Potensaphelenchus stammeri (Körner, 1954) Gu, Liu, Abolafia & Pedram, 2021 (Nematoda: Aphelenchoididae) from Pinus pinea Linnaeus, 1753 in Portugal
Abstract
1. Introduction
2. Materials and Methods
2.1. Nematodes Extraction and Culture Establishment
2.2. Morphological and Morphometric Characterisation
2.3. DNA Extraction and Amplification of the D2-D3 LSU rDNA Region
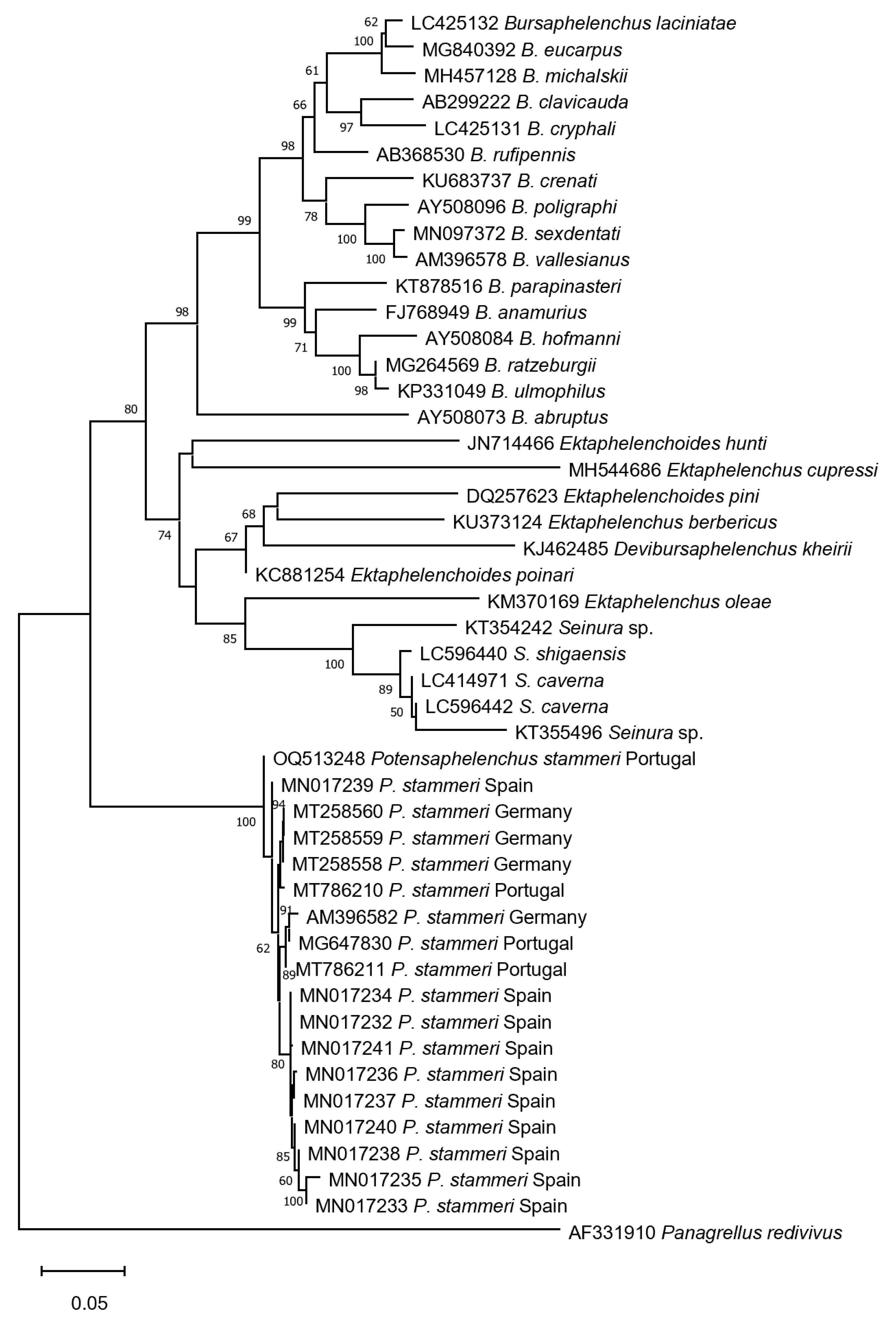
2.4. Sequencing and Phylogenetic Analysis
3. Results
3.1. Morphological and Morphometric Characterisation
3.2. Molecular Identification
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Körner, H. Die Nematoden fauna des vergehenden Holzes und ihre Beziehungen zu den Insekten. Zool. Jahrbücher Abt. Für Syst. Ökologie Und Geogr. Der Tiere 1954, 82, 245–353. [Google Scholar]
- Gu, J.; Liu, L.; Abolafia, J.; Pedram, M. A revision of the taxonomy of Aphelenchoides stammeri Körner, 1954 (Rhabditida: Aphelenchoididae) and proposal for a new genus. Nematology 2021, 23, 215–228. [Google Scholar] [CrossRef]
- Fuchs, G. Neue parasitich und halbparasitische Nematoden bei Borkenkäfern und einige andere Nematoden. Zool. Jb. 1937, 70, 291–442. [Google Scholar]
- Goodey, J.B. The classification of the Aphelenchoididae Fuch, 1937. Nematologica 1960, 5, 111–126. [Google Scholar] [CrossRef]
- Huang, R.E.; Ye, J.R. Seinura lii n. sp. and S. wuae n. sp. (Nematoda: Seinuridae) from pine wood in China. Nematology 2006, 8, 749–759. [Google Scholar] [CrossRef]
- Kanzaki, N.; Ekino, T.; Masuya, H. Seinura caverna n. sp. (Tylenchomorpha: Aphelenchoididae), na androdioecious species isolated from bat guano in a calcareous cave. Nematology 2019, 21, 207–225. [Google Scholar] [CrossRef]
- Braasch, H. Aphelenchoides stammeri Körner, 1954–ein in Deutschland weit verbreiteter Holznematode. Nachrichtenbl. Deut. Pflanzenschutzd. 1998, 50, 317–319. [Google Scholar]
- Urek, G.; Širca, S.; Geric, B. Morphometrical and molecular characterization of Bursaphelenchus species from Slovenia. Helminthologia 2007, 44, 37–42. [Google Scholar] [CrossRef]
- Dayi, M.; Uludamar, E.B.K.; Akbulut, S.; Elekcioğlu, I.H. First record of Aphelenchoides stammeri (Nematoda: Aphelenchoididae) from Turkey. J. Nematol. 2019, 51, e2019-70. [Google Scholar] [CrossRef] [PubMed]
- Whitehead, A.G.; Hemming, J.R. A comparison of some quantitative methods of extracting small vermiform nematodes from soil. Ann. Appl. Biol. 1965, 55, 25–38. [Google Scholar] [CrossRef]
