Field Supervisory Test of DREB-Transgenic Populus: Salt Tolerance, Long-Term Gene Stability and Horizontal Gene Transfer
Abstract
:1. Introduction
2. Materials and Methods
2.1. Characterization of Transformed Plants and Experimental Field Design


2.2. Materials
2.3. Screening for Kanamycin-Resistant Organisms
2.4. DNA Isolation and PCR Analyses
2.5. Total RNA Extraction and Real-Time Quantitative PCR (qRT-PCR)
2.6. Transgenic Populus Tissue Culture
3. Results and Discussion
3.1. Stability of the Foreign Gene after a Seven-Year Field Test

3.2. Expression of AhDREB1 mRNA after Three and Seven Years

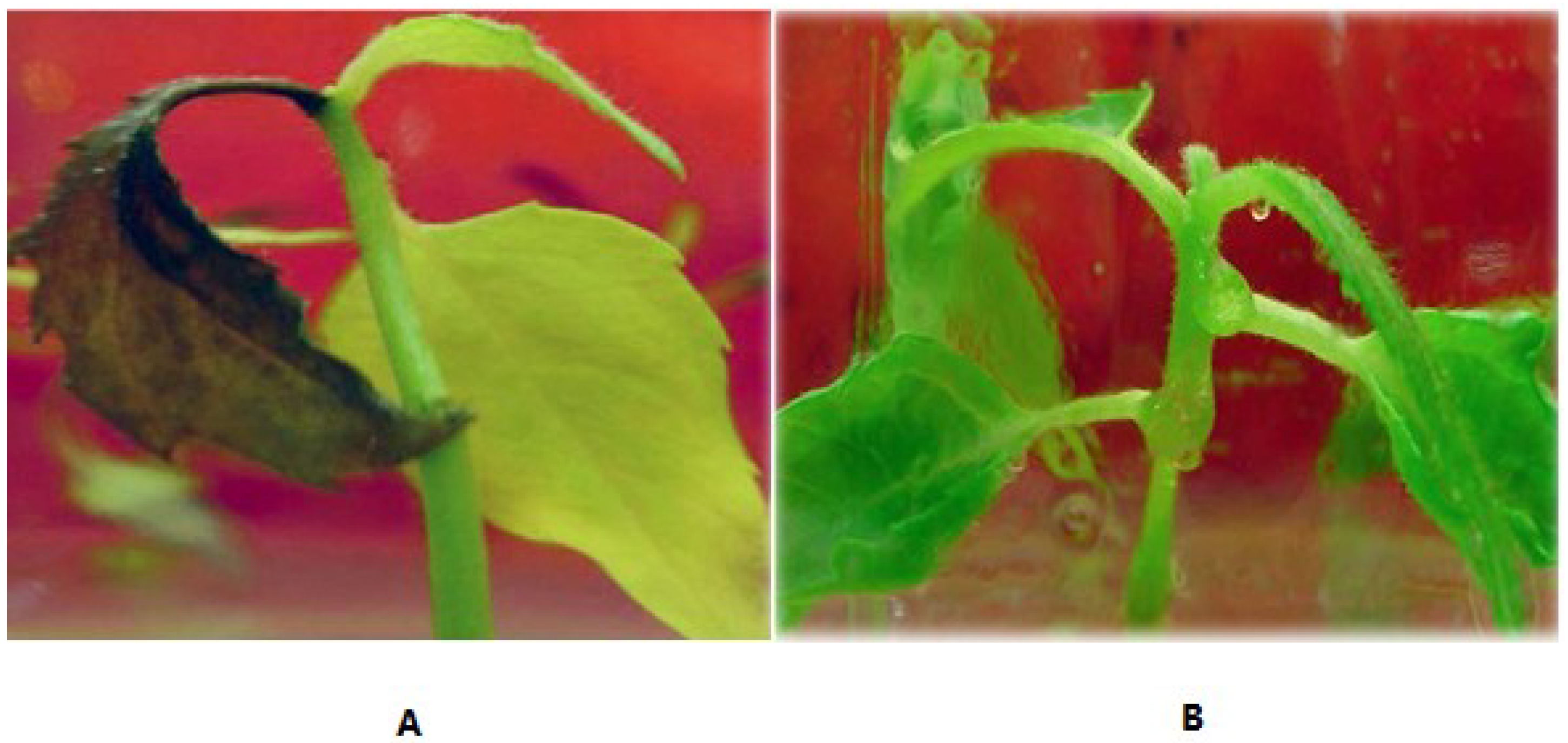
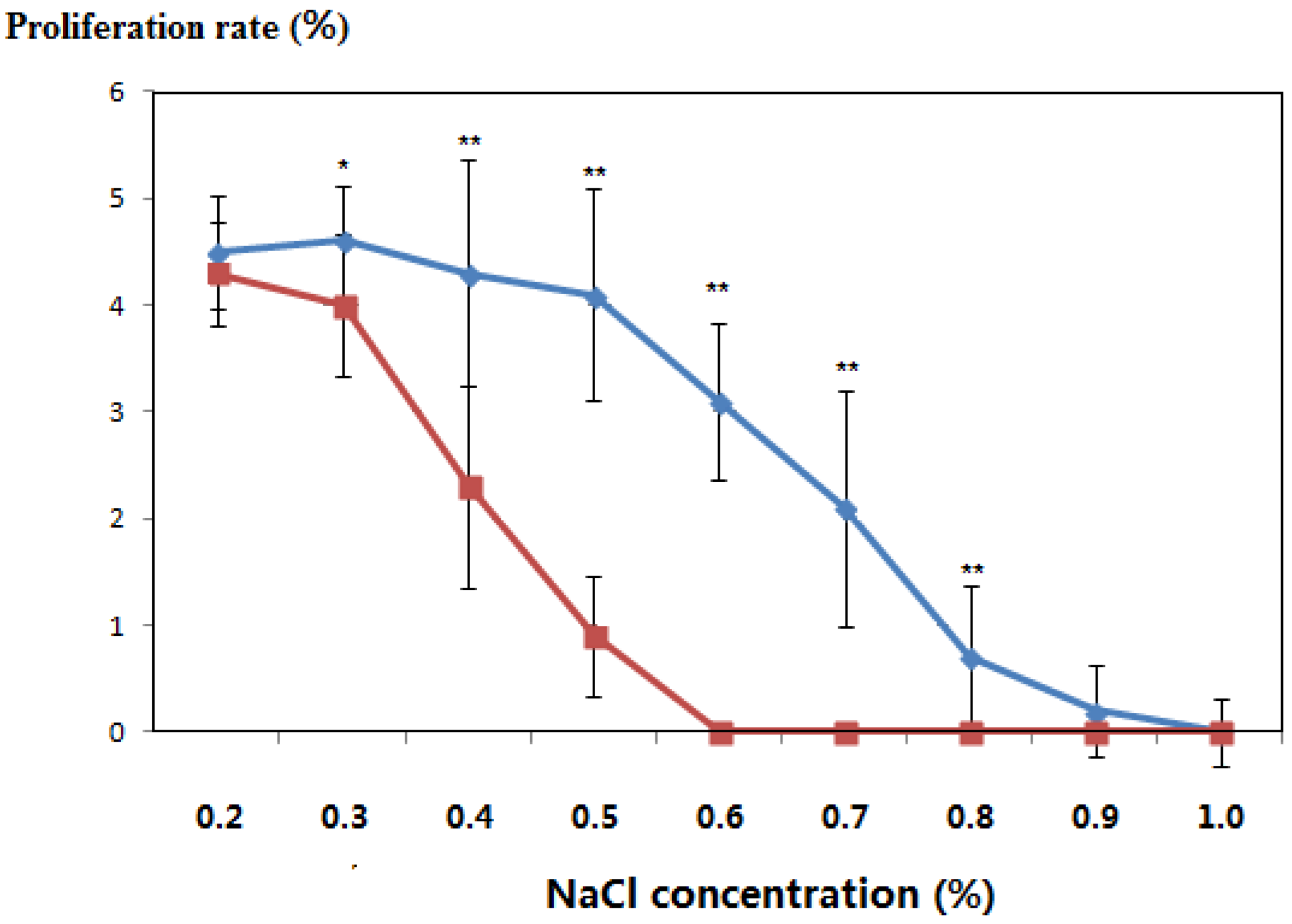
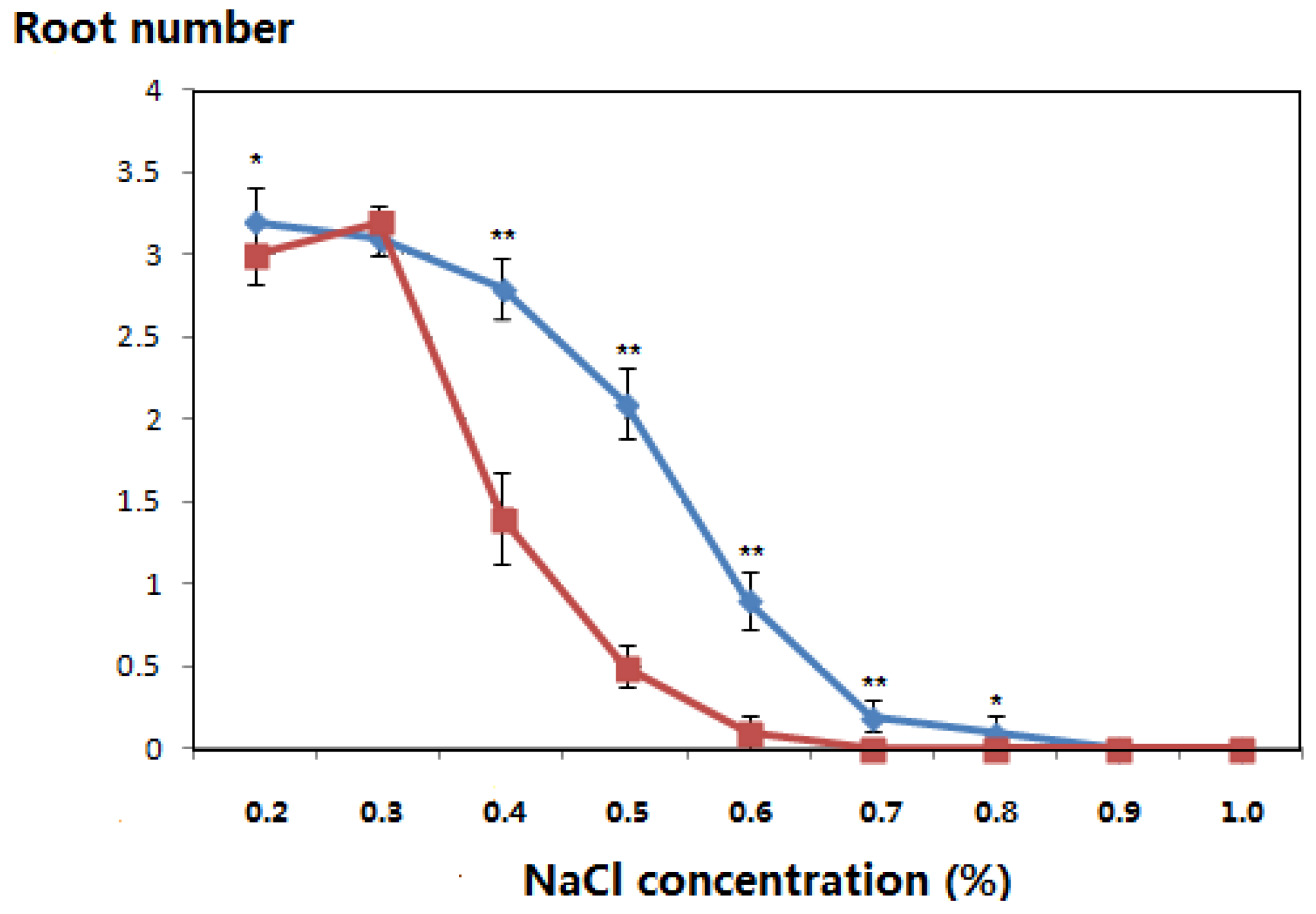
3.3. Salt-Tolerant Phenotype of the Transgenic Plants
| NaCl concentration (%) | Incubation Time (Days) | Leaf Color | Differentiation Rate (%) | ||
|---|---|---|---|---|---|
| Transgenic Populus | Controls | Transgenic Populus | Controls | ||
| 0.2 | 21 | Green | Green | 2.3 | 2.2 |
| 28 | Green | Green | 3.6 | 3.3 | |
| 35 | Green | Green | 4.5 | 4.3 | |
| 0.3 | 21 | Green | Green | 2.4 | 2.2 |
| 28 | Green | Green | 3.3 | 3.5 | |
| 35 | Green | Green | 4.6 | 4.2 | |
| 0.4 | 21 | Green | Green | 2.5 | 1.9 |
| 28 | Green | Green | 3.0 | 2.3 | |
| 35 | Green | Yellow/green | 4.3 | 2.7 | |
| 0.5 | 21 | Green | Green | 2.2 | 0.7 |
| 28 | Green | Green | 3.2 | 0.9 | |
| 35 | Green | Yellow | 4.1 | 1.1 | |
| 0.6 | 21 | Green | Green | 2.1 | - |
| 28 | Green | Yellow/green | 2.8 | - | |
| 35 | Yellow/green | Yellow | 3.1 | - | |
| 0.7 | 21 | Green | Yellow | 1.6 | - |
| 28 | Yellow/green | Yellow | 1.9 | - | |
| 35 | Yellow/green | Withered | 2.1 | - | |
| 0.8 | 21 | Yellow/green | Yellow | 0.6 | - |
| 28 | Yellow/green | Withered | 1.0 | - | |
| 35 | Yellow | Dead | 0.7 | - | |
| 0.9 | 21 | Yellow/green | Withered | 0.2 | - |
| 28 | Yellow | Withered | 0.4 | - | |
| 35 | Yellow | Dead | 0.2 | - | |
