1. Introduction
Biomass is an important characteristic of forest ecological systems. The accurate quantification of tree biomass is critical and essential for studying carbon storage, climate change, forest health, forest productivity, fuel (and bioenergy), nutrient cycling, etc. [
1,
2,
3,
4,
5]. Although direct measurement of the actual weight of each tree component (i.e., stem, branch, foliage, and root) is undoubtedly the most accurate method, it is destructive, time-consuming, and costly. Therefore, developing biomass models is considered a better approach for estimating forest biomass [
6,
7,
8].
To date, hundreds of biomass equations have been developed for more than 100 species worldwide [
9,
10,
11,
12,
13,
14]. However, most studies have focused on aboveground biomass, while relatively few studies have attempted to study belowground (or root) biomass, because excavating tree roots is extremely difficult and expensive [
15,
16,
17].
In forest ecosystems, the biomass of small trees (diameter at breast height <5 cm) constitutes an essential component of the total forest biomass. Unfortunately, in most empirical studies, the forest biomass calculations have ignored small trees, consequently underestimating the totality of the overall values [
18,
19]. Researchers have recently reported that small-diameter trees can play a substantial role in the estimation of total biomass and bioenergy, because these small-diameter trees compose a significant proportion of the tree population and can grow more rapidly than larger-diameter trees [
2,
20,
21]. However, there is no study that investigated the development of biomass equations including small-diameter trees in the temperate forests of the eastern Daxing’an Mountains in northeast China.
In general, tree diameter at breast height is the commonly used and reliable sole predictor of total, subtotal, and component biomass in most biomass equations [
5,
15,
22]. Furthermore, depending on research goals, other tree variables (e.g., tree height, crown length, and crown width) have been investigated as potential predictors for tree biomass modeling [
23,
24,
25,
26]. Despite these tree variables being much more difficult to obtain in practice, studies have shown that different tree height, crown length, and crown width for the same tree diameter at breast height clearly influence tree-level biomass equations. Including tree height, crown length, and/or crown width into biomass equations as the additional predictors can significantly improve model fitting and performance, explain more sources of variation in the data, and avoid potential limitations [
5,
8,
24,
26,
27,
28,
29].
The biomass equations for estimating the total, subtotal, and component biomass of trees can be classified as non-additive or additive in nature. Non-additive biomass equations fit the total, subtotal, and component biomass data separately. Consequently, the sum of model predictions from the component biomass models may not be equal to the model prediction from the total biomass models. On the other hand, additive biomass equations fit the total, subtotal, and component biomass data simultaneously to account for the inherent correlations among biomass components measured on the same sample trees [
6,
22,
24]. Thus, the sum of biomass predictions from the component biomass equations is equal to the biomass predictions from the total or subtotal biomass equations [
30,
31]. To ensure the additivity for a system of biomass equations, various model specification and parameter estimation methods have been proposed for both linear and nonlinear biomass equations [
6,
30,
31,
32]. In particular, seemingly unrelated regression (SUR) and nonlinear seemingly unrelated regression (NSUR) are more general and flexible, and they have become more popular as the parameter estimation methods for linear and non-linear biomass equations. Several recent applications of the SUR method have been reported to ensure the additivity in the nonlinear simultaneous equation systems of tree biomass [
6,
22,
24,
33,
34].
Tree biomass data often show heteroscedasticity in model residuals. To overcome this problem, a weighted regression or logarithmic transformation should be defined and used for each biomass model. With respect to logarithmic regression, a fitted logarithmic model provides the prediction of log(Y). However, the anti-log transformation from log(Y) to Y introduces bias, and the correction on the bias is necessary. To obtain the desired prediction of Y, the predicted values of Y should be adjusted in some manner, such as being multiplied by a correction factor. Several correction factors have been proposed and applied over the last decades [
35,
36,
37]. When the model error term is sufficiently small, the correction factor may not be necessary, but the correction factor cannot be ignored when the model error term is relatively large [
38,
39]. In general, logarithmic regression is simple and may be better for developing biomass equations when the sample sizes are small. Unfortunately, after a correction factor is applied to the additive system of logarithmic equations, the property of additivity may be damaged among the total, subtotal, and component biomass equations [
22]. When the sample sizes are relatively large, weighted regression can be used to directly overcome the heteroscedasticity in the residuals of total and component biomass models [
7,
24].
Since the cold temperate forests in the eastern Daxing’an Mountains in northeast China were excessively harvested from the 1960s to 2000s, many young and middle-aged forests now exist in the region. The current cold temperate forests play an important role not only in the national carbon storage and budget, but also in the recovery of biodiversity following harvest or wildfires. Dahurian larch (
Larix gmelinii Rupr.) and white birch (
Betula platyphylla Suk.) are the two dominant species in the natural forests of the eastern Daxing’an Mountains. However, the Chinese National Forest Inventory lacked accurate systems of tree biomass models for Dahurian larch and white birch, and only a few studies have investigated the aboveground and belowground biomass of these species in China. Wang [
15] and Mu et al. [
40] developed biomass equations for both species in northeast China, but their biomass data were collected from a limited forest region and originated from a relatively small sample size (e.g., two dominant, three codominant, three intermediate, and two suppressed trees) for both species. Moreover, the established biomass equations were non-additive. Dong et al. [
8,
22] constructed two additive systems of biomass models for several conifer and hardwood species (including Dahurian larch and white birch) in the Xiaoxing’an Mountains, but did not include trees <5.4 cm. Meng et al. [
28] used data from 48 trees to construct additive biomass models for white birch in the western Daxing’an Mountains (eastern Inner Mongolia), but the developed models were not applicable to belowground (root) biomass. In addition, few studies have focused on biomass equations for Dahurian larch and white birch in the eastern Daxing’an Mountains. Overall, accurate systems of biomass equations for cold temperate forests across the eastern Daxing’an Mountains in northeast China have not been developed.
