Dynamics of Fine Root Decomposition in Different Vegetation Types: Investigating the Impact of Soil Fungal Communities and Enzyme Activities
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Site
2.2. Sampling
2.3. Fine Root and Soil Physicochemical Properties
2.4. Soil Physicochemical Properties
2.5. Soil Enzyme Activities
2.6. Soil Fungal Molecular Analysis
2.7. Calculations
- (1)
- Fine root decomposition rate was calculated from dry mass remaining and percent (%) chemical mass remaining is expressed as follows [40]:
- (2)
- Litter decomposition is a dynamic process, and there is a single exponential function relationship between litter residue and decomposition time (Olson 1963). Therefore, Olson proposed an exponential attenuation model:
- —initial mass of plant litter, kg;
- —residual mass of litter after t-time decomposition, kg;
- k—litter decomposition constant;
- t—decomposition time.
2.8. Statistical Analyses
3. Results
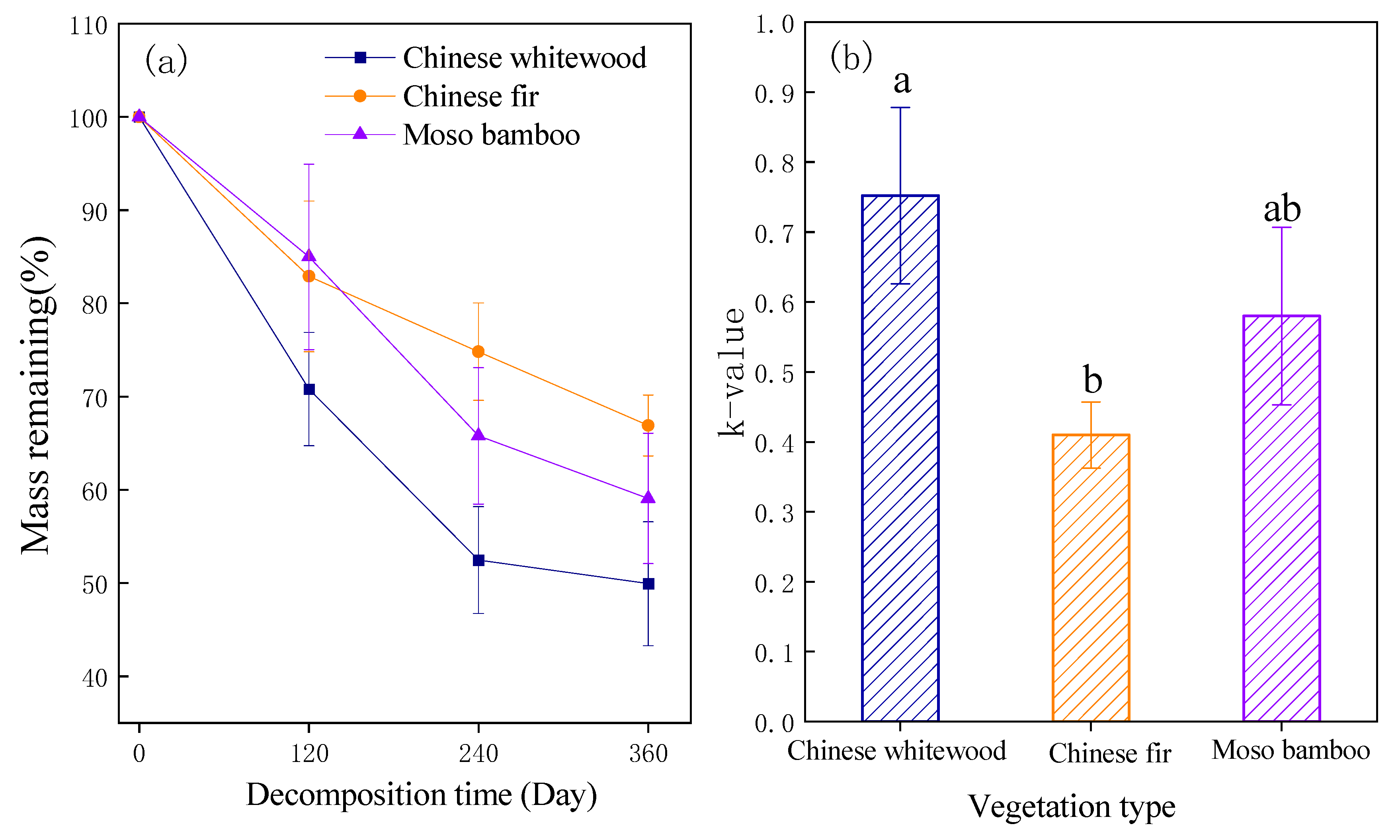
3.1. Fine Root Mass Loss and k Value of Decomposition Rate
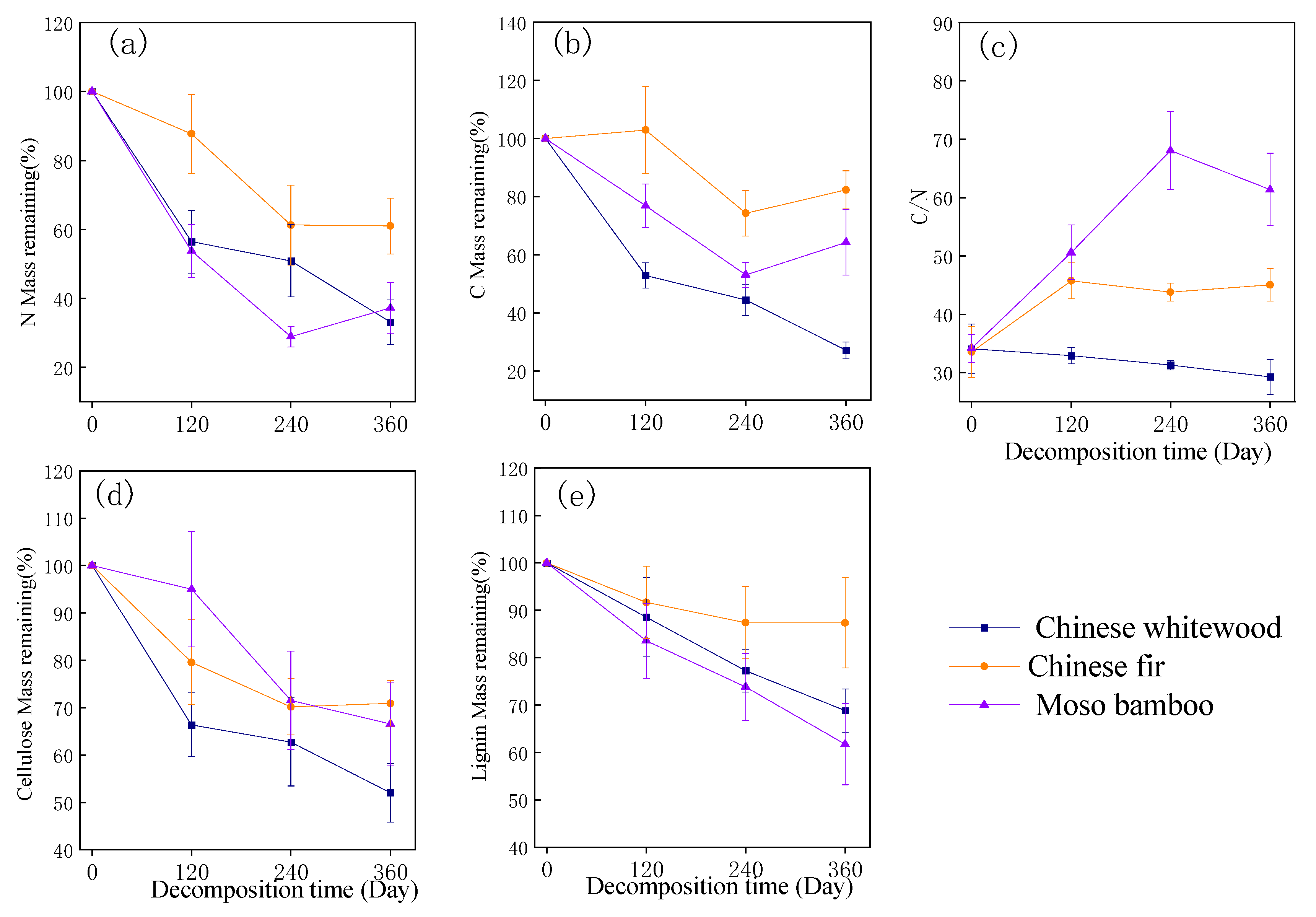
3.2. Changes in Chemical Quality of Fine Root Litter during Fine Root Decomposition
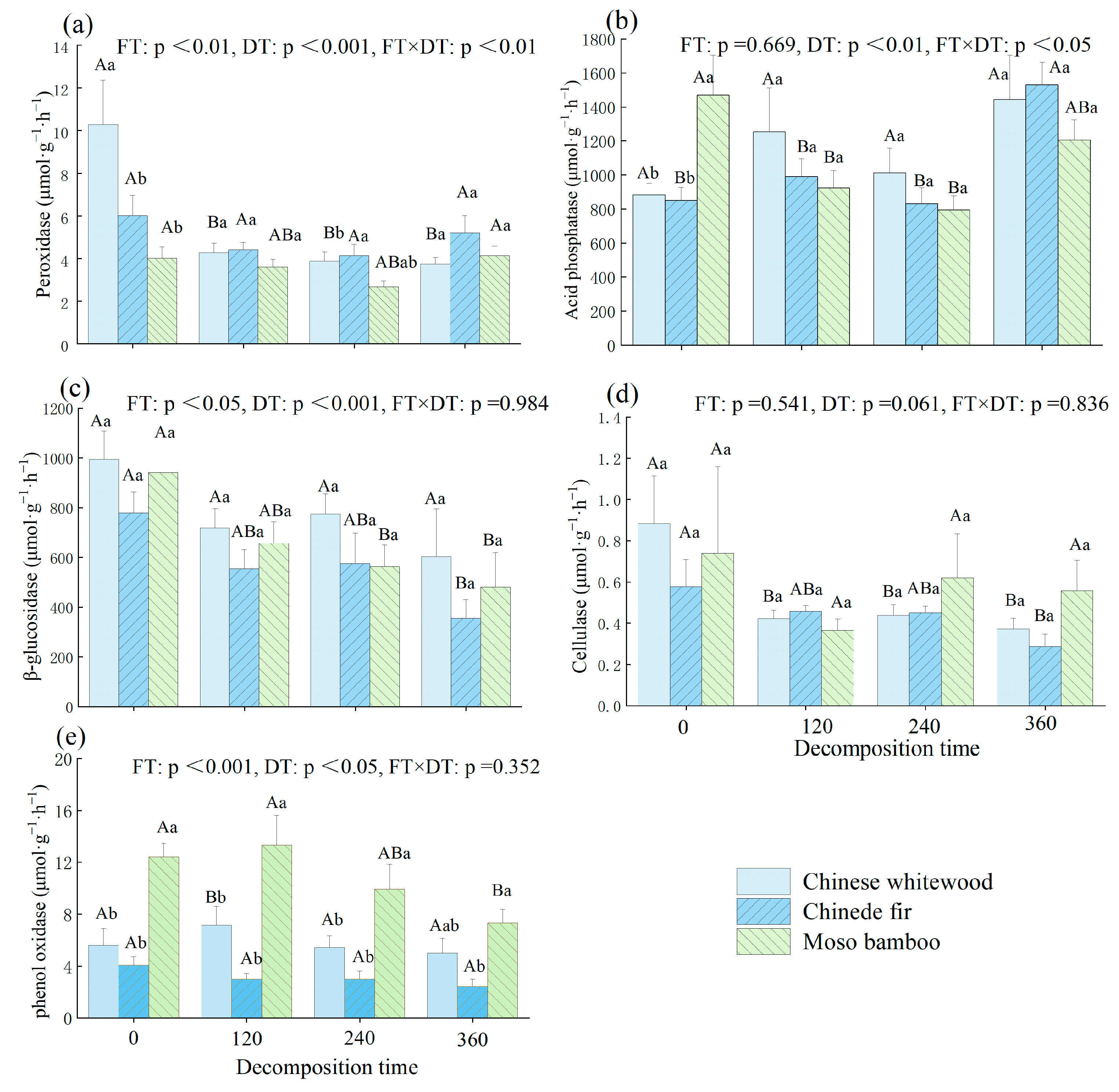
3.3. Dynamic Changes in Soil Enzyme Activities during the Decomposition of Fine Root Litter
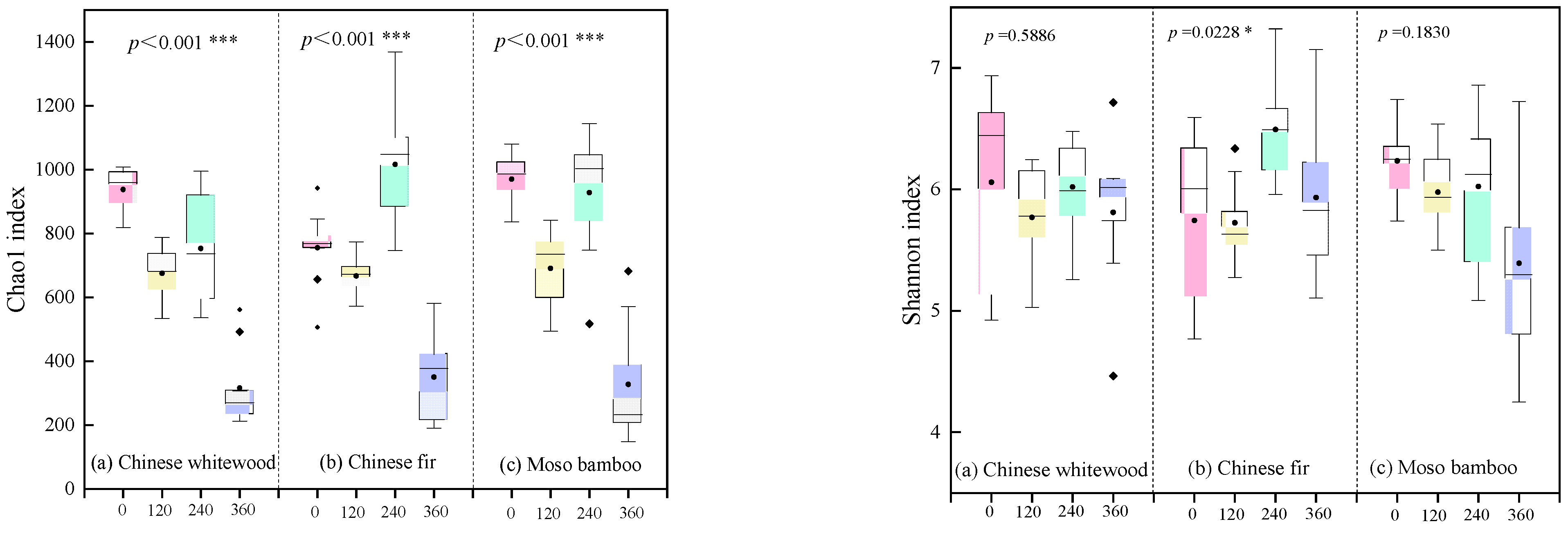
3.4. Soil Fungal Community Diversity during the Decomposition of Fine Root Litters
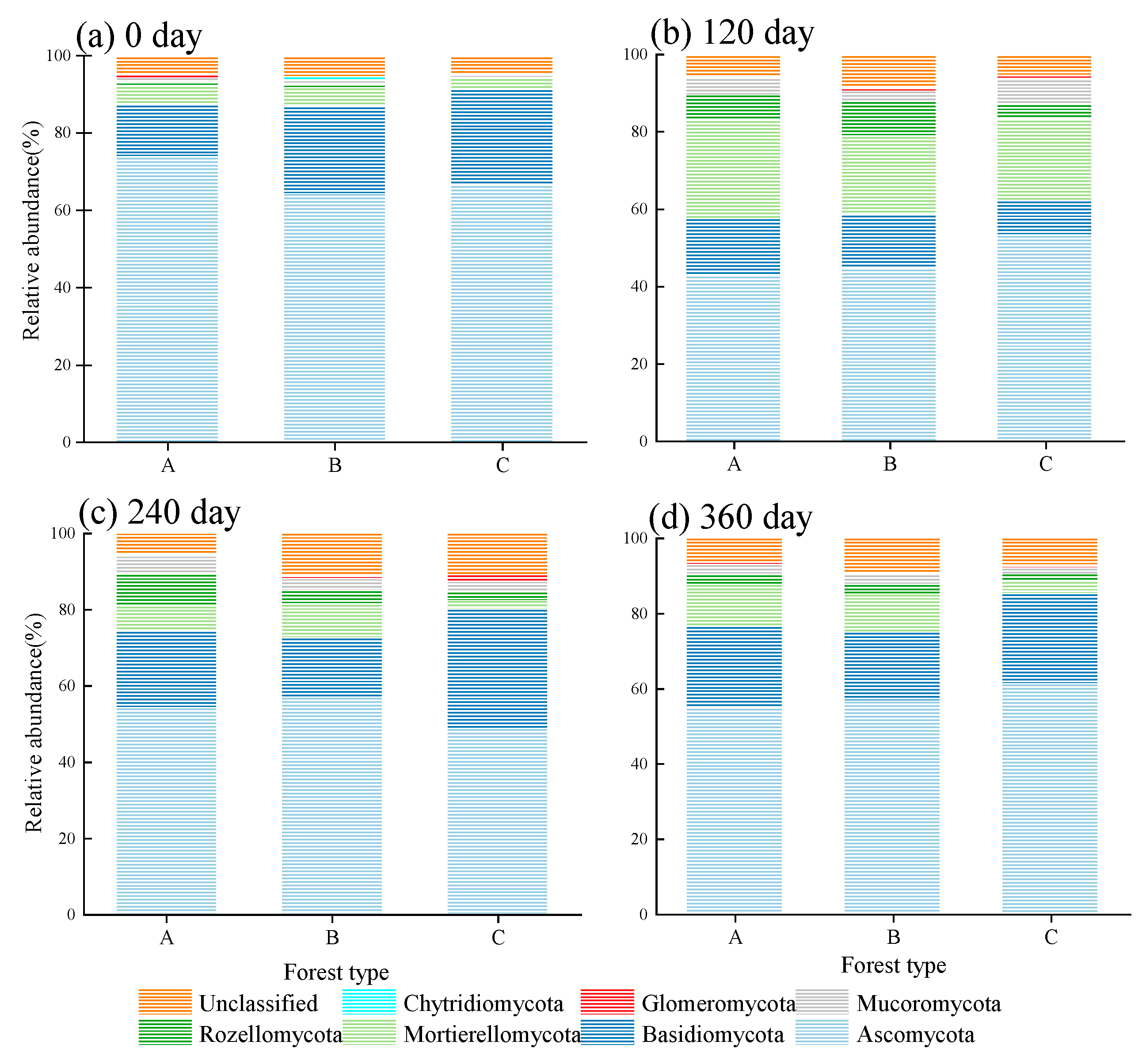
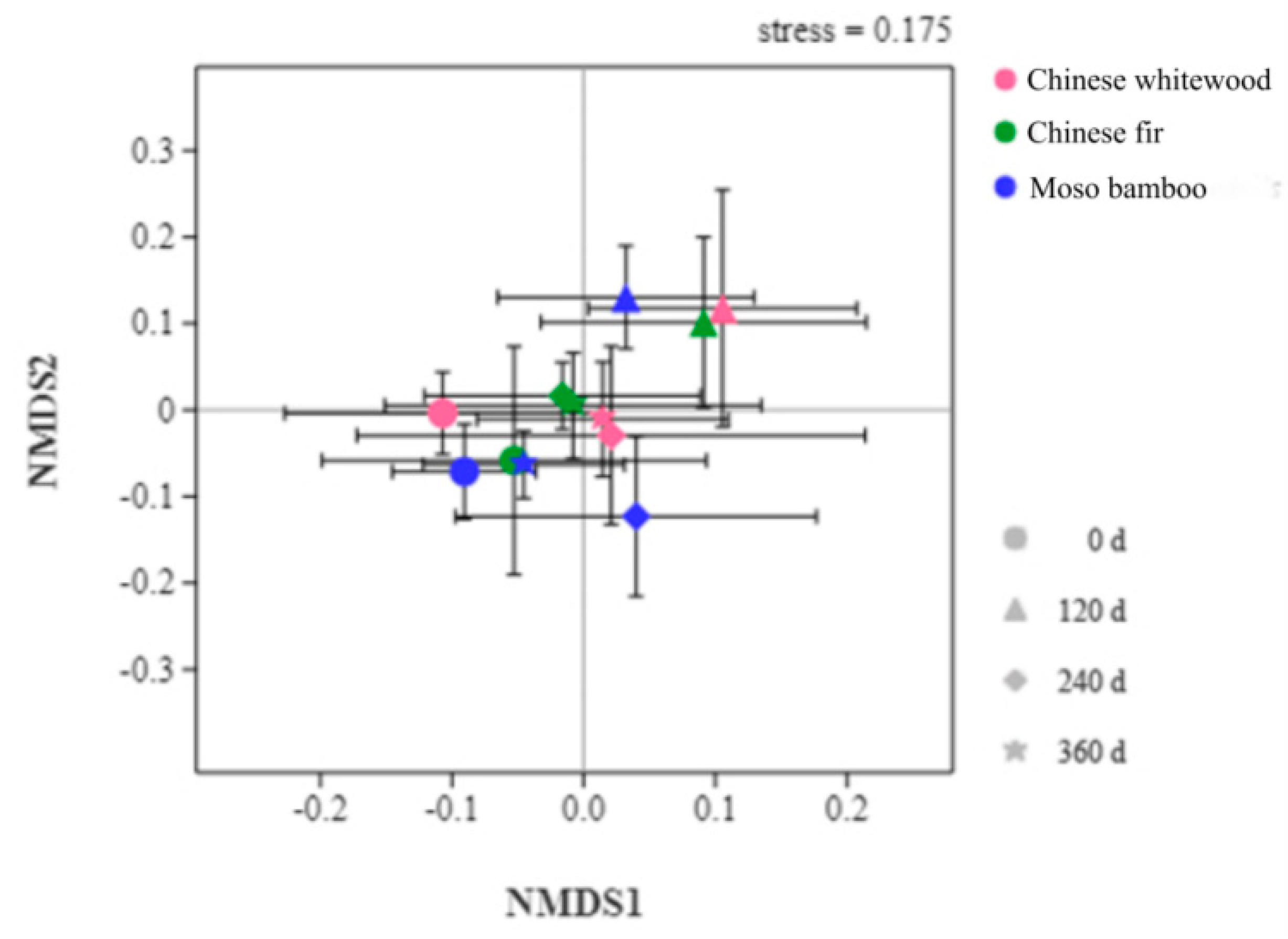
3.5. Soil Fungal Community Structure during the Decomposition of Fine Root Litters
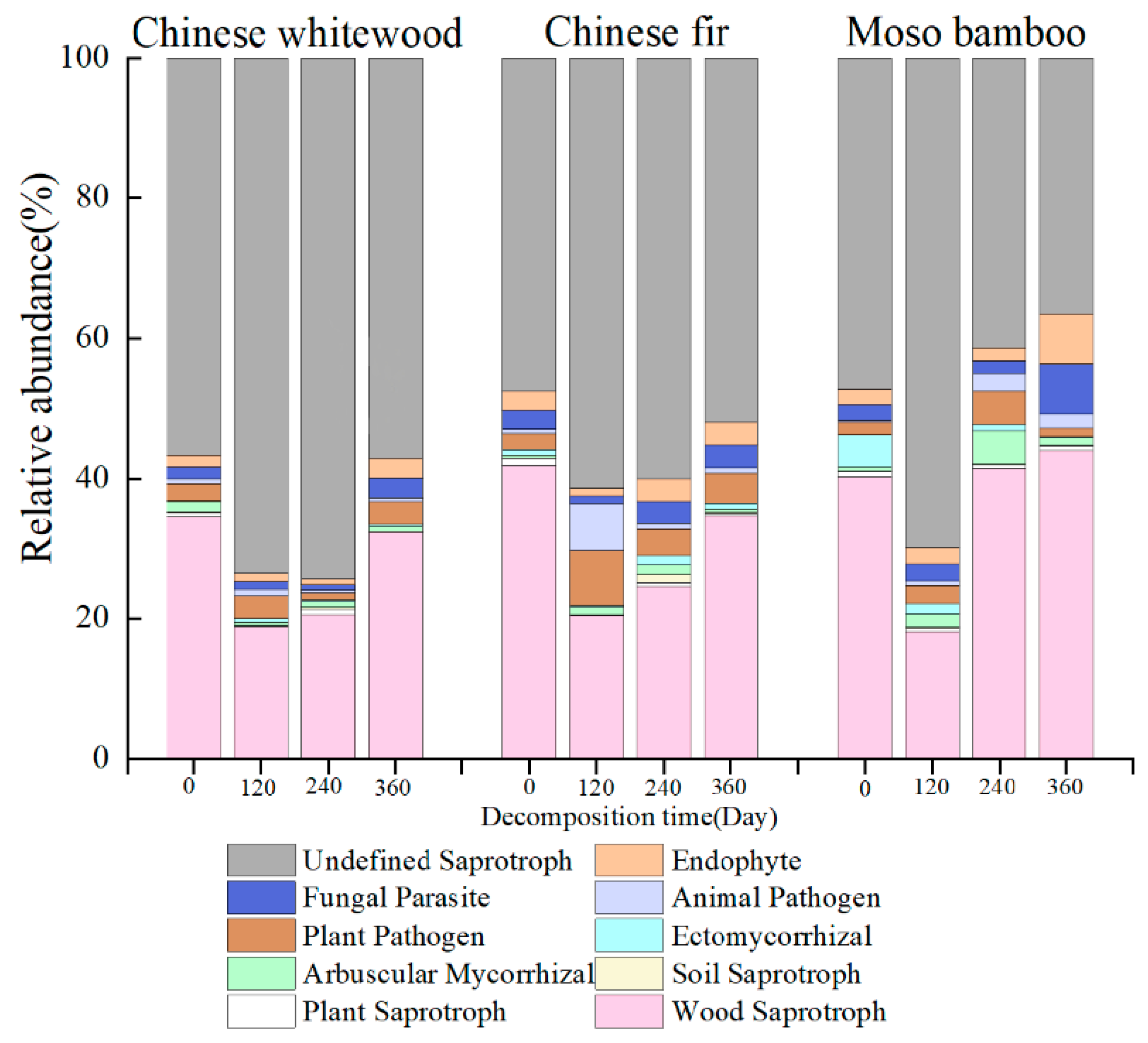
3.6. Fungal Guilds during the Decomposition of Fine Root Litter
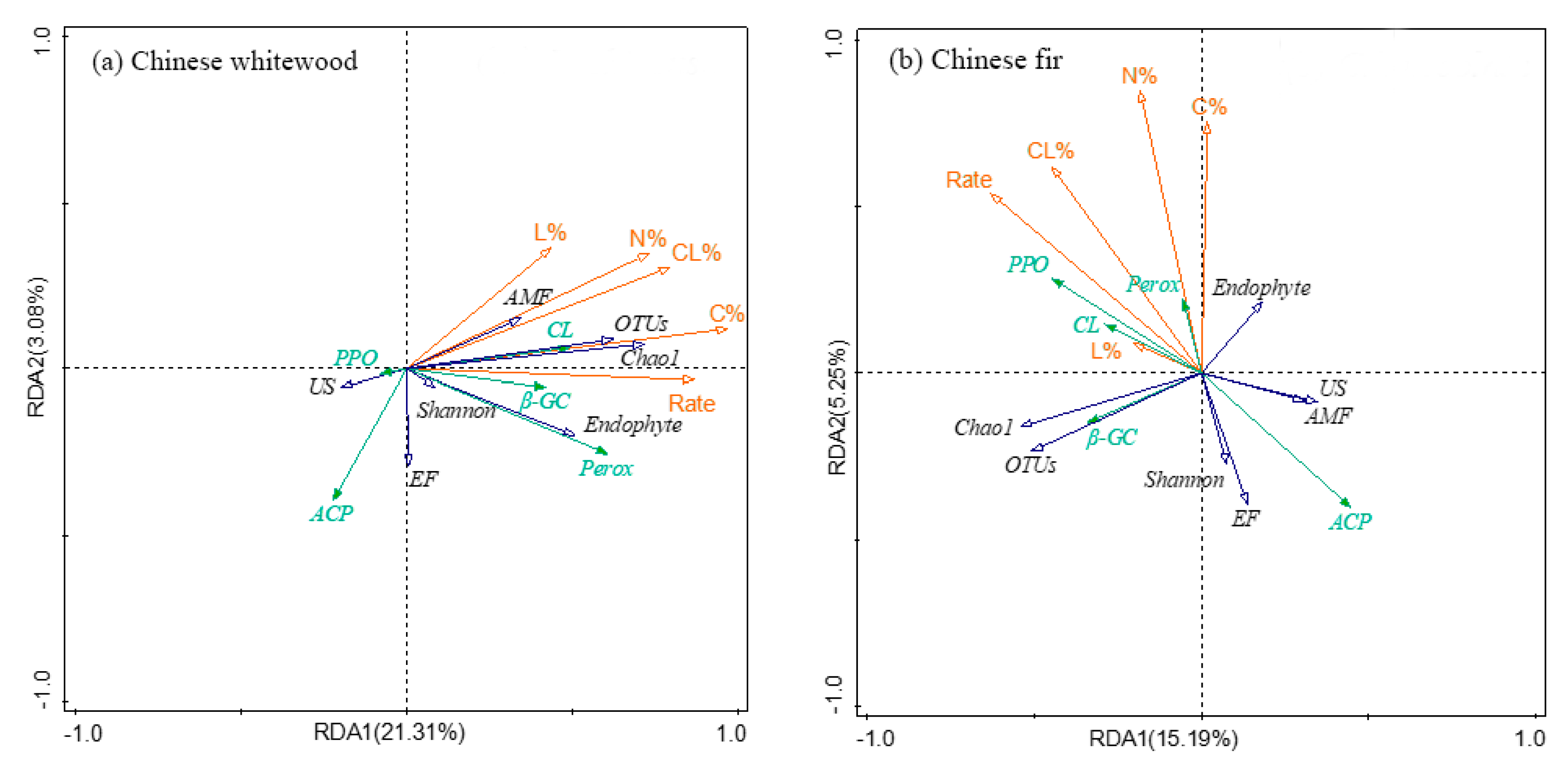
3.7. The Relationship between Fine Root Decomposition and Root Chemistry, Soil Fungal Community, and Soil Enzyme Activity
3.8. SEM Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Sun, T.; Dong, L.; Wang, Z.; Lü, X.; Mao, Z. Effects of long-term nitrogen deposition on fine root decomposition and its extracellular enzyme activities in temperate forests. Soil Biol. Biochem. 2016, 93, 50–59. [Google Scholar] [CrossRef]
- Larreguy, C.; Carrera, A.L.; Bertiller, M.B. Production and turnover rates of shallow fine roots in rangelands of the Patagonian Monte, Argentina. Ecol. Res. 2011, 27, 61–68. [Google Scholar] [CrossRef]
