Classification of Individual Tree Species Using UAV LiDAR Based on Transformer
Abstract
1. Introduction
2. Materials and Methods
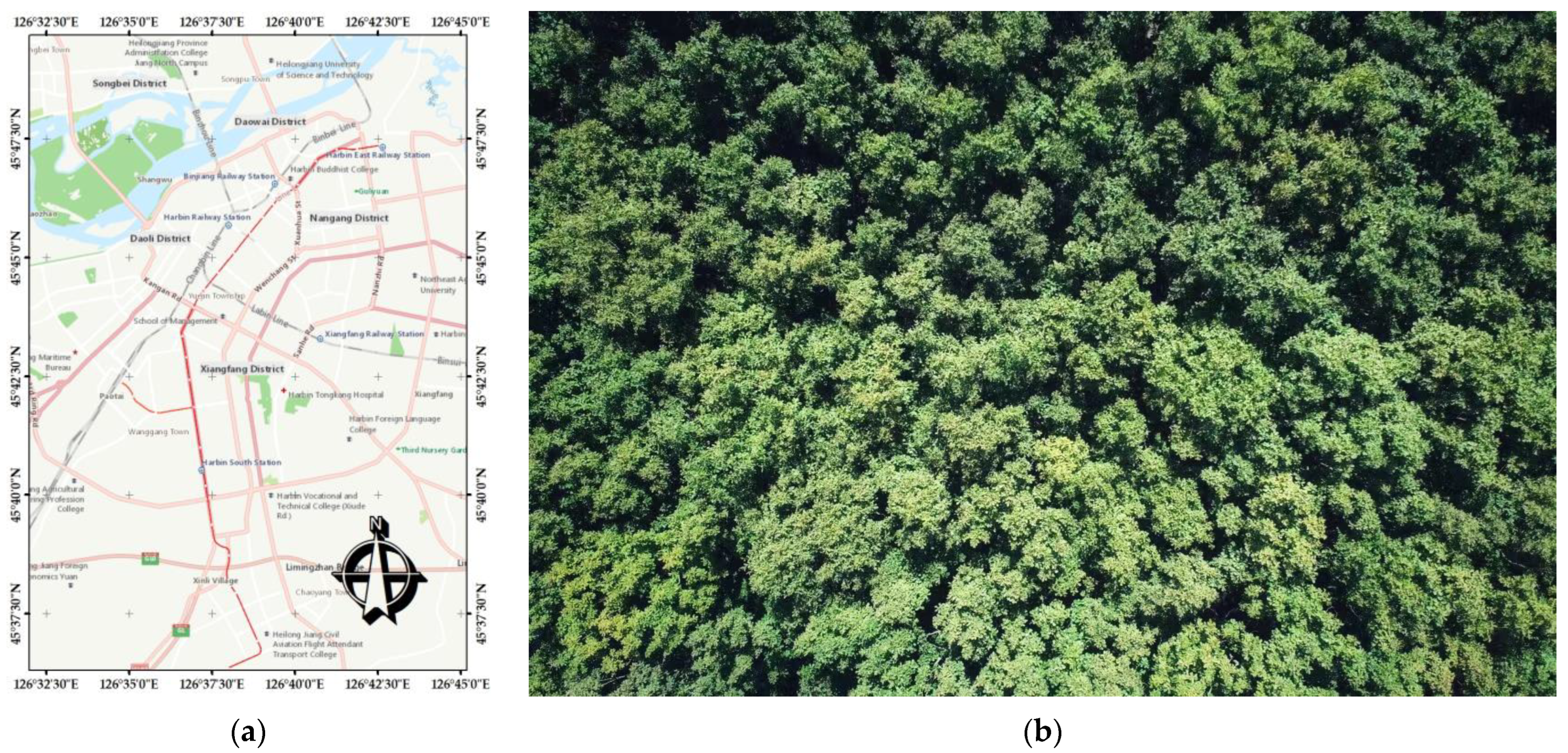
2.1. Study Area
2.2. Data Acquisition
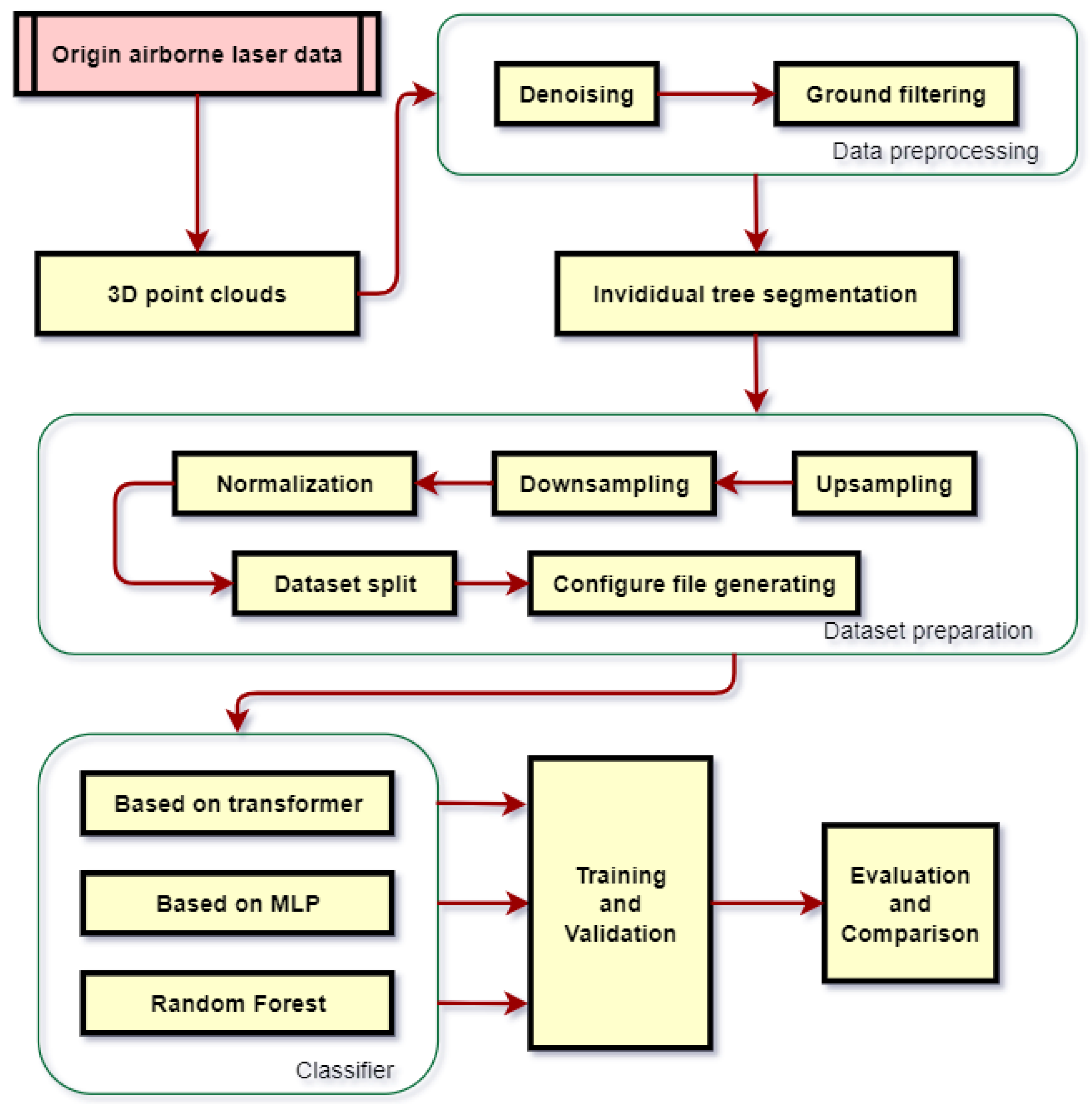