The objectives of this study were as follows: (1) compare biomass models based on different predictors of total, aboveground, root, stem, crown, branch, and foliage biomass for Dahurian larch and white birch trees in the eastern Daxing’an Mountains; (2) use SUR to construct four additive systems (i.e., one-, two-, three- and best-variable systems) of biomass equations with three constraints and overcome the heteroscedasticity problem; (3) use the jackknife technique to validate the performance of the biomass models; (4) evaluate the different methods of quantifying tree-level biomass; and (5) compare the newly developed additive biomass equation system against the biomass equations published in the literature.
2. Materials and Methods
2.1. Study Sites
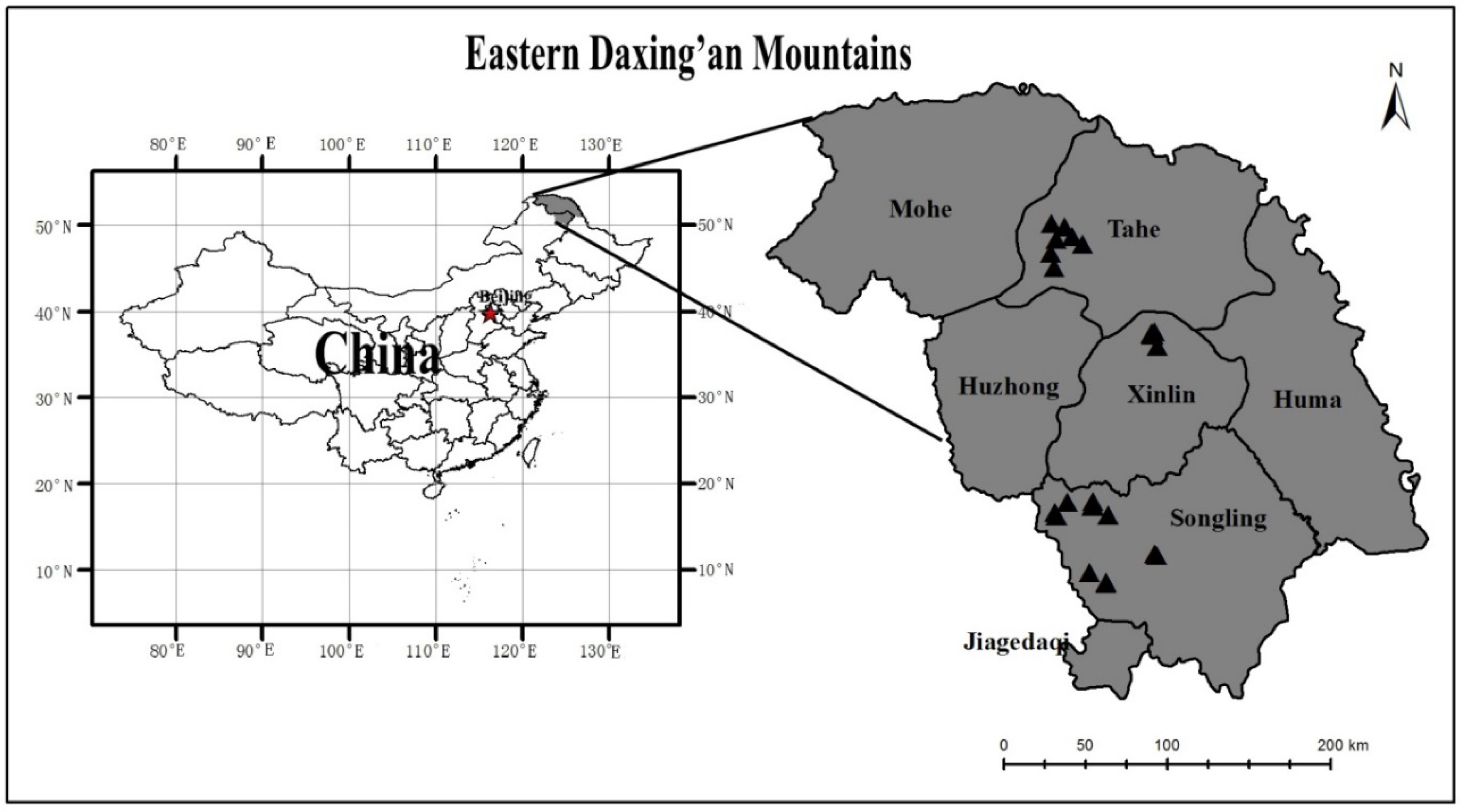
This study was conducted in the eastern Daxing’an Mountains in northeast China (from 121°12′ E to 127°00′ E and from 50°10′ N to 53°33′ N). The total area of the region is 83,000 km
2, and the altitude ranges from 300 to 1520 m above the sea level (
Figure 1). The soils are mostly Umbri-Gelic Cambosols according to the Chinese taxonomic system [
41]. This area is associated with a distinct cold temperate continental monsoon climate: the mean annual air temperature ranges from −1 to −2.8 °C; the mean air temperatures in January and July are −28 °C and +20 °C, respectively; and the mean annual rainfall ranges from 500 to 750 mm. Dahurian larch (
Larix gmelinii) and white birch (
Betula platyphylla) are the dominant tree species, accompanied by aspen (
Populus davidiana Dode.) and Mongolian pine (
Pinus sylvestris var.
mongolica). The understory and ground vegetation are dominated with blueberry (
Vaccinium angustifolium Aiton),
Rhododendron dahuricum L., and
Ledum palustre L., among others.
2.2. Plot Measurements and Biomass Estimation
The data used in this study were selected from a large data set of tree biomass. The tree species included Dahurian larch and white birch in the young and middle-aged natural forests of the eastern Daxing’an Mountains in northeast China. The sample plots were established in August of 2012, 2013, 2014, 2015, and 2016. A total of 31 plots were selected, including 18 plots of Dahurian larch forest, 4 plots of white birch forest, and 9 plots of Dahurian larch and white birch mixed forest. Each plot was 20 × 50 m in size. The characteristics of the forest types are listed in
Table 1. For each of the 31 plots, at least one sample tree for each of the dominant, intermediate, and suppressed trees were targeted. In addition, some small sample trees (diameter at breast height <5 cm) were selected outside of the 31 plots. In total, 138 Dahurian larch trees and 108 white birch trees were destructively sampled (
Table 2).
