S. Molecular biological studies on phytoplasmas. J. Gen. Plant Pathol. 2002, 68, 257–259. [Google Scholar] [CrossRef]
- Bayliss, K.L.; Saqib, M.; Dell, B.; Jones, M.G.K.; Hardy, G.E.S.J. First record of ‘candidatus phytoplasma australiense’ in paulownia trees. Australas Plant Pathol. 2005, 34, 123–124. [Google Scholar] [CrossRef]
- Bertaccini, A.; Duduk, B.; Paltrinieri, S.; Contaldo, N. Phytoplasmas and phytoplasma diseases: A severe threat to agriculture. Am. J. Plant Sci. 2014, 5, 1763–1788. [Google Scholar] [CrossRef]
- Rid, M.; Mesca, C.; Ayasse, M.; Gross, J. Apple proliferation phytoplasma influences the pattern of plant volatiles emitted depending on pathogen virulence. Front. Ecol. Environ. 2016, 3, 152. [Google Scholar] [CrossRef]
- Margaria, P.; Abbã, S.; Palmano, S. Novel aspects of grapevine response to phytoplasma infection investigated by a proteomic and phospho-proteomic approach with data integration into functional networks. BMC Genom. 2013, 14, 38. [Google Scholar] [CrossRef] [PubMed]
- Sugio, A.; Kingdom, H.N.; Maclean, A.M.; Grieve, V.M.; Hogenhout, S.A. Phytoplasma protein effector SAP11 enhances insect vector reproduction by manipulating plant development and defense hormone biosynthesis. Proc. Natl. Acad. Sci. USA 2011, 108, E1254–E1263. [Google Scholar] [CrossRef] [PubMed]
- Gai, Y.P.; Li, Y.Q.; Guo, F.Y.; Yuan, C.Z.; Mo, Y.Y.; Zhang, H.L.; Wang, H.; Ji, X.L. Analysis of phytoplasma-responsive sRNAs provide insight into the pathogenic mechanisms of mulberry yellow dwarf disease. Sci. Rep. 2014, 4, 5378. [Google Scholar] [CrossRef] [PubMed]
- Mollayi, S.; Zadali, R.; Farzaneh, M.; Ghassempour, A. Metabolite profiling of mexican lime (Citrus aurantifolia) leaves during the progression of witches’ broom disease. Phytochem. Lett. 2015, 13, 290–296. [Google Scholar] [CrossRef]
- Nejat, N.; Cahill, D.M.; Vadamalai, G.; Ziemann, M.; Rookes, J.; Naderali, N. Transcriptomics-based analysis using RNA-seq of the coconut (Cocos nucifera) leaf in response to yellow decline phytoplasma infection. Mol. Gene Genom. 2015, 290, 1899–1910. [Google Scholar] [CrossRef] [PubMed]
- Djamitchatchou, A.T.; Sananmishra, N.; Ntushelo, K.; Dubery, I.A. Functional roles of microRNAs in agronomically important plants—Potential as targets for crop improvement and protection. Front. Plant. Sci. 2017, 8, 378. [Google Scholar] [CrossRef]