2.3. Methodology
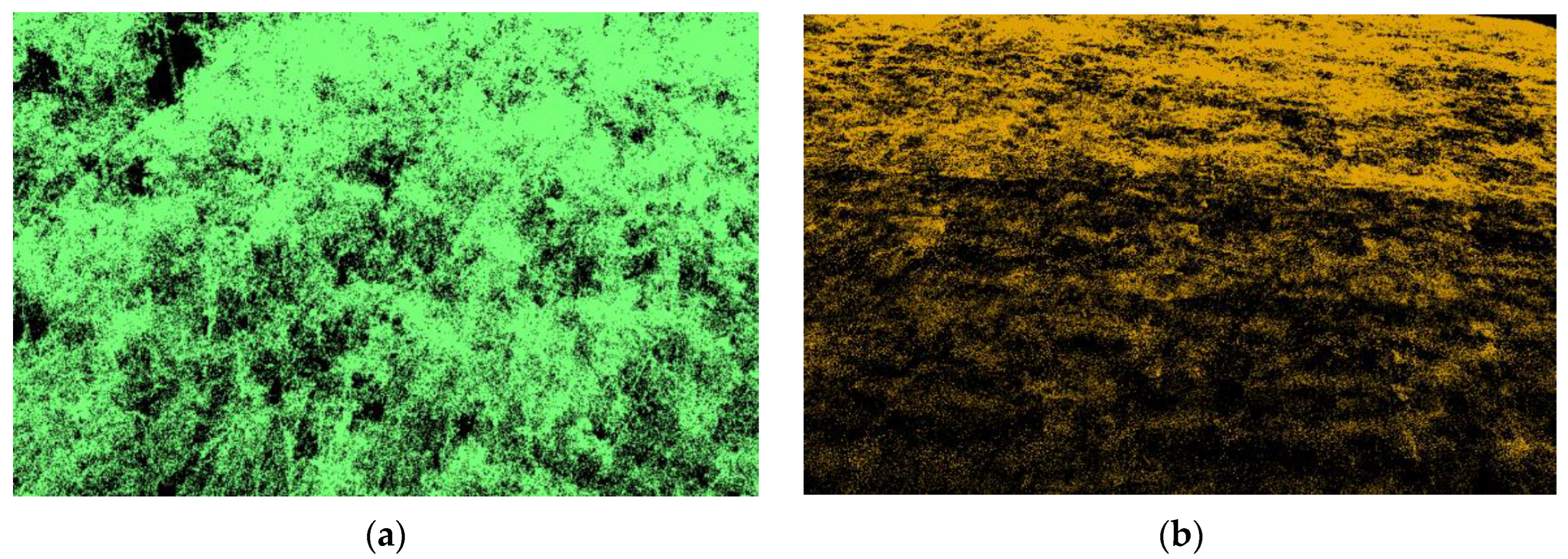
2.3.1. Preprocessing
- Denoising
- 2.