- Rodtassana, C.; Tanner, E.V.J. Litter removal in a tropical rain forest reduces fine root biomass and production but litter. Ecology 2018, 99, 735–742. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Fahey, T.J.; Pawlowska, T.E.; Fisk, M.C.; Burtis, J. Fine root decomposition, nutrient mobilization and fungal communities in a pine forest ecosystem. Soil Biol. Biochem. 2015, 83, 76–83. [Google Scholar] [CrossRef]
- Herzog, C.; Hartmann, M.; Frey, B.; Stierli, B.; Rumpel, C.; Buchmann, N.; Brunner, I. Microbial succession on decomposing root litter in a drought-prone Scots pine forest. ISME J. 2019, 13, 2346–2362. [Google Scholar] [CrossRef]
- Fu, Y.; Feng, F.; Zhang, X.; Qi, D. Changes in fine root decomposition of primary Pinus koraiensis forest after clear cutting and restoration succession into secondary broad-leaved forest. Appl. Soil Ecol. 2021, 158, 103785. [Google Scholar] [CrossRef]
- Mendez-Millan, M.; Dignac, M.F.; Rumpel, C.; Rasse, D.P.; Derenne, S. Molecular dynamics of shoot vs. root biomarkers in an agricultural soil estimated by natural abundance 13C labelling. Soil Biol. Biochem. 2010, 42, 169–177. [Google Scholar] [CrossRef]
- Cao, C.; Liu, S.Q.; Ma, Z.B.; Lin, Y.; Su, Q.; Chen, H.; Wang, J.J. Dynamics of multiple elements in fast decomposing vegetable residues. Sci. Total Environ. 2018, 616–617, 614–621. [Google Scholar] [CrossRef]
- Solly, E.F.; Schöning, I.; Boch, S.; Kandeler, E.; Marhan, S.; Michalzik, B.; Müller, J.; Zscheischler, J.; Trumbore, S.E.; Schrumpf, M. Factors controlling decomposition rates of fine root litter in temperate forests and grasslands. Plant Soil 2014, 382, 203–218. [Google Scholar] [CrossRef]
- Cotrufo, M.F.; Wallenstein, M.D.; Boot, C.M.; Denef, K.; Paul, E. The Microbial Efficiency-Matrix Stabilization (MEMS) framework integrates plant litter decomposition with soil organic matter stabilization: Do labile plant inputs form stable soil organic matter? Glob. Chang. Biol. 2013, 19, 988–995. [Google Scholar] [CrossRef]
- Jacobs, L.M.; Sulman, B.N.; Brzostek, E.R.; Feighery, J.J.; Phillips, R.P. Interactions among decaying leaf litter, root litter and soil organic matter vary with mycorrhizal type. J. Ecol. 2018, 106, 502–513. [Google Scholar] [CrossRef]
- El Moujahid, L.; Le Roux, X.; Michalet, S.; Bellvert, F.; Weigelt, A.; Poly, F. Effect of plant diversity on the diversity of soil organic compounds. PLoS ONE 2017, 12, e0170494. [Google Scholar] [CrossRef]
- Voriskova, J.; Baldrian, P. Fungal community on decomposing leaf litter undergoes rapid successional changes. ISME J. 2013, 7, 477–486. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Yang, K.; Lyu, Z.; Zhu, J. Microbial groups and their functions control the decomposition of coniferous litter: A comparison with broadleaved tree litters. Soil Biol. Biochem. 2019, 133, 196–207. [Google Scholar] [CrossRef]