- Bester, R.; Burger, J.T.; Maree, H.J. Differential expression of mirnas and associated gene targets in grapevine leafroll-associated virus 3-infected plants. Arch. Virol. 2016, 162, 987–996. [Google Scholar] [CrossRef] [PubMed]
- Jeyaraj, A.; Zhang, X.; Hou, Y.; Shangguan, M.; Gajjeraman, P.; Li, Y.; Wei, C. Genome-wide identification of conserved and novel micrornas in one bud and two tender leaves of tea plant (Camellia sinensis) by small rna sequencing, microarray-based hybridization and genome survey scaffold sequences. BMC Plant. Biol. 2017, 17, 212. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Yu, H.; Tang, G.; Huang, T. Small but powerful: Function of microRNAs in plant development. Plant Cell Rep. 2018, 37, 515–528. [Google Scholar] [CrossRef] [PubMed]
- Shao, F.; Zhang, Q.; Liu, H.; Lu, S.; Qiu, D. Genome-wide identification and analysis of microRNAs involved in Witches’-Broom phytoplasma response in Ziziphus jujuba. PLoS ONE 2016, 11, e0166099. [Google Scholar] [CrossRef] [PubMed]
- Ehya, F.; Monavarfeshani, A.; Mohseni, F.E.; Karimi, F.L.; Khayam, N.M.; Mardi, M.; Salekdeh, G.H. Phytoplasma-responsive microRNAs modulate hormonal, nutritional, and stress signalling pathways in Mexican lime trees. PLoS ONE 2013, 8, e66372. [Google Scholar] [CrossRef] [PubMed]
- Gai, Y.P.; Zhao, H.N.; Zhao, Y.N.; Zhu, B.S.; Yuan, S.S.; Li, S.; Guo, F.Y.; Ji, X.L. MiRNA-seq-based profiles of miRNAs in mulberry phloem sap provide insight into the pathogenic mechanisms of Mulberry yellow dwarf disease. Sci. Rep. 2018, 8, 812. [Google Scholar] [CrossRef] [PubMed]
- Snyman, M.C.; Solofoharivelo, M.C.; Souzarichards, R.; Stephan, D.; Murray, S.; Burger, J.T. The use of high-throughput small RNA sequencing reveals differentially expressed micrornas in response to aster yellows phytoplasma-infection in Vitis vinifera cv. ‘Chardonnay’. PLoS ONE 2017, 12, e0182629. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Dong, Y.; Fan, G.; Zhao, Z.; Deng, M.; Cao, X.; Niu, S. Discovery of genes related to witches’ broom disease in Paulownia tomentosa × Paulownia fortunei by a de novo assembled transcriptome. PLoS ONE 2013, 8, e80238. [Google Scholar] [CrossRef] [PubMed]
- Mou, H.Q.; Lu, J.; Zhu, S.F.; Lin, C.L.; Tian, G.Z.; Xu, X.; Zhao, W.J. Transcriptomic analysis of Paulownia infected by Paulownia witches’ broom Phytoplasma. PLoS ONE 2013, 8, e77217. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Fan, G.; Deng, M.; Zhao, Z.; Dong, Y. Identification of genes related to Paulownia witches’ broom by AFLP and MSAP. Int. J. Mol. Sci. 2014, 15, 14669–14683. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Fan, G.; Zhao, Z.; Deng, M.; Dong, Y. Morphological changes of Paulownia seedlings infected phytoplasmas reveal the genes associated with witches’ broom through AFLP and MSAP. PLoS ONE 2014, 9, e112533. [Google Scholar] [CrossRef] [PubMed]