- Fonseca, L.; Santos, M.C.V.; Santos, M.S.; Curtis, R.H.C.; Abrantes, I.M.O. Morpho-biometrical characterisation of Portuguese Bursaphelenchus xylophilus isolates with mucronate, digitate or round tailed females. Phytopathol. Mediterr. 2008, 47, 223–233. [Google Scholar]
- De Ley, P.; Felix, M.; Frisse, L.; Nadler, S.; Sternberg, P.; Thomas, W.K. Molecular and morphological characterisation of two reproductively isolated species with mirror-image anatomy (Nematoda: Cephalobidae). Nematology 1999, 1, 591–612. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.H.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA 11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 7, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The Neighbor-Joining method–A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406. [Google Scholar] [PubMed]
- Aliramaji, F.; Fouladvand, Z.M.; Pourjam, E.; Mortazavi, P.; Afshar, F.J.; Kanzaki, N.; Giblin-Davis, R.; Pedram, M. A new species of Basilaphelenchus Pedram, Kanzaki, Giblin-Davis & Pourjam, 2018 (Aphelenchoidea: Tylaphelenchinae), from natural forests of Golestan province, Iran. Nematology 2020, 22, 361–371. [Google Scholar]

| Character | Females | |||||
|---|---|---|---|---|---|---|
| Braasch, 1998 (n = 6) | Huang and Ye, 2006 (n = 14) | Urek et al., 2007 (n = 5) | Dayi, 2019 (n = 10) | Gu et al., 2021 (n = 15) | This Study (PsPt1) (n = 15) | |
| Linear (µm) | ||||||
| Body length (L) | 833.0 (730.0–910.0) | 885.0 ± 67.0 (763.0–1038.0) | 777.7 ± 98.2 (673.9–905.3) | 918.7 ± 46.2 (800.0–972.8) | 973.0 ± 45.4 (912.0–1075.0) | 768.8 ± 58.0 (690.0–856.4) |
| Greatest body width (GBW) | ----- | ----- | ----- | ----- | 28.9 ± 1.9 (25.8–32.4) | 27.2 ± 1.6 (24.6–30.0) |
| Stylet length | 16.0 (15.0–18.0) | 18.0 ± 0.8 (16.8–19.2) | 15.2 ± 1.0 (14.1–16.5) | 17.4 ± 0.4 (16.0–17.6) | 17.1 ± 1.4 (14.0–19.3) | 14.6 ± 0.3 (14.0–15.3) |
| Median bulb length | ----- | ----- | ----- | ----- | 20.8 ± 1.2 (18.8–23.0) | 19.7 ± 1.3 (18.1–22.6) |
| Median bulb diameter | ----- | ----- | ----- | ----- | 14.8 ± 1.1 (13.5–16.7) | 15.4 ± 0.8 (14.4–16.8) |
| Excretory pore to anterior end | ----- | ----- | ----- | ----- | 90.0 ± 4.5 (82.0–96.0) | 95.1 ± 4.5 (89.5–103.8) |
| Anterior end to end of median bulb (AEMB) | ----- | ----- | ----- | 80.8 ± 2.8 (76.8–84.8) | ----- | 74.6 ± 4.0 (67.8–83.1) |