- Lv, Y.; Wang, C.; Jia, Y.; Wang, W.; Ma, X.; Du, J.; Pu, G.; Tian, X. Effects of sulfuric, nitric, and mixed acid rain on litter decomposition, soil microbial biomass, and enzyme activities in subtropical forests of China. Appl. Soil Ecol. 2014, 79, 1–9. [Google Scholar] [CrossRef]
- Liu, X.; Li, C.; Meng, M.; Zhai, L.; Zhang, B.; Jia, Z.; Gu, Z.; Liu, Q.; Zhang, Y.; Zhang, J. Comparative effects of the recovery from sulfuric and nitric acid rain on the soil enzyme activities and metabolic functions of soil microbial communities. Sci. Total Environ. 2020, 714, 136788. [Google Scholar] [CrossRef]
- Otsing, E.; Barantal, S.; Anslan, S.; Koricheva, J.; Tedersoo, L. Litter species richness and composition effects on fungal richness and community structure in decomposing foliar and root litter. Soil Biol. Biochem. 2018, 125, 328–339. [Google Scholar] [CrossRef]
- Kohout, P.; Charvatova, M.; Stursova, M.; Masinova, T.; Tomsovsky, M.; Baldrian, P. Clearcutting alters decomposition processes and initiates complex restructuring of fungal communities in soil and tree roots. ISME J. 2018, 12, 692–703. [Google Scholar] [CrossRef] [PubMed]
- Fisk, M.C.; Fahey, T.J.; Sobieraj, J.H.; Staniec, A.C.; Crist, T.O. Rhizosphere disturbance influences fungal colonization and community development on dead fine roots. Plant Soil 2010, 341, 279–293. [Google Scholar] [CrossRef]
- Ayres, E.; Steltzer, H.; Simmons, B.L.; Simpson, R.T.; Steinweg, J.M.; Wallenstein, M.D.; Mellor, N.; Parton, W.J.; Moore, J.C.; Wall, D.H. Home-field advantage accelerates leaf litter decomposition in forests. Soil Biol. Biochem. 2009, 41, 606–610. [Google Scholar] [CrossRef]
- Allison, S.D.; Lu, Y.; Weihe, C.; Goulden, M.L.; Martiny, A.C.; Treseder, K.K.; Martiny, J.B. Microbial abundance and composition influence litter decomposition response to environmental change. Ecology 2013, 94, 714–725. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.-T.; Tu, L.-H.; Peng, Y.; Hu, H.-L.; Hu, T.-X.; Xu, Z.-F.; Liu, L.; Tang, Y. Effect of nitrogen additions on root morphology and chemistry in a subtropical bamboo forest. Plant Soil 2017, 412, 441–451. [Google Scholar] [CrossRef]
- Wang, C.; Han, S.; Zhou, Y.; Yan, C.; Cheng, X.; Zheng, X.; Li, M.H. Responses of fine roots and soil N availability to short-term nitrogen fertilization in a broad-leaved Korean pine mixed forest in northeastern China. PLoS ONE 2012, 7, e31042. [Google Scholar] [CrossRef] [PubMed]