| 1.0 | 21 | Yellow/green | Withered | 0.2 | - |
| 28 | Yellow | Dead | 0.2 | - | |
| 35 | Withered | Dead | 0.1 | - | |
| NaCl concentration (%) | Incubation Time (Days) | Leaf Color | Root Number | ||
|---|---|---|---|---|---|
| Transgenic Populus | Controls | Transgenic Populus | Controls | ||
| 0.2 | 21 | Green | Green | 1.5 | 1.6 |
| 28 | Green | Green | 2.4 | 2.3 | |
| 35 | Green | Green | 3.0 | 2.8 | |
| 42 | Green | Green | 3.2 | 3.0 | |
| 0.3 | 21 | Green | Green | 1.7 | 1.5 |
| 28 | Green | Green | 2.5 | 2.5 | |
| 35 | Green | Green | 2.7 | 2.9 | |
| 42 | Green | Green | 3.1 | 3.2 | |
| 0.4 | 21 | Green | Green | 1.4 | 1.1 |
| 28 | Green | Green | 2.0 | 1.2 | |
| 35 | Green | Yellow/green | 2.4 | 1.4 | |
| 42 | Green | Yellow/green | 2.8 | 1.4 | |
| 0.5 | 21 | Green | Green | 0.7 | 0.4 |
| 28 | Green | Green | 1.5 | 0.4 | |
| 35 | Green | Yellow/green | 1.5 | 0.5 | |
| 42 | Yellow/green | Yellow/green | 2.1 | 0.5 | |
| 0.6 | 21 | Green | Green | 0.6 | 0.2 |
| 28 | Yellow/green | Yellow/green | 0.8 | 0.1 | |
| 35 | Yellow/green | Yellow | 0.9 | 0.1 | |
| 42 | Yellow | Withered | 0.9 | 0.1 | |
| 0.7 | 21 | Green | Yellow | 0.2 | - |
| 28 | Yellow/green | Yellow | 0.2 | - | |
| 35 | Yellow/green | Withered | 0.2 | - | |
| 42 | Yellow | Withered | 0.2 | - | |
| 0.8 | 21 | Yellow/green | Yellow | 0.2 | - |
| 28 | Yellow/green | Withered | 0.0 | - | |
| 35 | Yellow | Dead | 0.1 | - | |
| 42 | Withered | Dead | 0.1 | - | |
| 0.9 | 21 | Yellow/green | Withered | 0.1 | - |
| 28 | Yellow | Withered | - | - | |
| 35 | Yellow | Dead | - | - | |
| 42 | Withered | Dead | - | - | |
| 1.0 | 21 | Yellow | Withered | - | - |
| 28 | Yellow | Dead | - | - | |
| 35 | Withered | Dead | - | - | |
| 42 | Withered | Dead | - | - | |
3.4. PCR Amplification of the NptII Gene in the Genomic DNA of Kanamycin-Resistant Microorganisms

3.5. Discussion