The targeted sample trees were cut at the ground. Tree measurements, including tree diameter at breast height (D), tree height (H), crown width (CW), and crown length (CL), were measured and recorded. Among them, H and CL were measured after cutting, while D and CW were measured before cutting. The live crown, which constitutes the first dead branch to the base of the terminal bud, was divided equally into three layers: top, middle, and bottom. All live branches within each crown layer were cut and weighed. Then, 1–2 branches were selected from each crown layer, and the branches and foliage were separated and weighed. The samples (approximately 50–100 g) of the branches and foliage were collected, weighed, and taken to the laboratory to determine their moisture content. The stems of each sample were cut into 1-m sections, after which, each section was weighed and recorded. At the end of each stem section, a 2–3-cm thick disc was cut, weighed, and taken to the laboratory to determine its moisture content and analyze its growth rings to determine tree age with WinDENDRO software (Regent Instruments, Inc., Québec, QC, Canada). The roots of large trees were excavated using a combination of chains (i.e., lifting equipment) and manual digging. However, the roots of small trees were removed manually. Because most fine roots were out of the working zones (approximately 3 m in radius), and excavating the fine roots is difficult, costly, and time consuming, the fine roots (diameter < 5 mm) were excluded in this study. The roots of the sampled trees were divided into large roots (diameter ≥ 5 cm), medium roots (diameter 2–5 cm), and small roots (5 mm < diameter < 2 cm). The root diameters were measured with a digital caliper. The root samples in each class were sampled (approximately 100–200 g), weighed, and then taken to the laboratory to determine their moisture content. All stem, branch, foliage, and root samples were oven-dried at 80 °C and then weighed. The dry biomass of each component was calculated by multiplying the fresh weight of each component by the dry weight-to-fresh weight ratio of that component. For each sampled tree, the sum of large, medium, and small root dry biomass yielded the root dry biomass. The sum of the branch dry biomass and foliage dry biomass yielded the crown dry biomass. The sum of the crown dry biomass and stem dry biomass equaled the aboveground biomass, and the sum of the aboveground dry biomass and the root dry biomass equaled the total tree biomass.
A summary of the descriptive statistics for D, H, CL, and CW as well as the total, subtotal, and component biomass of trees is shown in
Table 2. The stem, branch, foliage, and root biomass of all sample trees as well as their relationships with D, H, CL, and CW are shown in
Figure 2. We performed statistical tests (e.g., dummy variable regression) to confirm that there was no significant difference between young and middle-age trees for the total, stem, root, and foliage biomass of two species, with the exception of branch biomass. Therefore, the sampled trees from different diameter classes (or different ages) were combined for data analysis and model development in this study.
2.3. Systems of Additive Biomass Equations
2.3.1. Model Selection
Visual inspection of the stem, branch, foliage, and root biomass data indicated that the component biomass of trees can be modeled as a multivariable allometric model of tree variables (
Figure 2). The general tree biomass equation in this study is nonlinear with an additive error term as follows:
where W
i represents the root, stem, branch, foliage, aboveground, crown, and total tree biomass in kilograms (
i = r, s, b, f, a, c, and t for root, stem, branch, foliage, aboveground, crown, and total, respectively); X
j represents various tree variables (
j = 1, …, k), such as D, H, CL, and CW, respectively; β
ij represents the model parameters to be estimated; and ε
i is a model additive error term. From the general Equation (1), we proposed the following biomass models based on the different combinations of predictors of tree component biomass:
The total, subtotal (i.e., aboveground and crown), and component biomass of tree equations may have different combinations of predictor variables. We fit each of the above nonlinear equations to the total biomass, subtotal biomass, and component biomass (i.e., stems, branches, foliage, and roots) of trees, respectively. A “best model” was selected for each of the total, subtotal, and component biomass based on the model fitting statistics such as adjusted coefficient of determination (Ra2), root mean squared error (RMSE), and Akaike information criterion (AIC). Finally, we decided to use the best-, one-, two-, and three-predictor biomass models as the basic model formats to construct four candidate additive systems of biomass equations that each has three constraints. In four candidate additive systems, the biomass models of tree components were constrained to equal the (1) total tree biomass, (2) aboveground biomass, and (3) crown biomass.
2.3.2. Additive Biomass Equations
Following the model structure specified by Parresol [
31], the additive systems of seven equations with cross-equation constraints on the structural parameters and cross-equation error correlations for root, stem, branch, foliage, subtotal (i.e., crown and aboveground), and total biomass are specified as:
where W
r to W
t represent the vectors of the root, stem, branch, foliage, crown, aboveground, and total biomass in kilograms, respectively.
To overcome the heteroscedasticity in the residuals of the biomass models, a weight function should be defined and used for each biomass model, such as the total, subtotal, and tree component biomass equations. The error variance of the
ith observation is functionally related to one or more of the predictor variables and can be modeled as a power function of the predictor variables, as in
[
30,
42], where the power coefficient γ
ik can be obtained by stepwise regression using the logarithmic form of the error variance model
, in which
is the model residual of the unweighted model. Hence, we chose
as the weight function, and γ
ik was determined for each biomass model. In the computations, the weight function for heteroscedasticity
was multiplied and programmed using the PROC MODEL procedure in SAS by specifying resid.
[
43].