- Sariyildiz, T. Effects of tree species and topography on fine and small root decomposition rates of three common tree species (Alnus glutinosa, Picea orientalis and Pinus sylvestris) in Turkey. For. Ecol. Manag. 2015, 335, 71–86. [Google Scholar] [CrossRef]
- Dornbush, M.E.; Isenhart, T.M.; Raich, J.W. Quantifying fine-root decomposition: An alternative to buried litterbags. Ecology 2002, 83, 2985–2990. [Google Scholar] [CrossRef]
- State Forestry Administration of China. Forest Resources Report in China (2009–2013); National Forestry and Grassland Administration: Beijing, China, 2014. [Google Scholar]
- He, Z.; Yu, Z.; Huang, Z.; Davis, M.; Yang, Y. Litter decomposition, residue chemistry and microbial community structure under two subtropical forest plantations: A reciprocal litter transplant study. Appl. Soil Ecol. 2016, 101, 84–92. [Google Scholar] [CrossRef]
- Zhan, X.; Yu, G.; He, N.; Jia, B.; Zhou, M.; Wang, C.; Zhang, J.; Zhao, G.; Wang, S.; Liu, Y.; et al. Inorganic nitrogen wet deposition: Evidence from the North-South Transect of Eastern China. Environ. Pollut. 2015, 204, 1–8. [Google Scholar] [CrossRef]
- Yang, Y.; Xu, M.; Luo, Q.; Wang, J.; Li, H. De novo transcriptome analysis of Liriodendron chinense petals and leaves by Illumina sequencing. Gene 2014, 534, 155–162. [Google Scholar] [CrossRef]
- Van Soest, P.J.; Wine, R.H. Determination of Lignin and Cellulose in Acid-Detergent Fiber with Permanganate. J. Assoc. Off. Anal. Chem. 2020, 51, 780–785. [Google Scholar] [CrossRef]
- De Marco, A.; Spaccini, R.; Vittozzi, P.; Esposito, F.; Berg, B.; Virzo De Santo, A. Decomposition of black locust and black pine leaf litter in two coeval forest stands on Mount Vesuvius and dynamics of organic components assessed through proximate analysis and NMR spectroscopy. Soil Biol. Biochem. 2012, 51, 1–15. [Google Scholar] [CrossRef]
- Liu, X.; Cheng, X.; Wang, N.; Meng, M.; Jia, Z.; Wang, J.; Ma, S.; Tang, Y.; Li, C.; Zhai, L.; et al. Effects of Vegetation Type on Soil Shear Strength in Fengyang Mountain Nature Reserve, China. Forests 2021, 12, 490. [Google Scholar] [CrossRef]
- Rao, M.; Scelza, R.; Gianfreda, L. Soil enzymes. In Enzymes in Agricultural Sciences; OMICS Group: Foster City, CA, USA, 2014; pp. 1–18. [Google Scholar]
- Zhao, L.; Jiang, Y. Discussion on measurements of soil phosphates. Chin. J. Soil Sci. 1986, 17, 138–141. [Google Scholar]