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Zeng, F.; Qian, J.; Luo, W.; Zhan, Y.; Xin, Y.; Yang, C. Stability of transgenes in long-term micropropagation of plants of transgenic birch (Betula platyphylla). Biotechnol. Lett. 2010, 32, 151–156. [Google Scholar] [CrossRef]
- Ahuja, M.R. Transgene stability and dispersal in forest trees. Trees 2009, 23, 1125–1135. [Google Scholar] [CrossRef]
- Han, K.H.; Ma, C.; Strauss, S.H. Matrix attachment regions (MARs) enhance transformation frequency and transgene expression in poplar. Transgenic Res. 1997, 6, 415–420. [Google Scholar] [CrossRef]
- Bradshaw, H.; Ceulemans, R.; Davis, J.; Stettler, R. Emerging model systems in plant biology: Poplar (Populus) as a model forest tree. J. Plant Growth Regul. 2000, 19, 306–313. [Google Scholar] [CrossRef]
- Choi, H.; Lemaux, P.; Cho, M.J. Increased chromosomal variation in transgenic versus nontransgenic barley (L.) plants. Crop Sci. 2000, 40, 524–533. [Google Scholar] [CrossRef]
- Bizily, S.P.; Rugh, C.L.; Meagher, R.B. Phytodetoxification of hazardous organomercurials by genetically engineered plants. Nat. Biotechnol. 2000, 18, 213–217. [Google Scholar] [CrossRef]
- Chen, L.J.; Lee, D.S.; Song, Z.P.; Suh, H.S.; Lu, B.R. Gene flow from cultivated rice (Oryza sativa) to its weedy and wild relatives. Ann. Bot. 2004, 93, 67–73. [Google Scholar] [CrossRef]
- Chèvre, A.M.; Eber, F.; Baranger, A.; Renard, M. Gene flow from transgenic crops. Nature 1997, 389, 924. [Google Scholar]
- Gebhard, F.; Smalla, K. Monitoring field releases of genetically modified sugar beets for persistence of transgenic plant DNA and horizontal gene transfer. FEMS Microbiol. Ecol. 1999, 28, 261–272. [Google Scholar] [CrossRef]
- Schlüter, K.; Fütterer, J.; Potrykus, I. “Horizontal” gene transfer from a transgenic potato line to a bacterial pathogen (Erwinia chrysanthemi) occurs—If at all—At an Extremely Low Frequency. Nat. Biotechnol. 1995, 13, 1094–1098. [Google Scholar] [CrossRef]
- Gebhard, F.; Smalla, K. Transformation of Acinetobacter sp. strain BD413 by transgenic sugar beet DNA. Appl. Environ. Microbiol. 1998, 64, 1550–1554. [Google Scholar]