- Ground filtering
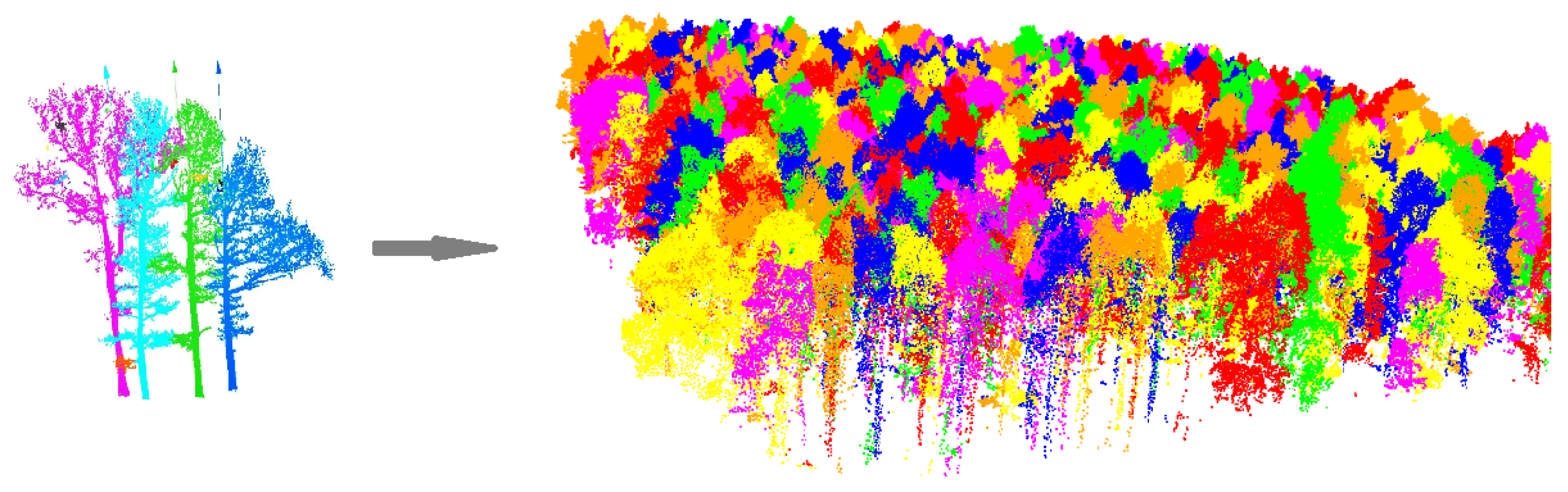
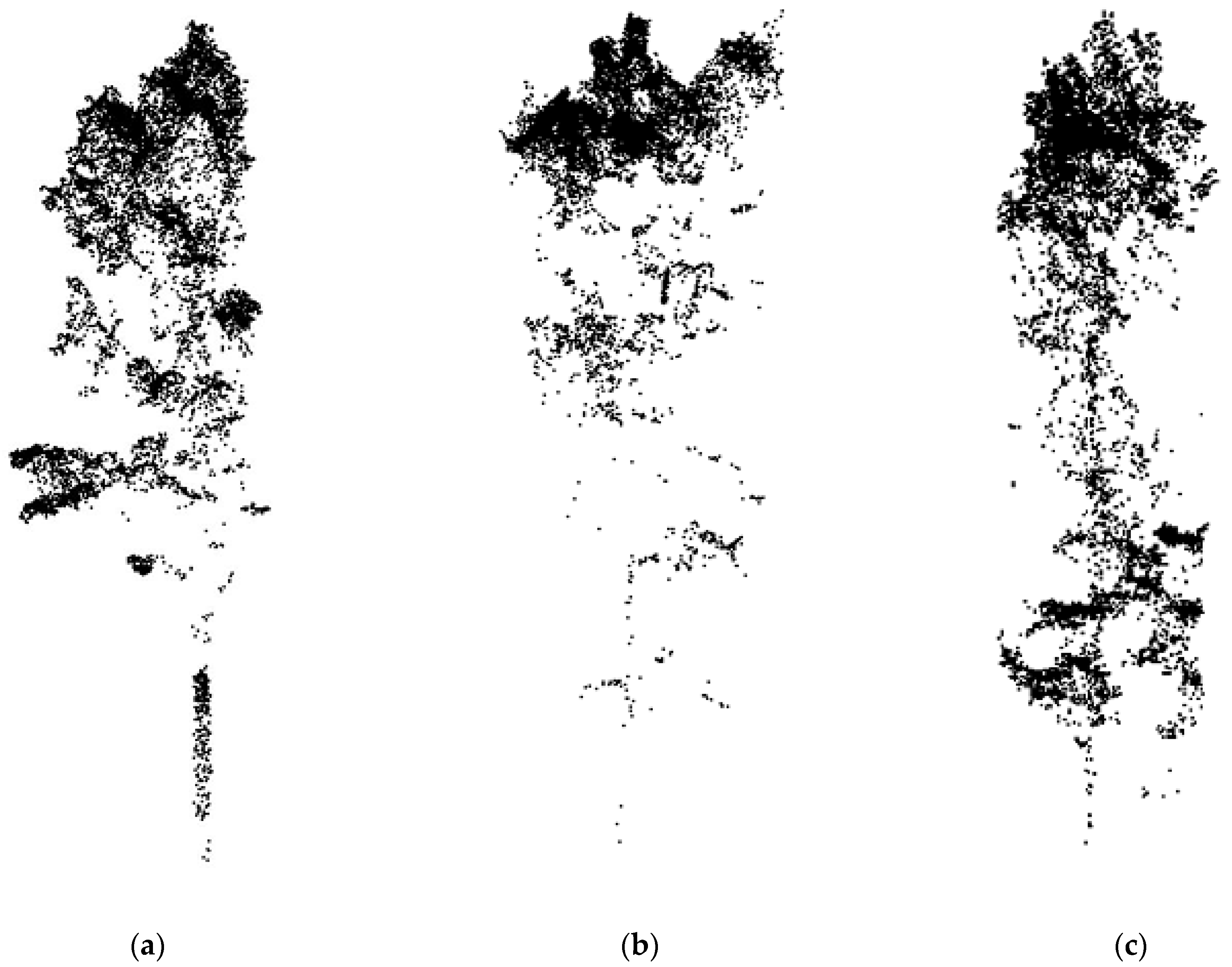