- Fan, G.; Dong, Y.; Deng, M.; Zhao, Z.; Niu, S.; Xu, E. Plant-pathogen interaction, circadian rhythm, and hormone-related gene expression provide indicators of phytoplasma infection in Paulownia fortunei. Int. J. Mol. Sci. 2014, 15, 23141–23162. [Google Scholar] [CrossRef] [PubMed]
- Fan, G.; Cao, X.; Niu, S.; Deng, M.; Zhao, Z.; Dong, Y. Transcriptome, microrna, and degradome analyses of the gene expression of paulownia with phytoplamsa. BMC Genom. 2015, 16, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.L.; Zhao, Z.L.; Fan, G.Q.; Cao, X.B. Effects of dimethyl sulphonate on the morphological changes of Paulownia fortunei seedlings with witches’ broom and their DNA base sequences. J. Henan Agric. Univ. 2011, 45, 287–291. [Google Scholar]
- Fan, G.; Zhao, G.; Zhai, X.; Cao, X. Morphological changes of Paulownia tomentosa × Paulownia fortunei seedlings with witches’ broom treated with dimethyl sulphonate and their SSR analyses. J. Northeast For. Univ. 2011, 39, 30–33. [Google Scholar]
- Fan, G.; Zhao, G.; Zhai, X.; Cao, X. Effect of dimethyl sulphate on phytoplasma of Paulownia tomentosa seedling infected by witches’ broom and its DNA loci at SSR level. J. Nanjing For. Univ. 2012, 36, 78–84. [Google Scholar]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef] [PubMed]
- Bonnet, E.; Wuyts, J.; Rouzé, P.; Peer, Y.V.D. Evidence that microRNA precursors, unlike other non-coding RNAs, have lower folding free energies than random sequences. Bioinformatics 2004, 20, 2911–2917. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.H.; Pan, X.P.; Cox, S.B.; Cobb, G.P.; Anderson, T.A. Evidence that miRNAs are different from other RNAs. Cell Mol. Life Sci. 2006, 63, 246–254. [Google Scholar] [CrossRef] [PubMed]
- Axtell, M.J. Classification and comparison of small RNAs from plants. Annu. Rev. Plant. Biol. 2013, 64, 137–159. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.S.; Zeng, H.Q.; Liu, Z.P.; Yang, Z.M. Genome-wide identification of Medicago truncatula microRNAs and their targets reveals their differential regulation by heavy metal. Plant Cell Environ. 2012, 35, 86–99. [Google Scholar] [CrossRef] [PubMed]