| Tail length (TL) | ----- | 57.0 ± 5.9 (52.0–69.0) | 49.4 ± 5.3 (44.9–59.8) | 63.8 ± 3.6 (56.0–68.8) | 61.0 ± 4.2 (52.0–69.0) | 58.0 ± 4.8 (49.9–65.0) |
| Body width at anus (BWA) | ----- | ----- | ----- | ----- | 16.0 ± 0.5 (15.1–16.8) | 14.2 ± 0.8 (12.9–15.9) |
| Vulva to anus | ----- | ----- | ----- | 230.0 ± 11.1 (214.4–240.0) | ----- | 182.6 ± 15.0 (154.0–200.4) |
| Ratio | ||||||
| a = L/GBW | 32.0 (19.0–34.0) | 40.0 ± 1.8 (36.4–42.2) | 38.3 ± 2.7 (34.9–42.3) | 31.9 ± 1.4 (30.4–33.0) | 33.6 ± 1.6 (30.2–36.4) | 28.2 ± 1.6 (26.1–31.5) |
| b1 = L/AEMB | ----- | ----- | ----- | ----- | ----- | 10.3 ± 0.9 (8.7–12.4) |
| c = L/TL | 16.0 (13.0–17.0) | 15.5 ± 2.3 (12.6–18.5) | 15.7 ± 1.1 (14.5–17.8) | 14.4 ± 1.2 (13.4–16.7) | 16.2 ± 1.3 (14.1–18.9) | 13.3 ± 1.4 (11.7–16.6) |
| c’ = TL/BWA | ----- | 4.3 ± 0.4 (3.8–5.1) | 4.1 ± 0.4 (3.7–4.6) | 4.0 ± 0.3 (3.3–4.5) | 3.8 ± 0.3 (3.3–4.3) | 4.1 ± 0.3 (3.5–4.5) |
| Percentage | ||||||
| V = Distance anterior end to vulva × 100/L | 69.0 (66.0–71.0) | 68.7 ± 0.9 (66.7–69.8) | 68.1 ± 0.6 (66.9–68.5) | 67.8 ± 1.6 (67.0–72.0) | 68.1 ± 0.7 (66.9–69.0) | 68.8 ± 1.4 (66.9–71.7) |
| Character | Males | |||||
|---|---|---|---|---|---|---|
| Braasch, 1998 (n = 6) | Huang and Ye, 2006 (n = 8) | Urek et al., 2007 (n = 5) | Dayi, 2019 (n = 10) | Gu et al., 2021 (n = 15) | This Study (PsPt1) (n = 15) | |
| Linear (µm) | ||||||
| Body length (L) | 853.0 (810.0–920.0) | 857.0 ± 26.1 (831.0–908.0) | 729.4 ± 53.8 (649.5–790.3) | 851.8 ± 61.8 (776.0–976.0) | 884.0 ± 44.8 (803.0–983.0) | 739.2 ± 34.6 (680.1–788.6) |
| Greatest body width (GBW) | ----- | ----- | ----- | ---- | 25.1 ± 1.6 (22.9–27.6) | 25.0 ± 1.8 (22.3–29.1) |
| Stylet length | 16.0 (15.0–18.0) | 17.1 ± 0.6 (16.4–18.0) | 14.7 ± 1.4 (13.2–16.6) | 17.1 ± 0.6 (16.0–17.6) | 16.6 ± 1.6 (14.1–18.8) | 14.7 ± 0.4 (14.0–15.3) |
| Median bulb length | ----- | ----- | ----- | ----- | 19.7 ± 0.8 (18.3–21.3) | 19.4 ± 1.2 (17.6–21.6) |
| Median bulb diameter | ----- | ----- | ----- | ----- | 13.5 ± 0.7 (12.5–14.9) | 14.6 ± 1.2 (12.6–17.2) |
| Excretory pore to anterior end | ----- | ----- | ----- | ----- | 88.0 ± 5.6 (81.0–93.0) | 100.5 ± 2.8 (96.3–103.6) |
| Anterior end to end of median bulb (AEMB) | ----- | ----- | ----- | 77.6 ± 2.5 (72.0–81.6) | ----- | 78.4 ± 4.7 (72.2–89.8) |
| Tail length (TL) | ----- | 43.0 ± 3.4 (42.0–50.0) | 41.0 ± 2.4 (39.0–43.6) | 40.0 ± 2.0 (38.4–43.2) | 47.0 ± 8.4 (43.0–55.0) | 47.1 ± 1.6 (44.7–50.3) |
| Body width at anus (BWA) | ----- | ----- | ----- | ----- | 16.9 ± 0.8 (15.5–18.7) | 16.3 ± 1.2 (13.8–18.5) |