- Widmer, F.; Seidler, R.; Donegan, K.; Reed, G. Quantification of transgenic plant marker gene persistence in the field. Mol. Ecol. 1997, 6, 1–7. [Google Scholar] [CrossRef]
- Kang, X.Y.; Liu, Z.M.; Li, S.G. Potential ecological risks of transgenic trees. Chin. J. Appl. Ecol. 2004, 15, 1281–1284. [Google Scholar]
- Peña, L.; Séguin, A. Recent advances in the genetic transformation of trees. TRENDS Biotechnol. 2001, 19, 500–506. [Google Scholar] [CrossRef]
- Strauss, S.H.; DiFazio, S.P.; Meilan, R. Genetically modified poplars in context. For. Chron. 2001, 77, 271–279. [Google Scholar] [CrossRef]
- Brunner, A.M.; Nilsson, O. Revisiting tree maturation and floral initiation in the poplar functional genomics era. New Phytol. 2004, 164, 43–51. [Google Scholar] [CrossRef]
- Li, J.; Meilan, R.; Ma, C.; Barish, M.; Strauss, S.H. Stability of herbicide resistance over 8 years of coppice in field-grown, genetically engineered poplars. West. J. Appl. For. 2008, 23, 89–93. [Google Scholar]
- Du, N.; Liu, X.; Li, Y.; Chen, S.; Zhang, J.; Ha, D.; Deng, W.; Sun, C.; Zhang, Y.; Pijut, P.M. Genetic transformation of Populus tomentosa to improve salt tolerance. Plant Cell Tissue Organ Cult. 2011, 108, 181–189. [Google Scholar]
- Frankenhuyzen, K.V.; Beardmore, T. Current status and environmental impact of transgenic forest trees. Can. J. For. Res. 2004, 34, 1163–1180. [Google Scholar] [CrossRef]
- Jouanin, L.; Goujon, T.; de Nadaı̈, V.; Martin, M.T.; Mila, I.; Vallet, C.; Pollet, B.; Yoshinaga, A.; Chabbert, B.; Petit-Conil, M. Lignification in transgenic poplars with extremely reduced caffeic acid O-methyltransferase activity. Plant Physiol. 2000, 123, 1363–1374. [Google Scholar] [CrossRef]
- Fladung, M.; Hoenicka, H.; Raj Ahuja, M. Genomic stability and long-term transgene expression in poplar. Transgenic Res. 2013, 22, 1167–1178. [Google Scholar] [CrossRef]