- Ni, Y.; Yang, T.; Zhang, K.; Shen, C.; Chu, H. Fungal Communities Along a Small-Scale Elevational Gradient in an Alpine Tundra Are Determined by Soil Carbon Nitrogen Ratios. Front. Microbiol. 2018, 9, 1815. [Google Scholar] [CrossRef] [PubMed]
- Sun, R.; Li, W.; Dong, W.; Tian, Y.; Hu, C.; Liu, B. Tillage Changes Vertical Distribution of Soil Bacterial and Fungal Communities. Front. Microbiol. 2018, 9, 699. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, Q.; Zhan, L.; Xu, X.; Bi, R.; Xiong, Z. Biochar addition stabilized soil carbon sequestration by reducing temperature sensitivity of mineralization and altering the microbial community in a greenhouse vegetable field. J. Environ. Manag. 2022, 313, 114972. [Google Scholar] [CrossRef]
- Manasa, M.R.K.; Katukuri, N.R.; Darveekaran Nair, S.S.; Haojie, Y.; Yang, Z.; Guo, R.B. Role of biochar and organic substrates in enhancing the functional characteristics and microbial community in a saline soil. J. Environ. Manag. 2020, 269, 110737. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, N.H.; Song, Z.; Bates, S.T.; Branco, S.; Tedersoo, L.; Menke, J.; Schilling, J.S.; Kennedy, P.G. FUNGuild: An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 2016, 20, 241–248. [Google Scholar] [CrossRef]
- Zhong, Y.; Yan, W.; Wang, R.; Shangguan, Z. Differential responses of litter decomposition to nutrient addition and soil water availability with long-term vegetation recovery. Biol. Fertil. Soils 2017, 53, 939–949. [Google Scholar] [CrossRef]
- Ågren, G.I.; Hyvönen, R.; Berglund, S.L.; Hobbie, S.E. Estimating the critical N:C from litter decomposition data and its relation to soil organic matter stoichiometry. Soil Biol. Biochem. 2013, 67, 312–318. [Google Scholar] [CrossRef]
- Melillo, J.M.; Aber, J.D.; Muratore, J.F. Nitrogen and Lignin Control of Hardwood Leaf Litter Decomposition Dynamics. Ecology 1982, 63, 621–626. [Google Scholar] [CrossRef]
- Zhang, C.; Li, S.; Zhang, L.; Xin, X.; Liu, X. Effects of Species and Low Dose Nitrogen Addition on Litter Decomposition of Three Dominant Grasses in Hulun Buir Meadow Steppe. J. Resour. Ecol. 2013, 4, 20–26. [Google Scholar] [CrossRef]
- Graaff, M.-A.d.; Six, J.; Jastrow, J.D.; Schadt, C.W.; Wullschleger, S.D. Variation in root architecture among switchgrass cultivars impacts root decomposition rates. Soil Biol. Biochem. 2013, 58, 198–206. [Google Scholar] [CrossRef]
- Boer, W.; Folman, L.B.; Summerbell, R.C.; Boddy, L. Living in a fungal world: Impact of fungi on soil bacterial niche development. FEMS Microbiol. Rev. 2005, 29, 795–811. [Google Scholar] [CrossRef] [PubMed]