- Audic, S.; Claverie, J.M. The significance of digital gene expression profiles. Genom. Res. 1997, 7, 986–995. [Google Scholar] [CrossRef]
- Chen, C.; Ridzon, D.A.; Broomer, A.J.; Zhou, Z.; Lee, D.H.; Nguyen, J.T.; Barbisin, M.; Xu, N.L.; Mahuvakar, V.R.; Andersen, M.R. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005, 33, e179. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔct method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Chen, C.Y.; Wang, K.C.; Luo, M.; Tai, R.; Yuan, L.; Zhao, M.; Yang, S.; Tian, G.; Cui, Y.; et al. Phytochrome interacting factor 3 associates with the histone deacetylase HDA15 in repression of chlorophyll biosynthesis and photosynthesis in etiolated Arabidopsis seedlings. Plant Cell 2013, 25, 1258–1273. [Google Scholar] [CrossRef] [PubMed]
- Ni, M.; Tepperman, J.M.; Quail, P.H. PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 1998, 95, 657–667. [Google Scholar] [CrossRef]
- Vranish, J.N.; Das, D.; Barondeau, D.P. Real-time kinetic probes support monothiol glutaredoxins as intermediate carriers in Fe-S cluster biosynthetic pathways. ACS Chem. Biol. 2016, 11, 3114–3121. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Luo, Q.; Yang, C.; Han, Y.; Li, W. A RAV-like transcription factor controls photosynthesis and senescence in soybean. Planta 2008, 227, 1389–1399. [Google Scholar] [CrossRef] [PubMed]
- De Lucas, M.; Prat, S. Pifs get brright: Phytochrome interacting factors as integrators of light and hormonal signals. New Phytol. 2014, 202, 1126–1141. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Feng, K.; Wang, M.; Wang, M.; Deng, P.; Song, W.; Nie, X. Genome-wide identification, phylogeny and expression analysis of AP2/ERF transcription factors family in Brachypodium distachyon. BMC Genom. 2016, 17, 636. [Google Scholar] [CrossRef] [PubMed]
- Iwase, A.; Mitsuda, N.; Koyama, T.; Hiratsu, K.; Kojima, M.; Arai, T.; Inoue, Y.; Seki, M.; Sakakibara, H.; Sugimoto, K. The AP2/ERF transcription factor wind1 controls cell dedifferentiation in Arabidopsis. Curr. Biol. 2011, 21, 508–514. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Hao, D.; Chen, L.; Lu, Q.; Zhang, Y.; Li, Y.; Duan, Y.; Li, W. Roles for a soybean RAV-like orthologue in shoot regeneration and photoperiodicity inferred from transgenic plants. J. Exp. Bot. 2012, 63, 3257–3270. [Google Scholar] [CrossRef] [PubMed]