2.3.3. Model Assessment and Validation
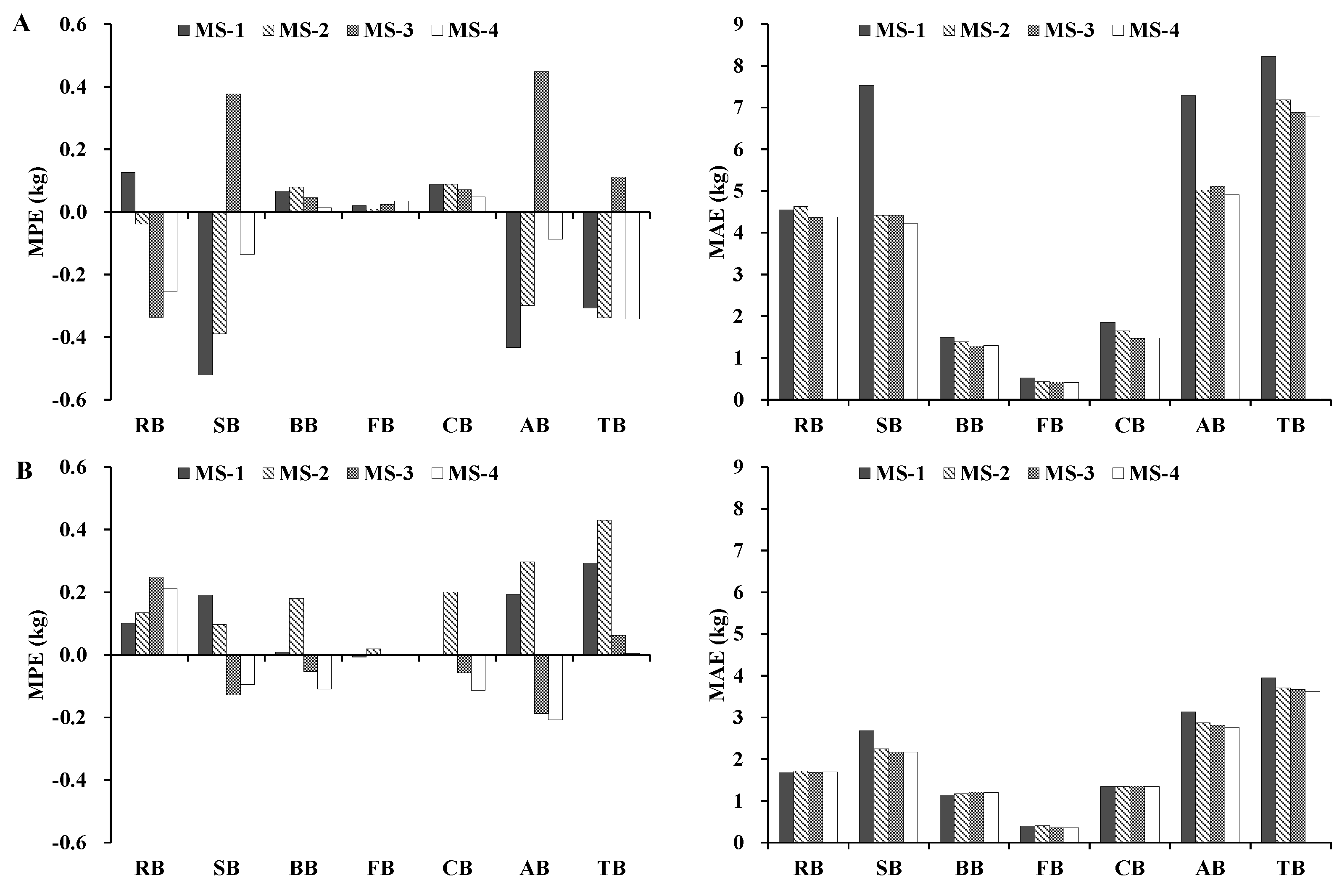
The four additive systems of biomass models were fitted to the entire data set, and the models were validated using the jackknife technique, in which a biomass model was constructed using all but one observation (sample size N − 1). Afterward, the fitted model was used to predict the value of the dependent variable for the excluded observation. Four statistics that were based on jackknifing and that were obtained for each system equation were used to evaluate the model fitting (Ra2 and RMSE) and performance (mean prediction error (MPE) and mean absolute error (MAE)) for each biomass prediction system, where MPE represents the average prediction error, and MAE represents the magnitude of prediction error.
The mathematical expression of the four statistics is displayed as follows:
where
is the
ith observed biomass value,
is the
ith predicted biomass value from the model that was fitted using all data (sample size N),
is the mean of the biomass value,
is the predicted value of the
ith observed value by the fitted model that was fitted by (N − 1) observations and that excluded the use of the
ith observation, and p is the number of model parameters.