- Kotowska, M.M.; Leuschner, C.; Triadiati, T.; Hertel, D. Conversion of tropical lowland forest reduces nutrient return through litterfall, and alters nutrient use efficiency and seasonality of net primary production. Oecologia 2016, 180, 601–618. [Google Scholar] [CrossRef]
- Krumins, J.A.; Long, Z.T.; Steiner, C.F.; Morin, P.J. Indirect effects of food web diversity and productivity on bacterial community function and composition. Funct. Ecol. 2006, 20, 514–521. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, W. The decomposition of fine and coarse roots: Their global patterns and controlling factors. Sci. Rep. 2015, 5, 9940. [Google Scholar] [CrossRef]
- Yan, B.; Sun, L.; Li, J.; Liang, C.; Wei, F.; Xue, S.; Wang, G. Change in composition and potential functional genes of soil bacterial and fungal communities with secondary succession in Quercus liaotungensis forests of the Loess Plateau, western China. Geoderma 2020, 364, 114199. [Google Scholar] [CrossRef]
- Deacon, L.J.; Janie Pryce-Miller, E.; Frankland, J.C.; Bainbridge, B.W.; Moore, P.D.; Robinson, C.H. Diversity and function of decomposer fungi from a grassland soil. Soil Biol. Biochem. 2006, 38, 7–20. [Google Scholar] [CrossRef]
- Osono, T.; Fukasawa, Y.; Takeda, H. Roles of diverse fungi in larch needle-litter decomposition. Mycologia 2003, 95, 820–826. [Google Scholar] [CrossRef]
- Worrall, J.J.; Anagnost, S.E.; Zabel, R.A. Comparison of wood decay among diverse lignicolous fungi. Mycologia 1997, 89, 199–219. [Google Scholar] [CrossRef]
- Yergeau, E.; Bokhorst, S.; Kang, S.; Zhou, J.; Greer, C.W.; Aerts, R.; Kowalchuk, G.A. Shifts in soil microorganisms in response to warming are consistent across a range of Antarctic environments. ISME J. 2012, 6, 692–702. [Google Scholar] [CrossRef] [PubMed]
- Violita, V.; Triadiati, T.; Anas, I.; Miftahudin, M. Fine Root Production and Decomposition in Lowland Rainforest and Oil Palm Plantations in Sumatra, Indonesia. HAYATI J. Biosci. 2016, 23, 7–12. [Google Scholar] [CrossRef]
- Cobb, R.C.; Rizzo, D.M. Litter Chemistry, Community Shift, and Non-additive Effects Drive Litter Decomposition Changes Following Invasion by a Generalist Pathogen. Ecosystems 2016, 19, 1478–1490. [Google Scholar] [CrossRef]
- Zeng, Q.; Liu, Y.; An, S. Impact of litter quantity on the soil bacteria community during the decomposition of Quercus wutaishanica litter. PeerJ 2017, 5, e3777. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, C.; Wang, M. Linkages of C: N: P stoichiometry between soil and leaf and their response to climatic factors along altitudinal gradients. J. Soils Sediments 2019, 19, 1820–1829. [Google Scholar] [CrossRef]