| Spicule length (curved median line) | 19.0 (18.0–20.0) | 20.7 ± 0.5 (20.0–21.5) | 20.2 ± 1.4 (18.1–21.6) | 20.3 ± 3.2 (12.8–24.0) | 17.3 ± 1.6 (15.3–21.0) | 18.5 ± 0.9 (16.4–19.7) |
| Ratio | ||||||
| a = L/GBW | 31.0 (27.0–36.0) | 42.0 ± 1.4 (40.4–44.0) | 37.4 ± 3.5 (34.4–43.0) | 33.5 ± 5.3 (21.0–40.6) | 35.3 ± 1.7 (31.4–37.6) | 29.7 ± 2.1 (25.5–33.6) |
| b1 = L/DAEMB | ----- | ----- | ----- | ----- | ----- | 9.4 ± 0.4 (8.5–10.1) |
| c = L/TL | 18.0 (16.0–21.0) | 20.0 ± 1.5 (18.4–21.5) | 17.8 ± 1.4 (16.3–19.6) | 21.1 ± 1.1 (19.4–22.5) | 18.5 ± 0.8 (17.5–19.6) | 15.7 ± 0.7 (14.5–17.6) |
| c’ = TL/BWA | ----- | 3.2 ± 0.3 (2.8–3.6) | 3.2 ± 0.2 (3.1–3.5) | 2.4 ± 0.1 (2.2–2.7) | 2.9 ± 0.2 (2.6–3.3) | 2.9 ± 0.2 (2.4–3.4) |
| Species | Query Cover | E Value | Percentage Identity | Accession Number | Country |
|---|---|---|---|---|---|
| Potensaphelenchus stammeri | 100% | 0.0 | 99.60% | MN017239.1 | Spain |
| P. stammeri | 100% | 0.0 | 99.19% | MN017236.1 | Spain |
| P. stammeri | 96% | 0.0 | 99.72% | MT258558.1 | Germany |
| P. stammeri | 96% | 0.0 | 99.72% | AM396582.1 | Germany |
| P. stammeri | 95% | 0.0 | 99.72% | MG647830.1 | Portugal |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Silva, H.; Cardoso, J.M.S.; da Costa, R.M.F.; Abrantes, I.; Fonseca, L. Potensaphelenchus stammeri (Körner, 1954) Gu, Liu, Abolafia & Pedram, 2021 (Nematoda: Aphelenchoididae) from Pinus pinea Linnaeus, 1753 in Portugal. Forests 2023, 14, 962. https://doi.org/10.3390/f14050962
Silva H, Cardoso JMS, da Costa RMF, Abrantes I, Fonseca L. Potensaphelenchus stammeri (Körner, 1954) Gu, Liu, Abolafia & Pedram, 2021 (Nematoda: Aphelenchoididae) from Pinus pinea Linnaeus, 1753 in Portugal. Forests. 2023; 14(5):962. https://doi.org/10.3390/f14050962
Chicago/Turabian StyleSilva, Hugo, Joana M. S. Cardoso, Ricardo M. F. da Costa, Isabel Abrantes, and Luís Fonseca. 2023. "Potensaphelenchus stammeri (Körner, 1954) Gu, Liu, Abolafia & Pedram, 2021 (Nematoda: Aphelenchoididae) from Pinus pinea Linnaeus, 1753 in Portugal" Forests 14, no. 5: 962. https://doi.org/10.3390/f14050962
APA StyleSilva, H., Cardoso, J. M. S., da Costa, R. M. F., Abrantes, I., & Fonseca, L. (2023). Potensaphelenchus stammeri (Körner, 1954) Gu, Liu, Abolafia & Pedram, 2021 (Nematoda: Aphelenchoididae) from Pinus pinea Linnaeus, 1753 in Portugal. Forests, 14(5), 962. https://doi.org/10.3390/f14050962