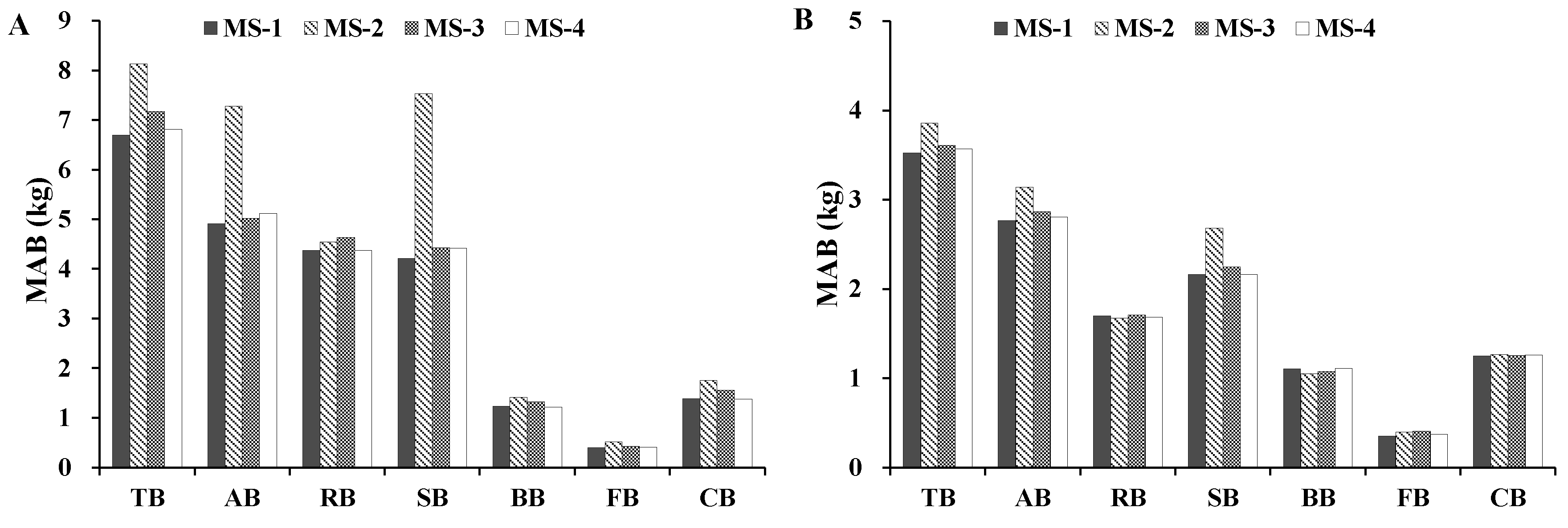
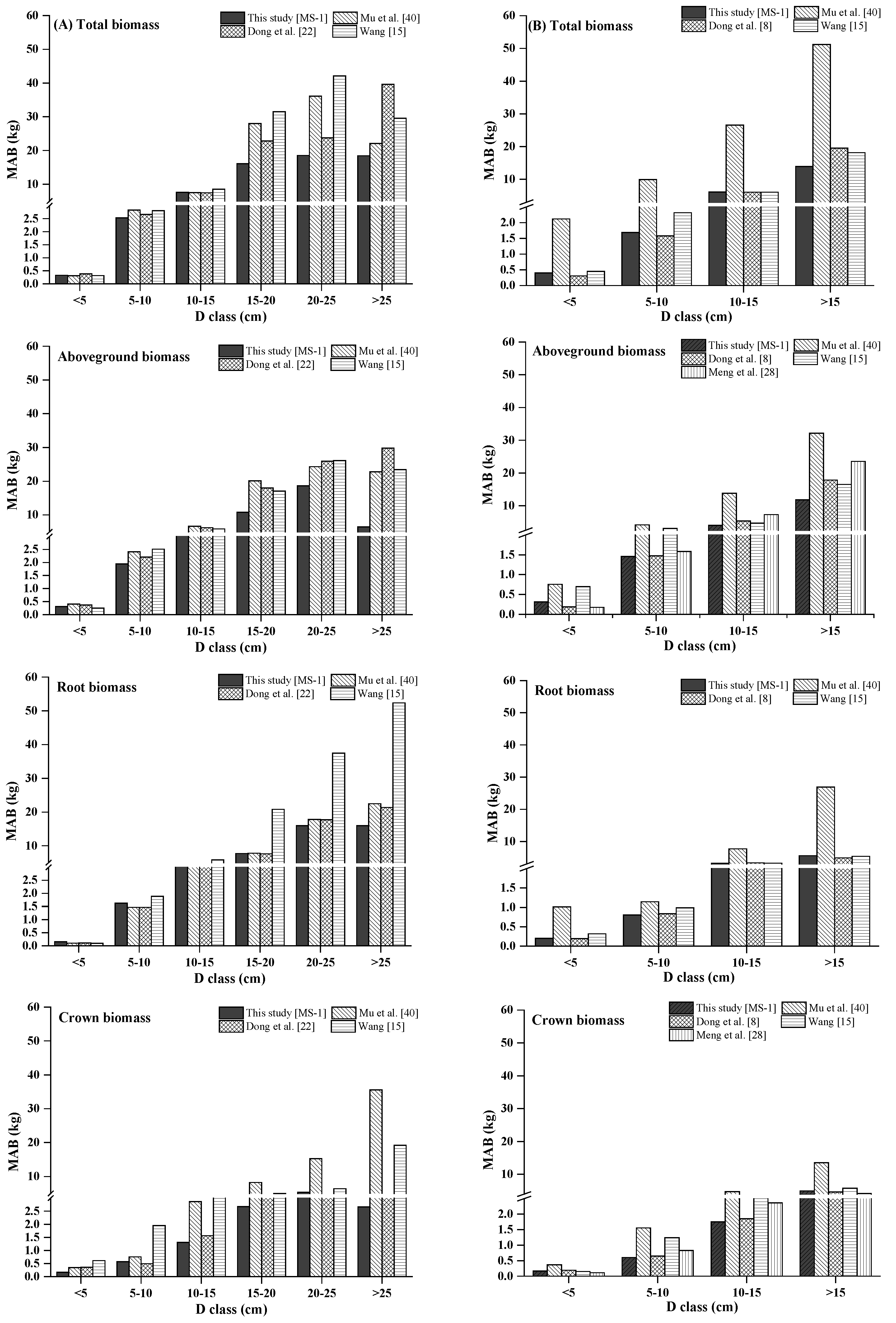
In addition, we calculated the mean absolute bias (MAB) to compare the newly developed additive biomass equation systems against the biomass models published in the literature.
2.3.4. Model Estimation and Data Analysis
For the above four additive systems of biomass equations, PROC MODEL procedure in SAS [
43] was used to simultaneously estimate the coefficients of the tree component biomass models. To analyze the partitioning of tree biomass into basic components, the fitted models were used to partition across 5-cm diameter classes. The analysis of variance (ANOVA) of the PROC GLM procedure in SAS software [
43] was used to test the differences between the four additive systems to estimate the total and component biomass at the individual tree level as well as at the plot level followed by the contrasts between the four additive systems.