- Marin, E.; Nussaume, L.; Quesada, A.; Gonneau, M.; Sotta, B.; Hugueney, P.; Frey, A.; Marion-Poll, A. Molecular identification of zeaxanthin epoxidase of Nicotiana plumbaginifolia, a gene involved in abscisic acid biosynthesis and corresponding to the ABA locus of Arabidopsis thaliana. EMBO J. 1996, 15, 2331–2342. [Google Scholar] [PubMed]
- Mase, K.; Ishihama, N.; Mori, H.; Takahashi, H.; Kaminaka, H.; Kodama, M.; Yoshioka, H. Ethylene-responsive AP2/ERF transcription factor MACD1 participates in phytotoxin-triggered programmed cell death. Mol. Plant Microbe Int. 2013, 26, 868–879. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Zhang, B.L.; Sun, S.; Xing, G.M.; Wang, F.; Li, M.Y.; Tian, Y.S.; Xiong, A.S. AP2/ERF transcription factors involved in response to tomato yellow leaf curly virus in tomato. Plant Genome 2016, 9, 82. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, S.; Himani; Sembi, J.K.; Upadhyay, S.K. Gene architecture and expression analyses provide insights into the role of glutathione peroxidases (GPXs) in bread wheat (Triticum aestivum L.). J. Plant Physiol. 2018, 223, 19–31. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Berkowitz, G.A. Ca2+ conduction by plant cyclic nucleotide gated channels and associated signaling components in pathogen defense signal transduction cascades. New Phytol. 2011, 190, 566–572. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J. Phospholipase D and phosphatidic acid in plant defence response: From protein–protein and lipid–protein interactions to hormone signalling. J. Exp. Bot. 2015, 66, 1721–1736. [Google Scholar] [CrossRef] [PubMed]
- Fillingame, R.H. Membrane sectors of F- and V-type H+-transporting ATPases. Curr. Opin. Struct. Biol. 1996, 6, 491–498. [Google Scholar] [CrossRef]
- Oshima, K.; Miyata, S.; Sawayanagi, T.; Kakizawa, S.; Nishigawa, H.; Jung, H.Y.; Furuki, K.; Yanazaki, M.; Suzuki, S.; Wei, W. Minimal set of metabolic pathways suggested from the genome of onion yellows phytoplasma. J. Gen. Plant Pathol. 2002, 68, 225–236. [Google Scholar] [CrossRef]
- Niu, S.; Fan, G.; Deng, M.; Zhao, Z.; Xu, E.; Cao, L. Discovery of microRNAs and transcript targets related to witches’ broom disease in Paulownia fortunei by high-throughput sequencing and degradome approach. Mol. Genet. Genom. 2016, 291, 181–191. [Google Scholar] [CrossRef] [PubMed]