- Pilate, G.; Guiney, E.; Holt, K.; Petit-Conil, M.; Lapierre, C.; Leplé, J.C.; Pollet, B.; Mila, I.; Webster, E.A.; Marstorp, H.G. Field and pulping performances of transgenic trees with altered lignification. Nat. Biotechnol. 2002, 20, 607–612. [Google Scholar] [CrossRef]
- Meilan, R.; Auerbach, D.; Ma, C.; DiFazio, S.; Strauss, S. Stability of herbicide resistance and GUS expression in transgenic hybrid poplars (Populus sp.) during four years of field trials and vegetative propagation. HortSci. 2002, 37, 277–280. [Google Scholar]
- Li, J.; Brunner, A.M.; Meilan, R.; Strauss, S.H. Stability of transgenes in trees: Expression of two reporter genes in poplar over three field seasons. Tree Physiol. 2009, 29, 299–312. [Google Scholar]
- Borejsza-Wysocka, E.; Norelli, J.L.; Aldwinckle, H.S.; Malnoy, M. Stable expression and phenotypic impact of attacin E transgene in orchard grown apple trees over a 12 year period. BMC Biotechnol. 2010, 10, 41. [Google Scholar] [CrossRef]
- Zeng, F.; Xin, X.; Li, B.; Zhan, Y.; Yang, C. The stability of transgene expression and effect of DNA methylation on post transcriptional gene silencing (PTGS) in birch. Afr. J. Biotechnol. 2013, 10, 8188–8193. [Google Scholar]
- Hawkins, S.; Lepl, J.C.; Cornu, D.; Jouanin, L.; Pilate, G. Stability of transgene expression in poplar: A model forest tree species. Ann. For. Sci. 2003, 60, 427–438. [Google Scholar] [CrossRef]
- Traore, S.B.; Carlson, R.E.; Pilcher, C.D.; Rice, M.E. Bt and non-Bt maize growth and development as affected by temperature and drought stress. Agron. J. 2000, 92, 1027–1035. [Google Scholar] [CrossRef]
- Sachs, E.; Benedict, J.; Stelly, D.; Taylor, J.; Altman, D.; Berberich, S.; Davis, S. Expression and segregation of genes encoding CryIA insecticidal proteins in cotton. Crop Sci. 1998, 38, 1–11. [Google Scholar] [CrossRef]