4. Discussion
Many studies have introduced typical allometric equations based on power-law models to increase biomass estimation accuracy. With respect to allometric equations, D is an essential predictor variable in forest growth and yield models as well as biomass models. Tree biomass models using D as the sole predictor are simple in structure and require only basic forest inventory data to apply in practice [
15,
44]. The results of the present study showed that D was the main predictor variable in the simple allometric model. Nevertheless, for a given D, large variation occurs among the component and total biomass values. Therefore, using D only in the biomass model was not sufficient for predicting the total, subtotal, or component biomass of trees; the use of an additional tree variable(s) was generally necessary to improve the predictive ability of most of the biomass component equations [
8,
22]. Adding H and crown attributes (CW and CL) as the additional predictors into biomass equations can significantly improve model fitting and predictive ability [
6,
7,
24,
26]. In the present study, our tree biomass data were collected across a relatively large geographical region in the eastern Daxing’an Mountains in northeast China. We tested six common formats of nonlinear biomass models (Equations (2)–(7)). Our results demonstrated that adding H and crown attributes to the biomass equations could improve most of the biomass equations for the two species, which were consistent with the literature [
6,
7,
24,
45]. For trees in an uneven-aged natural stand (such as in this study), age is not a meaningful tree variable (not a complete chronosequence) and is considered unimportant in modeling tree biomass or growth. Even though the ages of the sampled trees can be determined in the lab, tree ages are difficult to obtain in practice in uneven-aged forests. Therefore, it would be hard to apply the biomass models in which tree age is one of the predictors. This may be one of the reasons that almost no biomass models or other growth models included tree age as a predictor for uneven-aged stands. In addition, crown attributes (CW and CL) have been investigated as potential predictors for tree biomass models. However, not only are the measurements of crown attributes costly, but they add uncertainties to biomass estimation due to their measurement errors.
Biomass additivity is a desirable property of biomass equations for predicting total, subtotal, and components biomass. Moreover, many biomass equations reported in the literature are non-additive and were developed separately to estimate the total, subtotal, and component biomass of trees [