- Wang, W.; Zhang, X.; Tao, N.; Ao, D.; Zeng, W.; Qian, Y.; Zeng, H. Effects of litter types, microsite and root diameters on litter decomposition in Pinus sylvestris plantations of northern China. Plant Soil 2014, 374, 677–688. [Google Scholar] [CrossRef]
- Bryanin, S.; Abramova, E.; Makoto, K. Fire-derived charcoal might promote fine root decomposition in boreal forests. Soil Biol. Biochem. 2018, 116, 1–3. [Google Scholar] [CrossRef]
- Zhang, K.; Cheng, X.; Dang, H.; Ye, C.; Zhang, Y.; Zhang, Q. Linking litter production, quality and decomposition to vegetation succession following agricultural abandonment. Soil Biol. Biochem. 2013, 57, 803–813. [Google Scholar] [CrossRef]
- Yue, K.; Yang, W.; Peng, C.; Peng, Y.; Zhang, C.; Huang, C.; Tan, Y.; Wu, F. Foliar litter decomposition in an alpine forest meta-ecosystem on the eastern Tibetan Plateau. Sci. Total Environ. 2016, 566–567, 279–287. [Google Scholar] [CrossRef]
- Hossain, M.; Siddique, M.R.H.; Abdullah, S.M.R.; Saha, S.; Ghosh, D.C.; Rahman, M.S.; Limon, S.H. Nutrient Dynamics Associated with Leaching and Microbial Decomposition of Four Abundant Mangrove Species Leaf Litter of the Sundarbans, Bangladesh. Wetlands 2014, 34, 439–448. [Google Scholar] [CrossRef]
- Yin, H.; Wheeler, E.; Phillips, R.P. Root-induced changes in nutrient cycling in forests depend on exudation rates. Soil Biol. Biochem. 2014, 78, 213–221. [Google Scholar] [CrossRef]
- Cheeke, T.E.; Phillips, R.P.; Brzostek, E.R.; Rosling, A.; Bever, J.D.; Fransson, P. Dominant mycorrhizal association of trees alters carbon and nutrient cycling by selecting for microbial groups with distinct enzyme function. New Phytol. 2017, 214, 432–442. [Google Scholar] [CrossRef]
- Caldwell, B.A. Enzyme activities as a component of soil biodiversity: A review. Pedobiologia 2005, 49, 637–644. [Google Scholar] [CrossRef]
- Bae, K.; Fahey, T.J.; Yanai, R.D.; Fisk, M. Soil Nitrogen Availability Affects Belowground Carbon Allocation and Soil Respiration in Northern Hardwood Forests of New Hampshire. Ecosystems 2015, 18, 1179–1191. [Google Scholar] [CrossRef]
- Brzostek, E.R.; Dragoni, D.; Brown, Z.A.; Phillips, R.P. Mycorrhizal type determines the magnitude and direction of root-induced changes in decomposition in a temperate forest. New Phytol. 2015, 206, 1274–1282. [Google Scholar] [CrossRef] [PubMed]
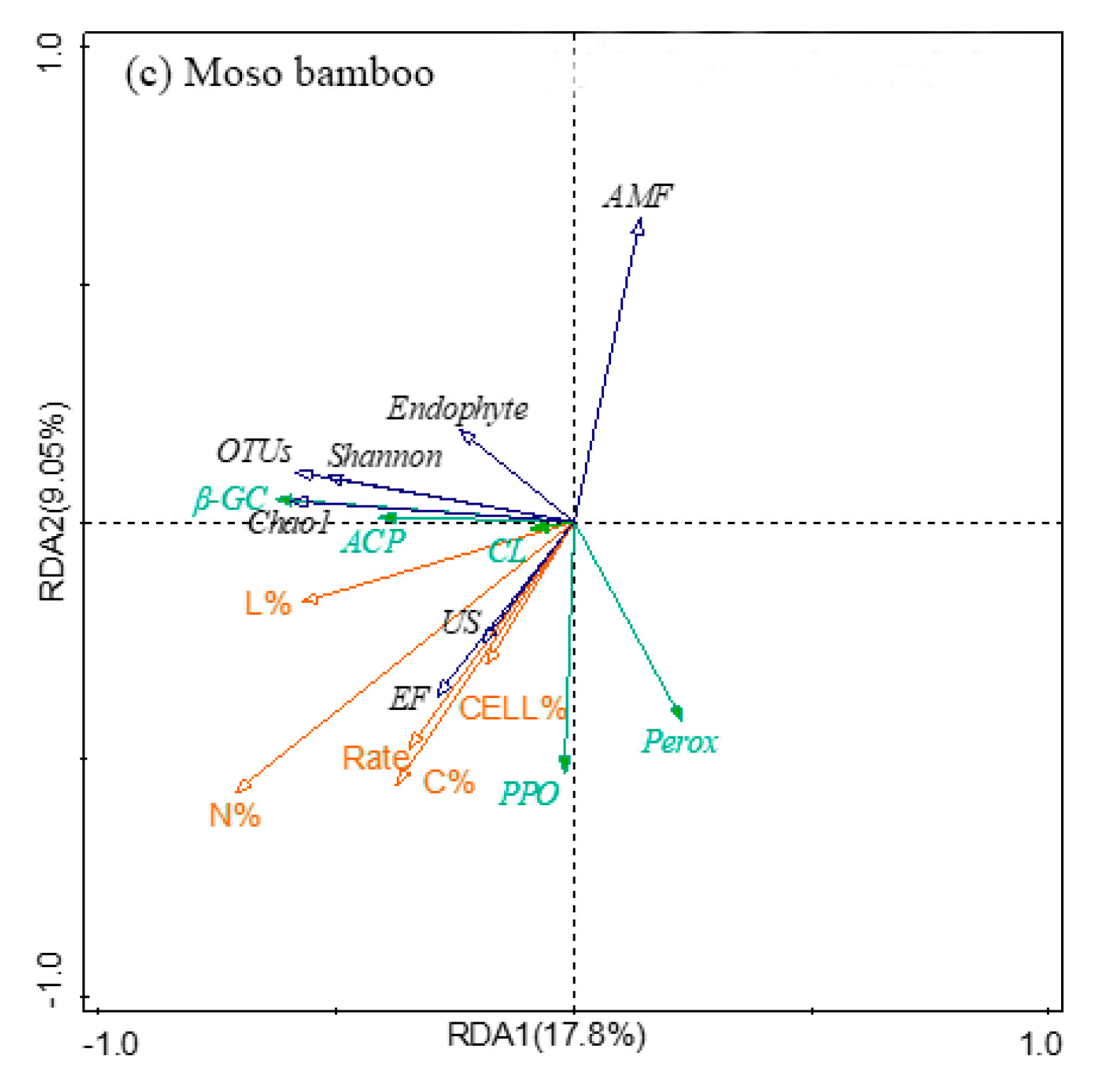