- Bonadei, M.; Zelasco, S.; Giorcelli, A.; Gennaro, M.; Calligari, P.; Quattrini, E.; Carbonera, D.; Balestrazzi, A. Transgene stability and agronomical performance of two transgenic Basta®-tolerant lines of Populus alba L. Plant Biosyst.-Int. J. Deal. Asp. Plant Biol. 2012, 146, 33–40. [Google Scholar]
- Zhang, C.; Hampp, R.; Nehls, U. Investigation of horizontal gene transfer in poplar/Amanita muscaria ectomycorrhizas. Environ. Biosaf. Res. 2005, 4, 235–242. [Google Scholar] [CrossRef]
- Kaldorf, M.; Fladung, M.; Muhs, H.J.; Buscot, F. Mycorrhizal colonization of transgenic aspen in a field trial. Planta 2002, 214, 653–660. [Google Scholar] [CrossRef]
- Hou, Y.J. Preliminary Study on the Ecological Safety Assessment of Transgenic Poplar. Ph.D. Thesis, Chinese Academy of Forestry, Beijing, China, 2008. [Google Scholar]
- Hoffmann, T.; Golz, C.; Schieder, O. Foreign DNA sequences are received by a wild-type strain of Aspergillus niger after co-culture with transgenic higher plants. Curr. Genet. 1994, 27, 70–76. [Google Scholar] [CrossRef]
- Won, H.; Renner, S.S. Horizontal gene transfer from flowering plants to Gnetum. Proc. Natl. Acad. Sci. 2003, 100, 10824–10829. [Google Scholar] [CrossRef]
- Kondo, N.; Nikoh, N.; Ijichi, N.; Shimada, M.; Fukatsu, T. Genome fragment of Wolbachia endosymbiont transferred to X chromosome of host insect. Proc. Natl. Acad. Sci. 2002, 99, 14280–14285. [Google Scholar] [CrossRef]
- Kay, E.; Vogel, T.M.; Bertolla, F.; Nalin, R.; Simonet, P. In situ transfer of antibiotic resistance genes from transgenic (transplastomic) tobacco plants to bacteria. Appl. Environ. Microbiol. 2002, 68, 3345–3351. [Google Scholar] [CrossRef]
- Richter, B.; Smalla, K. Screening of rhizosphere and soil bacteria for transformability. Environ. Biosaf. Res. 2007, 6, 91–99. [Google Scholar] [CrossRef]
- Monier, J.M.; Bernillon, D.; Kay, E.; Faugier, A.; Rybalka, O.; Dessaux, Y.; Simonet, P.; Vogel, T.M. Detection of potential transgenic plant DNA recipients among soil bacteria. Environ. Biosaf. Res. 2007, 6, 71–83. [Google Scholar] [CrossRef]