- Fan, G.; Niu, S.; Zhao, Z.; Deng, M.; Xu, E.; Wang, Y.; Lu, Y. Identification of micrornas and their targets in Paulownia fortunei plants free from phytoplasma pathogen after methyl methane sulfonate treatment. Biochimie 2016, 127, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Fan, G.; Niu, S.; Xu, T.; Deng, M.; Zhao, Z.; Wang, Y.; Cao, L.; Wang, Z. Plant-pathogen interaction-related microRNAs and their targets provide indicators of phytoplasma infection in Paulownia tomentosa × Paulownia fortunei. PLoS ONE 2015, 10, e0140590. [Google Scholar] [CrossRef] [PubMed]
- Zhai, X.Q.; Cao, X.B.; Fan, G.Q. Growth of Paulownia witches’ broom seedlings treated with methylmethane sulphonate and SSR analysis. Sci. Silv. Sin. 2010, 46, 176–181. [Google Scholar]
- Fan, G.; Li, Y.; Zheng, J.; Zhai, X. SDS-page of proteins related to Paulownia witches’ broom. Sci. Silv. Sin. 2003, 29, 119–122. [Google Scholar]
- Cao, X.; Fan, G.; Dong, Y.; Zhao, Z.; Deng, M.; Wang, Z.; Liu, W. Proteome profiling of Paulownia seedlings infected with phytoplasma. Front. Plant Sci. 2017, 8, 342. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.; Wang, Z.; Li, X.; Zhao, Z.; Deng, M.; Dong, Y.; Cao, X.; Fan, G. Comparative proteomic analysis of paulownia fortunei response to phytoplasma infection with dimethyl sulfate treatment. Int. J. Genom. 2017, 2017, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Gai, Y.P.; Han, X.J.; Li, Y.Q.; Yuan, C.Z.; Mo, Y.Y.; Guo, F.Y.; Liu, Q.X.; Ji, X.L. Metabolomic analysis reveals the potential metabolites and pathogenesis involved in mulberry yellow dwarf disease. Plant Cell Environ. 2014, 37, 1474–1490. [Google Scholar] [CrossRef] [PubMed]
- Bertamini, M.; Grando, M.S.; Nedunchezhian, N. Effects of phytoplasma infection on pigments, chlorophyll-protein complex and photosynthetic activities in field grown apple leaves. Biol. Plant. 2003, 47, 237–242. [Google Scholar] [CrossRef]
- Stephenson, P.G.; Fankhauser, C.; Terry, M.J. PIF3 is a repressor of chloroplast development. Proc. Natl. Acad. Sci. USA 2009, 106, 7654–7659. [Google Scholar] [CrossRef] [PubMed]
- Toledoortiz, G.; Huq, E.; Rodríguezconcepción, M. Direct regulation of phytoene synthase gene expression and carotenoid biosynthesis by phytochrome-interacting factors. Proc. Natl. Acad. Sci. USA 2010, 107, 11626–11631. [Google Scholar] [CrossRef] [PubMed]
- Moulin, M.; Mccormac, A.C.; Terry, M.J.; Smith, A.G. Tetrapyrrole profiling in Arabidopsis seedlings reveals that retrograde plastid nuclear signaling is not due to Mg-protoporphyrin IX accumulation. Proc. Natl. Acad. Sci. USA 2008, 105, 15178–15183. [Google Scholar] [CrossRef] [PubMed]