25,
46,
47]. The additivity of biomass equations has not always been addressed when predicting the total and component biomass of trees. In this study, we developed four sets of system equations based on the best combinations of the predictors, D alone, D and H, and D, H, and crown attributes (namely, MS-1, MS-2, MS-3, and MS-4, respectively). The four additive systems of biomass equations accurately predicted different component biomass values. Compared with MS-2 and MS-3 (which were the additive biomass systems that used D alone or D and H as the predictors) MS-1 and MS-4 (the latter of which included D, H, and crown attributes (CW and CL)) further strengthened the most component biomass predictions (
Figure 5). Despite crown attributes being both difficult to accurately measure and costly in terms of labor and time, the use of MS-1 or MS-4 in conjunction with individual growth models is very useful for accurate predictions of Dahurian larch and white birch growth in response to changes in stand conditions, such as thinning and temperature and precipitation fluctuations, and is appropriate for use in many ecological and forest management studies [
24]. Moreover, we evaluated different additive systems of biomass equations for quantifying tree- and plot-level biomass. The results indicated that, in general, the MS-1 and MS-4 performed better. However, if the variables of crown attributes are not available, MS-3 can be used to predict the total, subtotal, and component biomass of trees more accurately.
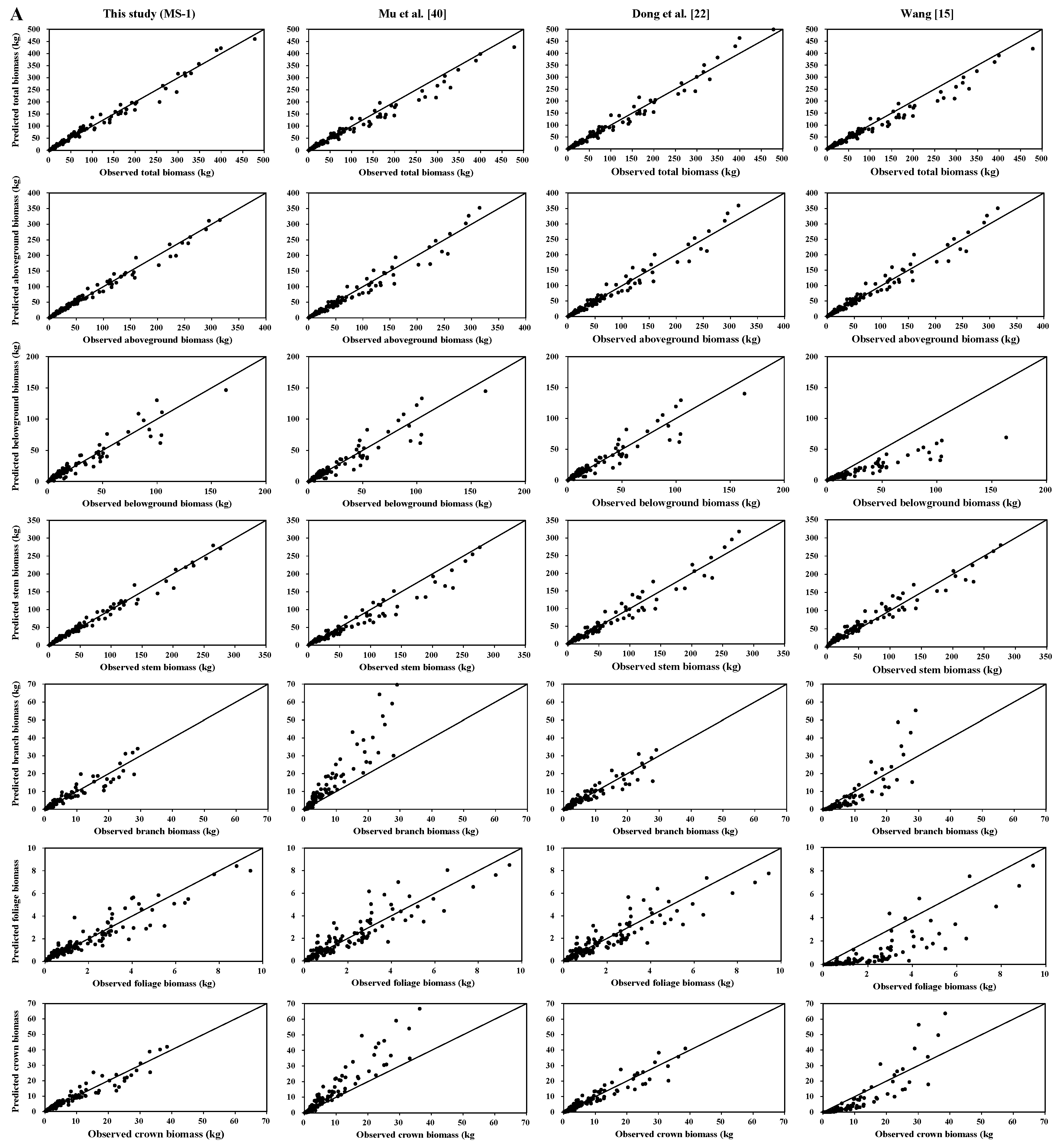
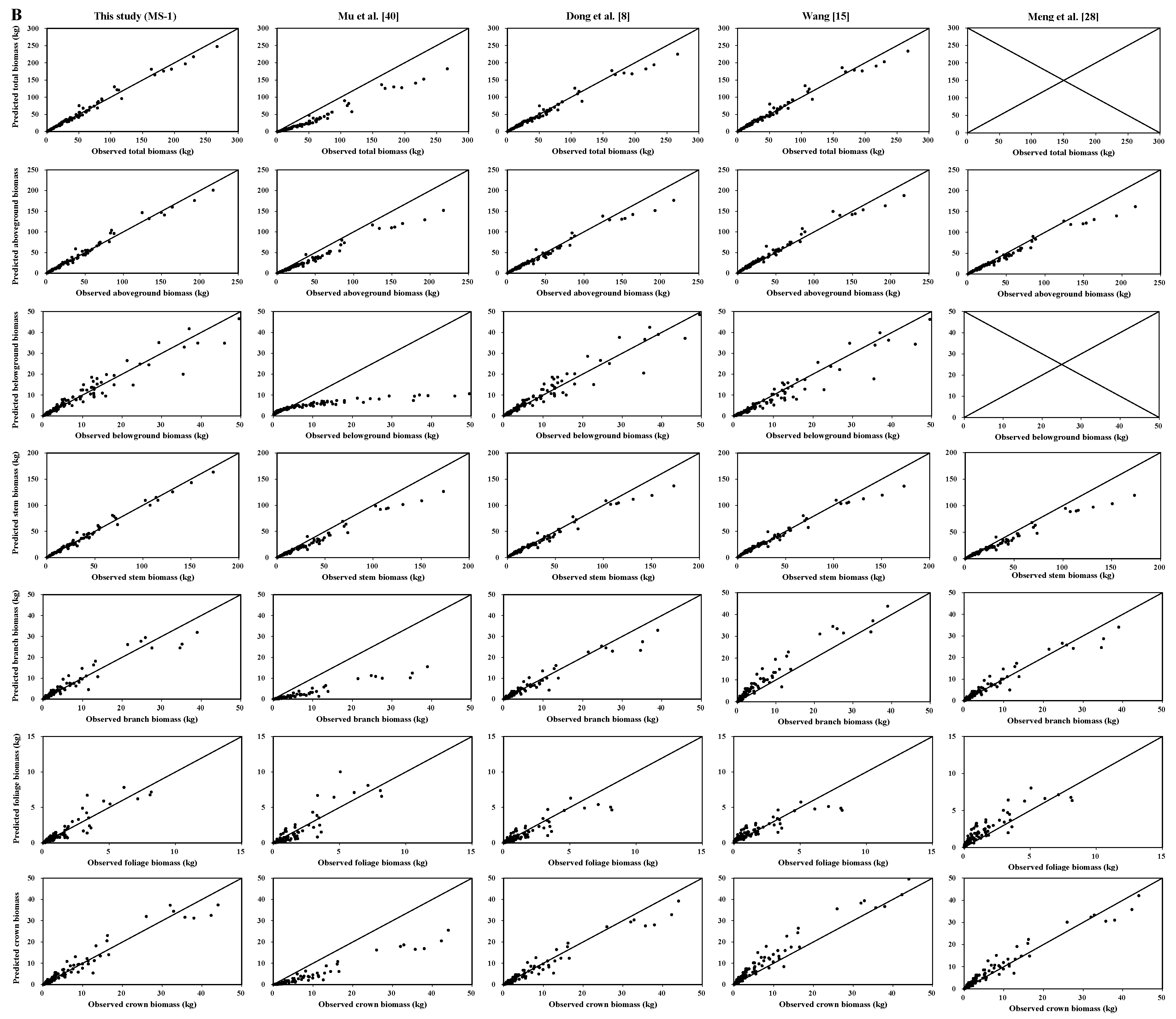
Total, aboveground, and stem biomass were better predicted than crown biomass components by our new biomass equation systems and the previously published biomass models. Our new equation systems, however, provide better prediction of each component biomass than those in the literature. Our new systems greatly improved the prediction of total, subtotal, and component biomass. There are several possible reasons: (1) the data in those four studies came from different sampling sites; (2) each species in those four studies came from different forests; and (3) the sample numbers and sample size ranges differed. These reasons could lead to differences in tree root morphological features, soil conditions, and growth processes [
6,
28,
48,
49]. Overall, the results of the graphical analysis and comparisons of MAB suggested that, for predicting biomass, our MS-1 performed better than the biomass models of Wang [
15], Mu et al. [
40], Dong et al. [
8,
22], and Meng et al. [
28].