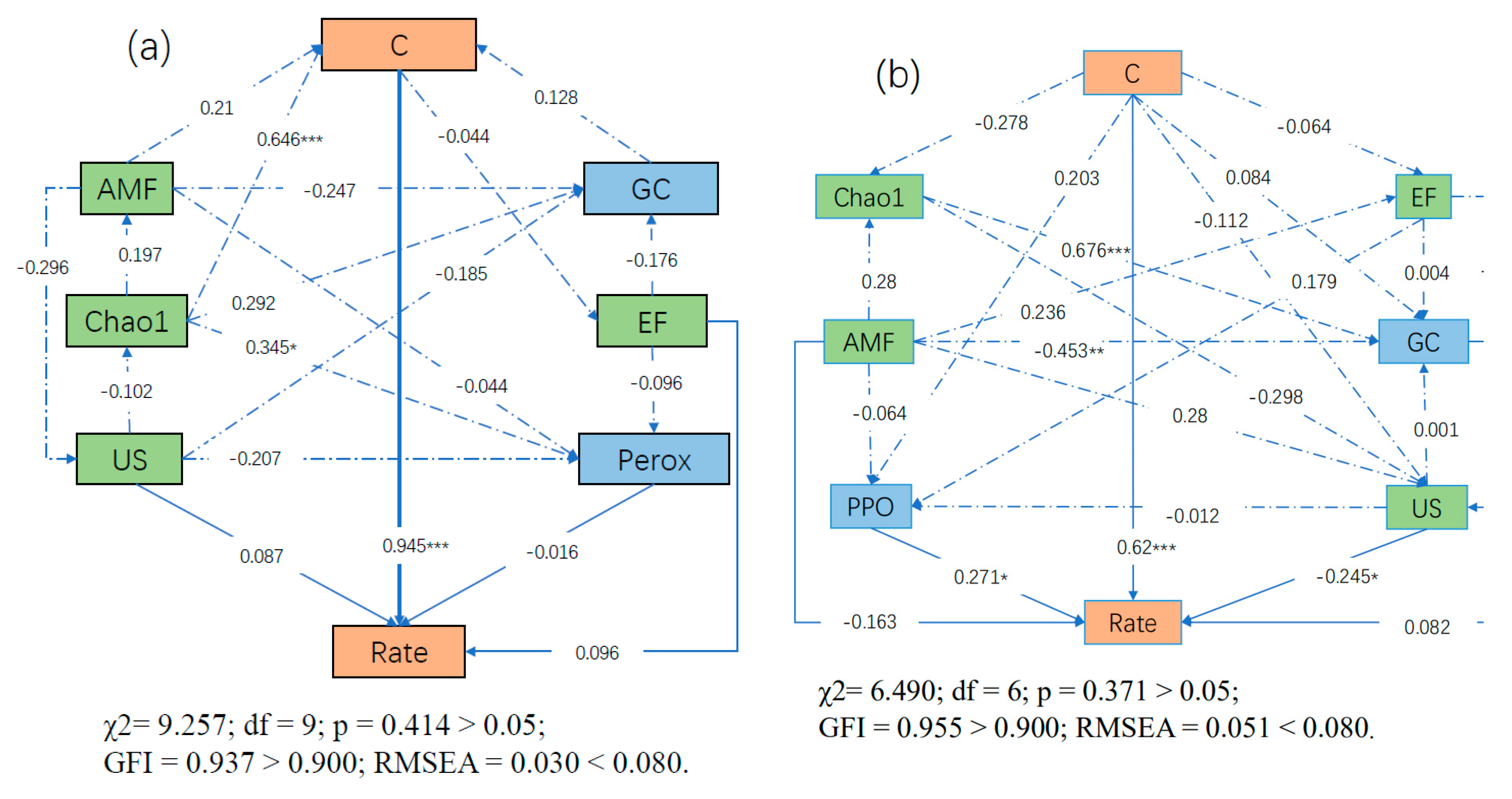
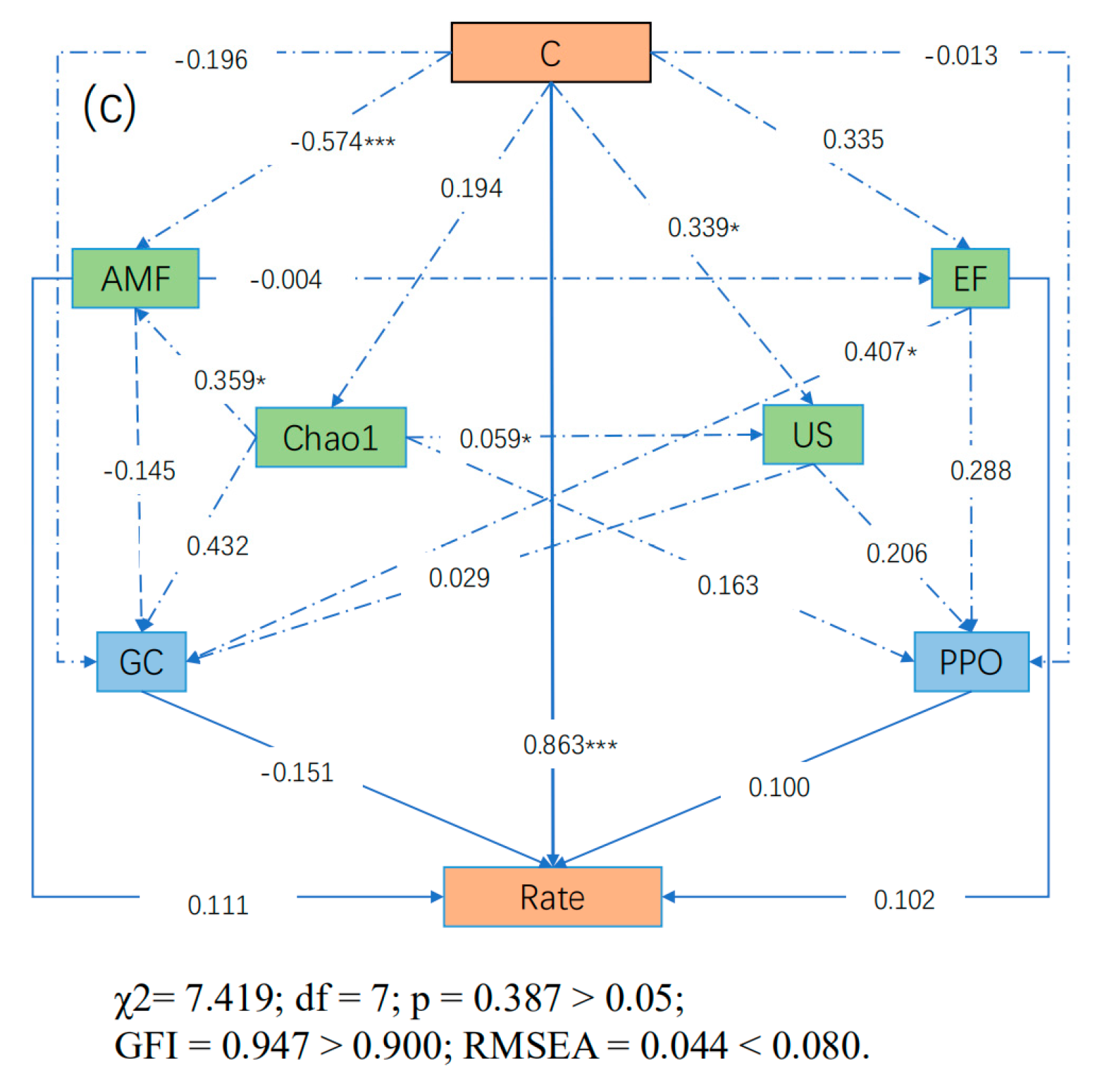
| Differences | R-Value | p-Value | Significance | |
|---|---|---|---|---|
| Decomposition time | Chinese whitewood | 0.2118 | 0.001 | ** |
| Chinese fir | 0.1265 | 0.012 | ** | |
| Moso bamboo | 0.3687 | 0.001 | ** | |
| Forest type | 0 day | 0.071 | 0.063 | NS |
| 120 days | 0.0357 | 0.201 | NS | |
| 240 days | 0.1744 | 0.018 | * | |
| 360 days | 0.0617 | 0.125 | NS | |
| C% | AMF | Chao1 | EF | US | PPO | Perox | GC | ||
|---|---|---|---|---|---|---|---|---|---|
| Rate (Chinese Whitewood) | Direct Effects | 0.945 | 0.000 | 0.000 | 0.096 | 0.087 | - | −0.016 | 0.000 |
| Indirect Effects | −0.003 | 0.169 | 0.671 | −0.020 | −0.087 | - | 0.000 | 0.120 | |
| Total Effects | 0.942 | 0.169 | 0.671 | 0.076 | −0.001 | - | −0.016 | 0.120 | |
| Rate (Chinese fir) | Direct Effects | 0.620 | −0.163 | 0.000 | 0.000 | −0.245 | 0.271 | - | 0.082 |
| Indirect Effects | 0.048 | −0.068 | 0.130 | 0.084 | 0.000 | −0.003 | - | 0.000 | |
| Total Effects | 0.668 | −0.231 | 0.130 | 0.084 | −0.249 | 0.267 | - | 0.082 | |
| Rate (Moso bamboo) | Direct Effects | 0.863 | 0.111 | 0.000 | 0.102 | 0.000 | 0.100 | - | −0.151 |
| Indirect Effects | −0.019 | 0.022 | 0.280 | −0.033 | 0.016 | 0.000 | - | 0.000 | |
| Total Effects | 0.844 | 0.132 | 0.280 | 0.069 | 0.016 | 0.100 | - | −0.151 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, X.; Jiang, S.; El-Naggar, A.; Tang, Y.; Liu, X.; Zhang, J. Dynamics of Fine Root Decomposition in Different Vegetation Types: Investigating the Impact of Soil Fungal Communities and Enzyme Activities. Forests 2023, 14, 1321. https://doi.org/10.3390/f14071321
Cheng X, Jiang S, El-Naggar A, Tang Y, Liu X, Zhang J. Dynamics of Fine Root Decomposition in Different Vegetation Types: Investigating the Impact of Soil Fungal Communities and Enzyme Activities. Forests. 2023; 14(7):1321. https://doi.org/10.3390/f14071321
Chicago/Turabian StyleCheng, Xuefei, Siyuan Jiang, Ali El-Naggar, Yingzhou Tang, Xin Liu, and Jinchi Zhang. 2023. "Dynamics of Fine Root Decomposition in Different Vegetation Types: Investigating the Impact of Soil Fungal Communities and Enzyme Activities" Forests 14, no. 7: 1321. https://doi.org/10.3390/f14071321
APA StyleCheng, X., Jiang, S., El-Naggar, A., Tang, Y., Liu, X., & Zhang, J. (2023). Dynamics of Fine Root Decomposition in Different Vegetation Types: Investigating the Impact of Soil Fungal Communities and Enzyme Activities. Forests, 14(7), 1321. https://doi.org/10.3390/f14071321