- Papenbrock, J.; Pfündel, E.; Mock, H.P.; Grimm, B. Decreased and increased expression of the subunit CHl I diminishes Mg Chelatase activity and reduces chlorophyll synthesis in transgenic tobacco plants. Plant J. 2000, 22, 155–164. [Google Scholar] [CrossRef] [PubMed]
- Mittal, A.; Jiang, Y.; Ritchie, G.L.; Burke, J.J.; Rock, C.D. AtRAV1 and at RAV2 overexpression in cotton increases fiber length differentially under drought stress and delays flowering. Plant Sci. 2015, 241, 78–95. [Google Scholar] [CrossRef] [PubMed]
- Giri, M.K.; Swain, S.; Gautam, J.K.; Singh, S.; Singh, N.; Bhattacharjee, L.; Nandi, A.K. The Arabidopsis thaliana at4g13040 gene, a unique member of the AP2/EREBP family, is a positive regulator for salicylic acid accumulation and basal defense against bacterial pathogens. J. Plant Physiol. 2014, 171, 860–867. [Google Scholar] [CrossRef] [PubMed]
- Dautermann, O.; Lohr, M. A functional zeaxanthin epoxidase from red algae shedding light on the evolution of light-harvesting carotenoids and the xanthophyll cycle in photosynthetic eukaryotes. Plant J. 2017, 92, 879–891. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.Y.; Huang, Y.H.; Lin, C.P.; Lin, Y.Y.; Hsu, H.C.; Wang, C.N.; Liu, L.Y.; Shen, B.N.; Lin, S.S. MicroR396-targeted short vegetative phase is required to repress flowering and is related to the development of abnormal flower symptoms by the phyllody symptoms1 effector. Plant Physiol. 2015, 168, 1702–1716. [Google Scholar] [CrossRef] [PubMed]
- Doğramaci, M.; Anderson, J.V.; Chao, W.S.; Horvath, D.P.; Hernandez, A.G.; Mikel, M.A.; Foley, M.E. Foliar glyphosate treatment alters transcript and hormone profiles in crown buds of leafy spurge and induces dwarfed and bushy phenotypes throughout its perennial lifecycle. Plant Genome 2017, 10, 879–891. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Chu, C. MicroRNAs in crop improvement: Fine-tuners for complex traits. Nat. Plants 2017, 30, 17077. [Google Scholar] [CrossRef] [PubMed]
- Kaur, H.; Yadav, C.B.; Alatar, A.A.; Faisal, M.; Jyothsna, P.; Malathi, V.G.; Praveen, S. Gene expression changes in tomato during symptom development in response to leaf curl virus infection. J. Plant Biochem. Biotechnol. 2014, 24, 347–354. [Google Scholar] [CrossRef]
- Hasunuma, T.; Fukusaki, E.; Kobayashi, A. Expression of fungal pectin methylesterase in transgenic tobacco leads to alteration in cell wall metabolism and a dwarf phenotype. J. Biotechnol. 2004, 111, 241–251. [Google Scholar] [CrossRef] [PubMed]

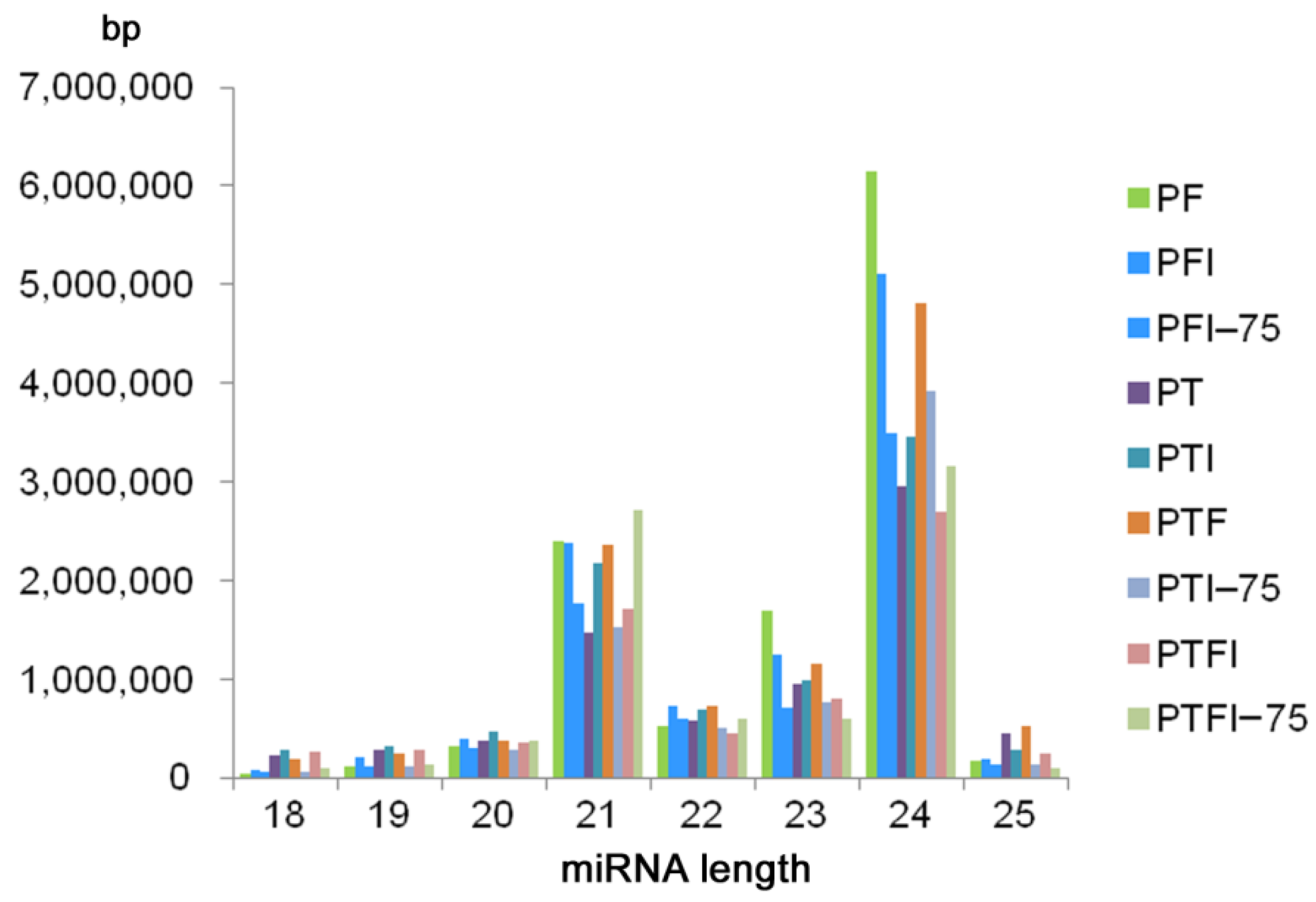