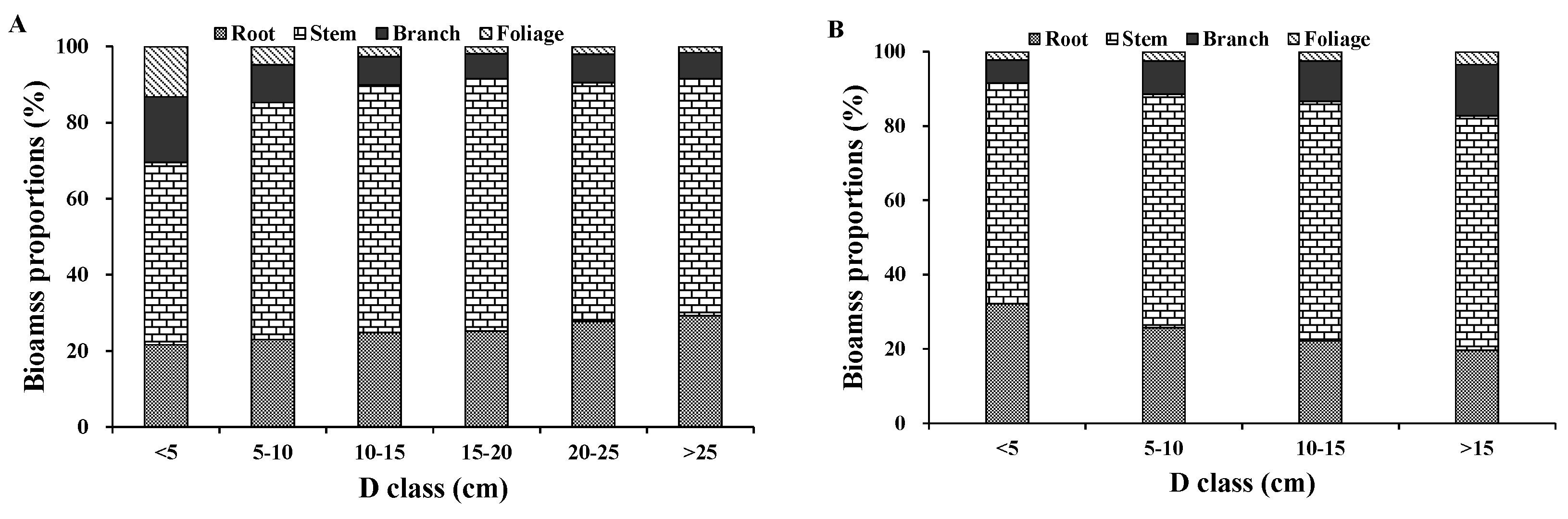
We observed a diameter-related pattern in the changes of biomass partitioning among individual aboveground tree components. For both Dahurian larch and white birch, the results demonstrated that the relative proportion of stem biomass was greater in medium- and large-diameter trees than in small-diameter trees. However, the patterns of the relative proportions of root and crown biomass were different between Dahurian larch and white birch. With respect to Dahurian larch, the crown biomass in relatively larger trees was smaller than that in relatively smaller trees, whereas for white birch, the crown biomass in relatively larger trees was greater than that in relatively smaller trees; these results occurred mostly because large white birch trees are usually forked and have thick branches, whereas Dahurian larch trees usually have a small canopy and relatively thin branches. The allocation of root biomass depends on tree root morphology (e.g., shallow root or deep root), growth processes, and soil conditions [
18,
50,
51]. However, total root excavation was impossible for both species in this study because of the propensity of the roots to grow clonally via lateral roots. This phenomenon may introduce errors in root biomass estimation, which can then influence root biomass partitioning. The increase or decrease in root, stem, branch, and foliage biomass across different diameter classes observed in this study supports previous findings. In addition, some researchers have reported clear differences in the partitioning of different biomass components; our results showed that the aboveground biomass was approximately 75% of the total biomass, and belowground biomass was approximately 25%. Consequently, our biomass partitioning results were consistent with those in the literature [
8,
52,
53].
Finally, because of excessive harvesting during the past 50 years, many young and middle-aged forests currently exist in the eastern Daxing’an Mountains. Thus, the sample trees from those young and middle-aged forests have relatively small D, H, CL, and CW values. Many researchers have reported that small diameters can significantly affect the estimation of total biomass. In this study, the smallest and the largest D values across both species ranged from 1.4 to 1.7 cm and from 20.5 to 28.4 cm, respectively. If the newly developed biomass equations in this study were used to estimate the biomass outside of our data range (e.g., D > 30 cm), the models could produce larger prediction errors. In addition, if our models were used in other regions, caution should be taken because different environmental and growth conditions may yield different allometric relationships between tree biomass and variables. Therefore, the biomass equations developed in this study are best suited to the eastern Daxing’an Mountains in northeast China.
5. Conclusions
In this study, four additive systems of biomass equations were developed for Dahurian larch and white birch in the eastern Daxing’an Mountains in northeast China to estimate the total, aboveground, root, stem, branch, foliage, and crown biomass. As expected, the accuracy of the biomass component equations differed among the four additive systems across both species: the model Ra2 was >0.86 for MS-1, >0.81 for MS-2, >0.81 for MS-3, and >0.84 for MS-4. The model RMSE was relatively small for the total, aboveground, and stem biomass equations, but was larger for the root, branch, foliage, and crown biomass equations. Overall, adding H and crown attributes to a system of biomass equations can significantly improve model fitting and performance.
In addition, we analyzed the biomass partitioning of the aboveground and belowground components of the two species. Our results were consistent with the literature in that stem biomass accounted for the largest proportion of total biomass. We also evaluated different additive systems of biomass equations for quantifying individual biomass. The results indicated that, in general, MS-1 and MS-4 were the best predictors of the majority of the biomass components. The overall ranking of the four additive systems of biomass equations followed the order of MS-1 > MS-4 > MS-3 > MS-2. Overall, the tree biomass data in this study was widely distributed throughout the eastern Daxing’an Mountains in northeast China. Thus, these established biomass equations can be used to estimate individual tree biomass in the Chinese National Forest Inventory. However, caution should be taken when using the newly developed systems of equations to predict biomass of trees outside the range of the data and region.