- Simpson, D.J.; Dawson, L.F.; Fry, J.C.; Rogers, H.J.; Day, M.J. Influence of flanking homology and insert size on the transformation frequency of Acinetobacter baylyi BD413. Environ. Biosaf. Res. 2007, 6, 55–69. [Google Scholar] [CrossRef]
- De Vries, J.; Wackernagel, W. Integration of foreign DNA during natural transformation of Acinetobacter sp. by homology-facilitated illegitimate recombination. Proc. Natl. Acad. Sci. 2002, 99, 2094–2099. [Google Scholar] [CrossRef]
- Wei, B.; Li, Y.; Du, N.X.; Liu, X. Transgene stability of transgenic hybrid of Populus[(Populus tomentosa × P. bolleana) × P. tomentosa] and its effect on soil microorganism. J. Nucl. Agric. Sci. 2009, 23, 1054–1059. [Google Scholar]
- Amann, R.I.; Ludwig, W.; Schleifer, K.H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 1995, 59, 143–169. [Google Scholar]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Lu, N.; Wei, B.; Sun, Y.; Liu, X.; Chen, S.; Zhang, W.; Zhang, Y.; Li, Y. Field Supervisory Test of DREB-Transgenic Populus: Salt Tolerance, Long-Term Gene Stability and Horizontal Gene Transfer. Forests 2014, 5, 1106-1121. https://doi.org/10.3390/f5051106
Lu N, Wei B, Sun Y, Liu X, Chen S, Zhang W, Zhang Y, Li Y. Field Supervisory Test of DREB-Transgenic Populus: Salt Tolerance, Long-Term Gene Stability and Horizontal Gene Transfer. Forests. 2014; 5(5):1106-1121. https://doi.org/10.3390/f5051106
Chicago/Turabian StyleLu, Nan, Bing Wei, Yuhan Sun, Xin Liu, Shouyi Chen, Wanke Zhang, Yingzhi Zhang, and Yun Li. 2014. "Field Supervisory Test of DREB-Transgenic Populus: Salt Tolerance, Long-Term Gene Stability and Horizontal Gene Transfer" Forests 5, no. 5: 1106-1121. https://doi.org/10.3390/f5051106
APA StyleLu, N., Wei, B., Sun, Y., Liu, X., Chen, S., Zhang, W., Zhang, Y., & Li, Y. (2014). Field Supervisory Test of DREB-Transgenic Populus: Salt Tolerance, Long-Term Gene Stability and Horizontal Gene Transfer. Forests, 5(5), 1106-1121. https://doi.org/10.3390/f5051106