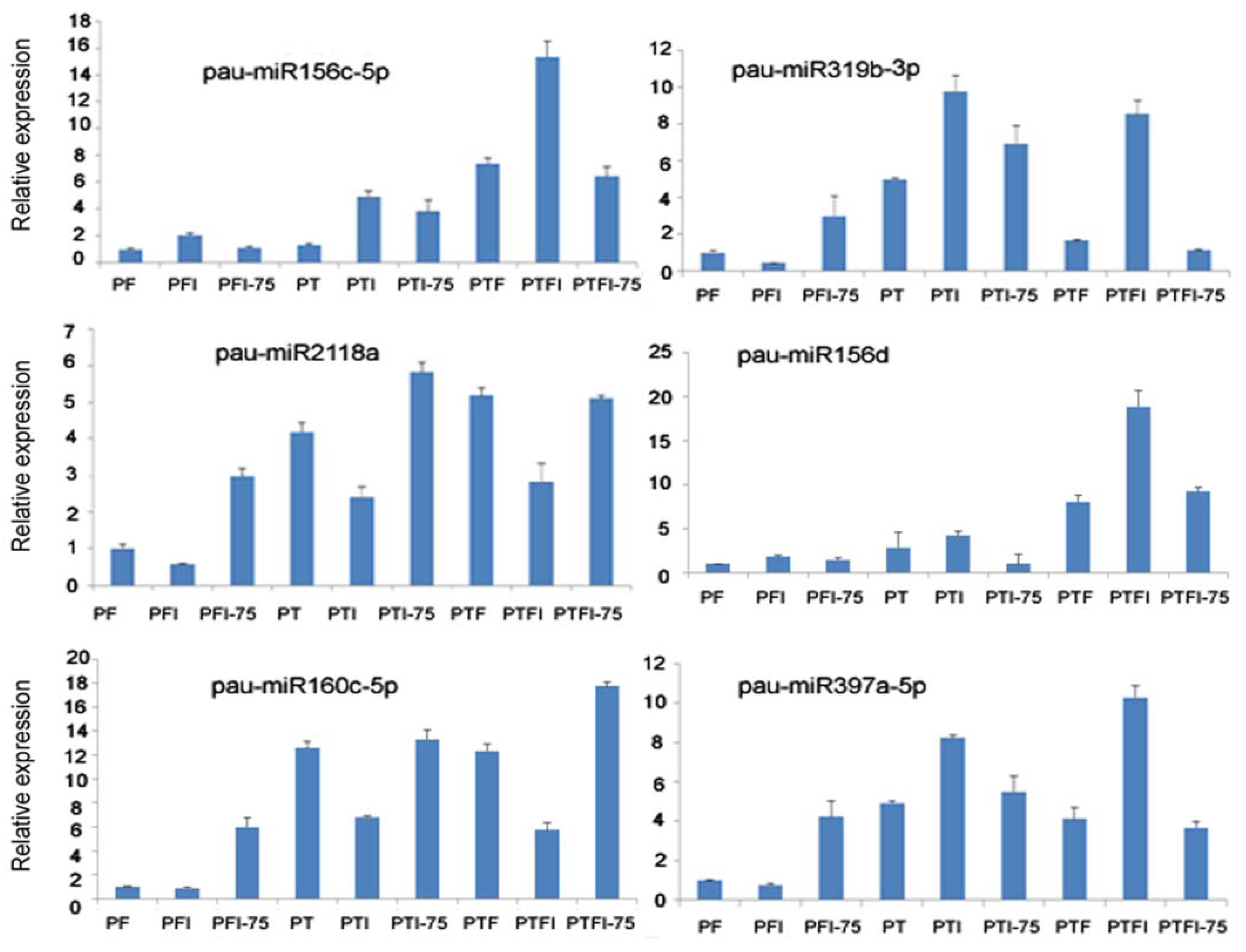
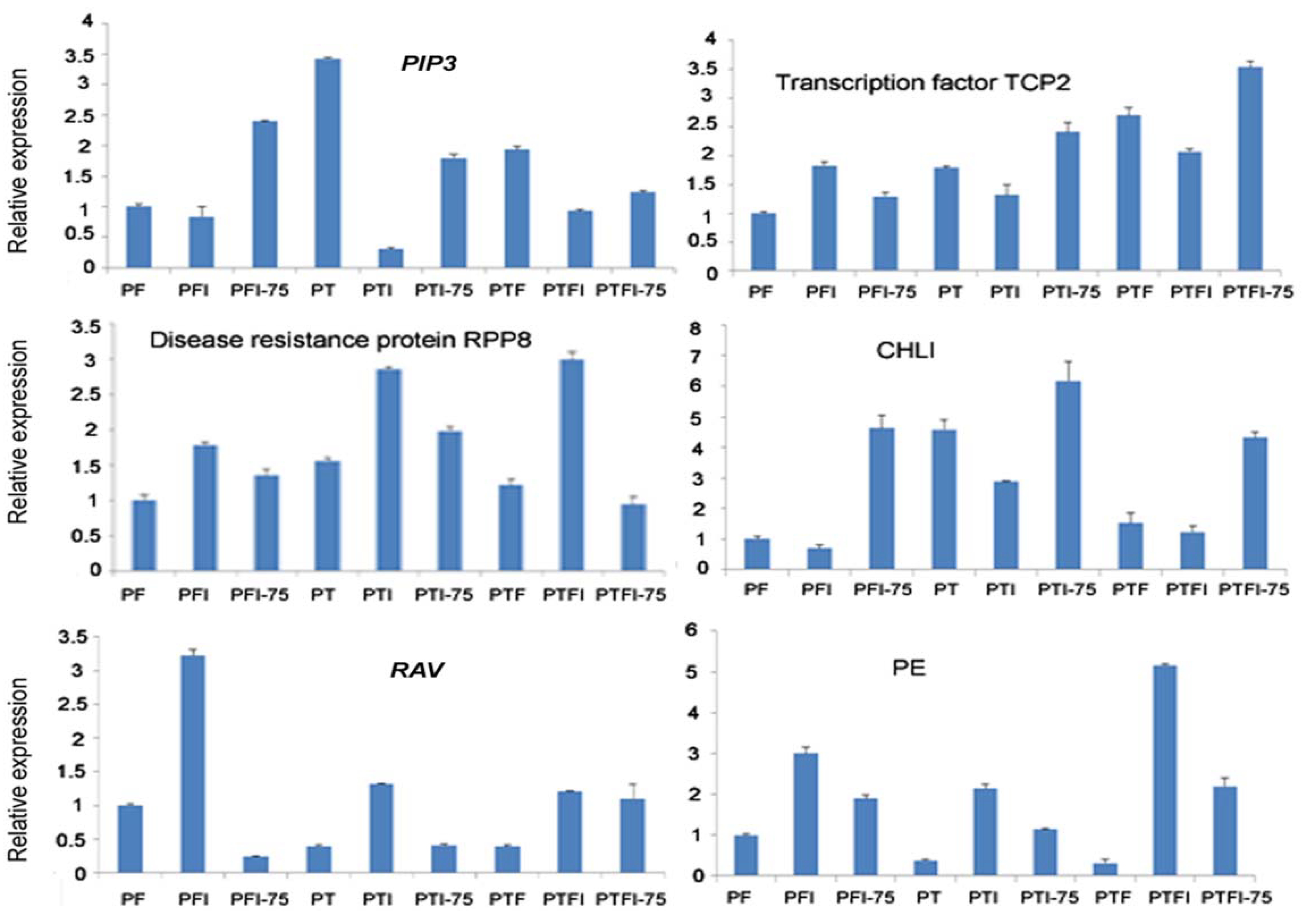
| Name | miR_seq | Length | Type |
|---|---|---|---|
| pau-mir154 | TCAAACGGTGAGGATCATGTCACG | 24 | 5′ |
| pau-miR156b-3p | GCTCACTCTCTGTCTGTCACC | 21 | 3′ |
| pau-miR156c-3p | GCTCACTCTCTGTCTGTCACC | 21 | 3′ |
| pau-miR156d | TTGACAGAAGAGAGAGAGCAC | 21 | 5′ |
| pau-miR156f-3p | GCTCTCTATGCTTCTGTCATC | 21 | 3′ |
| pau-miR156g-3p | GCTCACTTCTCTTTCTGTCAGC | 22 | 3′ |
| pau-miR159a-5p | AGCTGCTGATTTATGGATCCC | 21 | 5′ |
| pau-miR2118a | TTTCCGATGCCTCCCATACCGA | 22 | 3′ |
| pau-miR2118b | TTTCCGATGCCTCCCATACCGA | 22 | 3′ |
| pau-miR2916b | CGGATGTTGCTTTTAGGACTC | 21 | 3′ |
| pau-miR319a-5p | GCTGCCGACTCATTCATTCAA | 21 | 5′ |
| pau-miR319b-3p | TTGGACTGAAGGGAGCTCCT | 20 | 3′ |
| pau-miR319c-5p | GCTGCCGACTCATTCATTCAA | 21 | 5′ |
| pau-miR319d-3p | TTGAATCTTAAGCTTCCTGTT | 21 | 3′ |
| pau-miR391 | TACGCAGGAGAGATGATGCCC | 21 | 5′ |
| pau-miR397a-5p | TTGAGTGCAGCGTTGATGATA | 21 | 5′ |
| pau-miR397b-5p | TTGAGTGCAGCGTTGATGATA | 21 | 5′ |
| pau-miR398c | CATGTTCTCAGGTCGCCCCTG | 21 | 3′ |
| pau-miR408a-5p | ACGGGGACGAGGCAGAGCATG | 21 | 5′ |
| pau-mir42-3p | TCTTGATACCATCAATGGTGG | 21 | 3′ |
| pau-mir42-5p | ACCGTTGATGGTATCAAAATC | 21 | 5′ |
| pau-miR4414a-5p | AGCTGCTGACTCGTTGGTTCA | 21 | 5′ |
| pau-mir45 | TAACTTGAGATGGTCTAAGGT | 21 | 5′ |
| pau-mir48-5p | GGTGCAATGGGAGACGCCGAGA | 22 | 5′ |
| pau-miR396c-5p | TTCCACAGCTTTCTTGAACTG | 21 | 5′ |
| pau-miR396d-5p | TTCCACAGCTTTCTTGAACTG | 21 | 5′ |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, X.; Zhai, X.; Zhang, Y.; Cheng, Z.; Li, X.; Fan, G. Comparative Analysis of MicroRNA Expression in Three Paulownia Species with Phytoplasma Infection. Forests 2018, 9, 302. https://doi.org/10.3390/f9060302
Cao X, Zhai X, Zhang Y, Cheng Z, Li X, Fan G. Comparative Analysis of MicroRNA Expression in Three Paulownia Species with Phytoplasma Infection. Forests. 2018; 9(6):302. https://doi.org/10.3390/f9060302
Chicago/Turabian StyleCao, Xibing, Xiaoqiao Zhai, Yanfang Zhang, Zhiyuan Cheng, Xiyao Li, and Guoqiang Fan. 2018. "Comparative Analysis of MicroRNA Expression in Three Paulownia Species with Phytoplasma Infection" Forests 9, no. 6: 302. https://doi.org/10.3390/f9060302
APA StyleCao, X., Zhai, X., Zhang, Y., Cheng, Z., Li, X., & Fan, G. (2018). Comparative Analysis of MicroRNA Expression in Three Paulownia Species with Phytoplasma Infection. Forests, 9(6), 302. https://doi.org/10.3390/f9060302