2.3.2. Individual Tree Segmentation
2.3.3. Data Set Creation
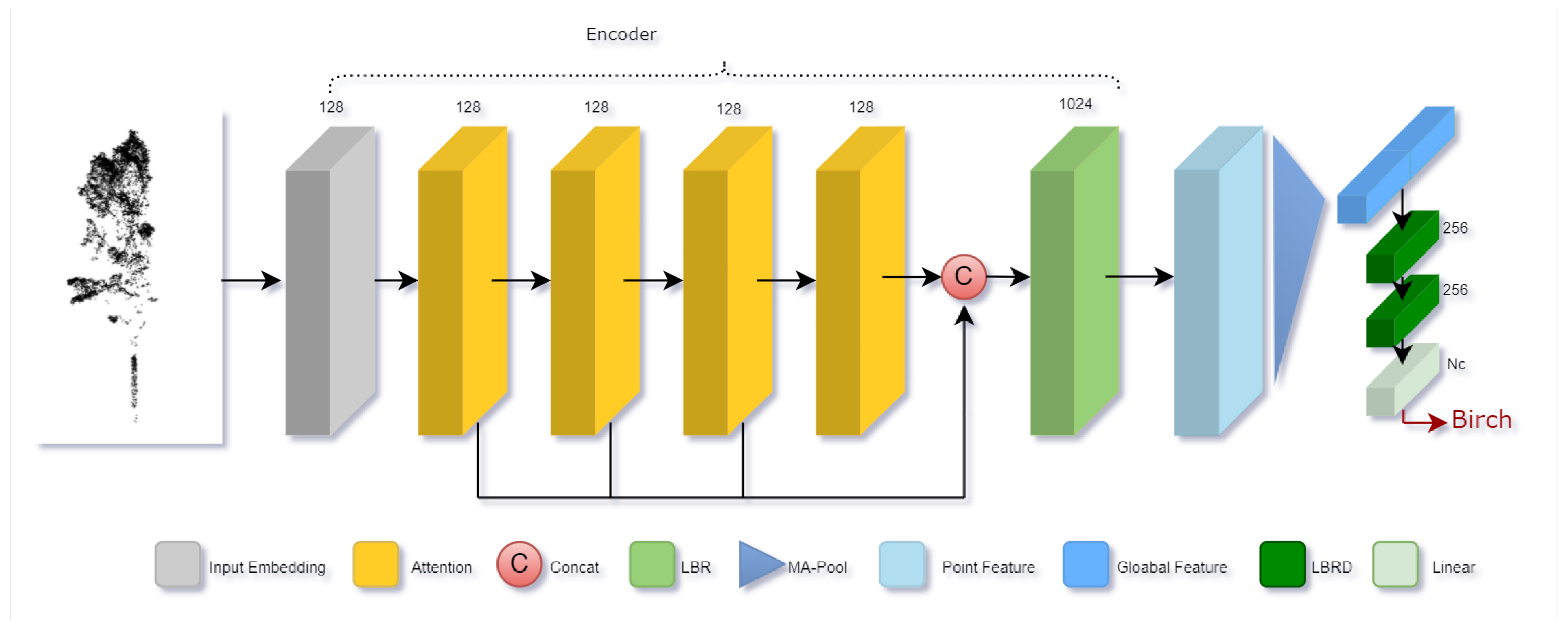
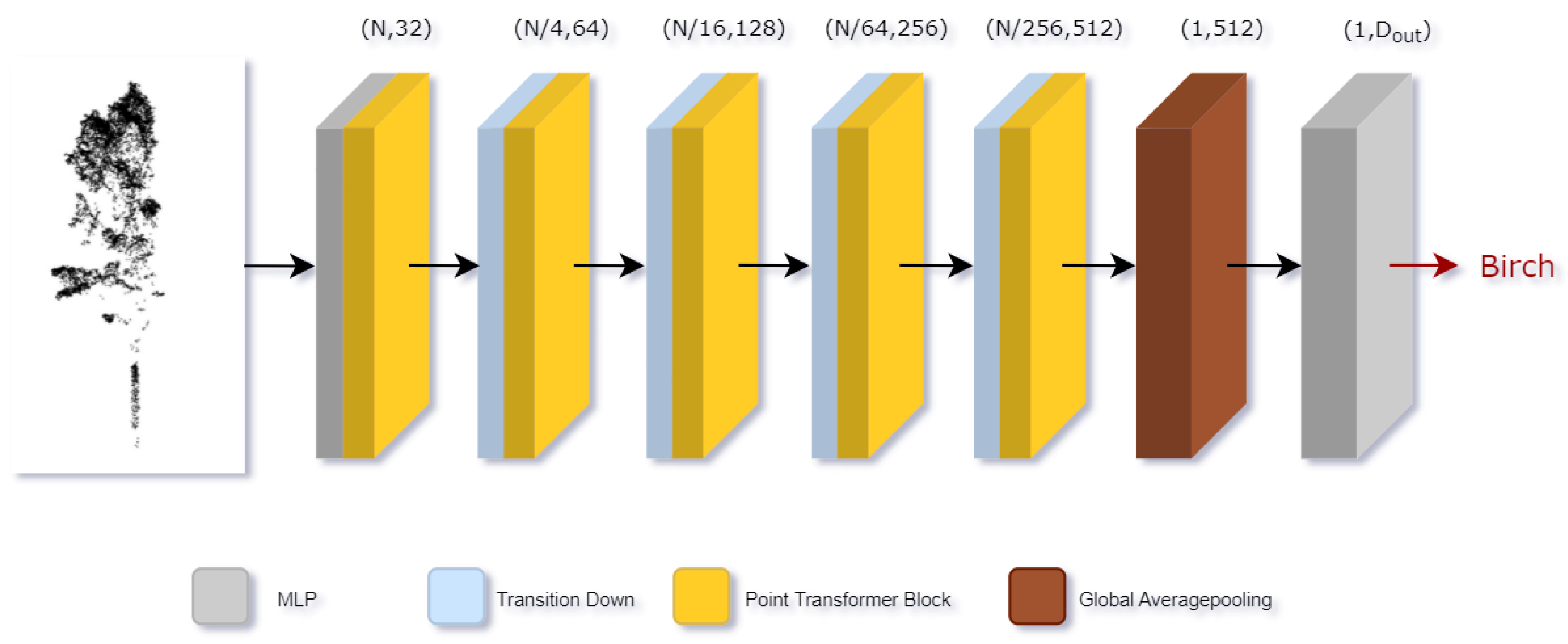
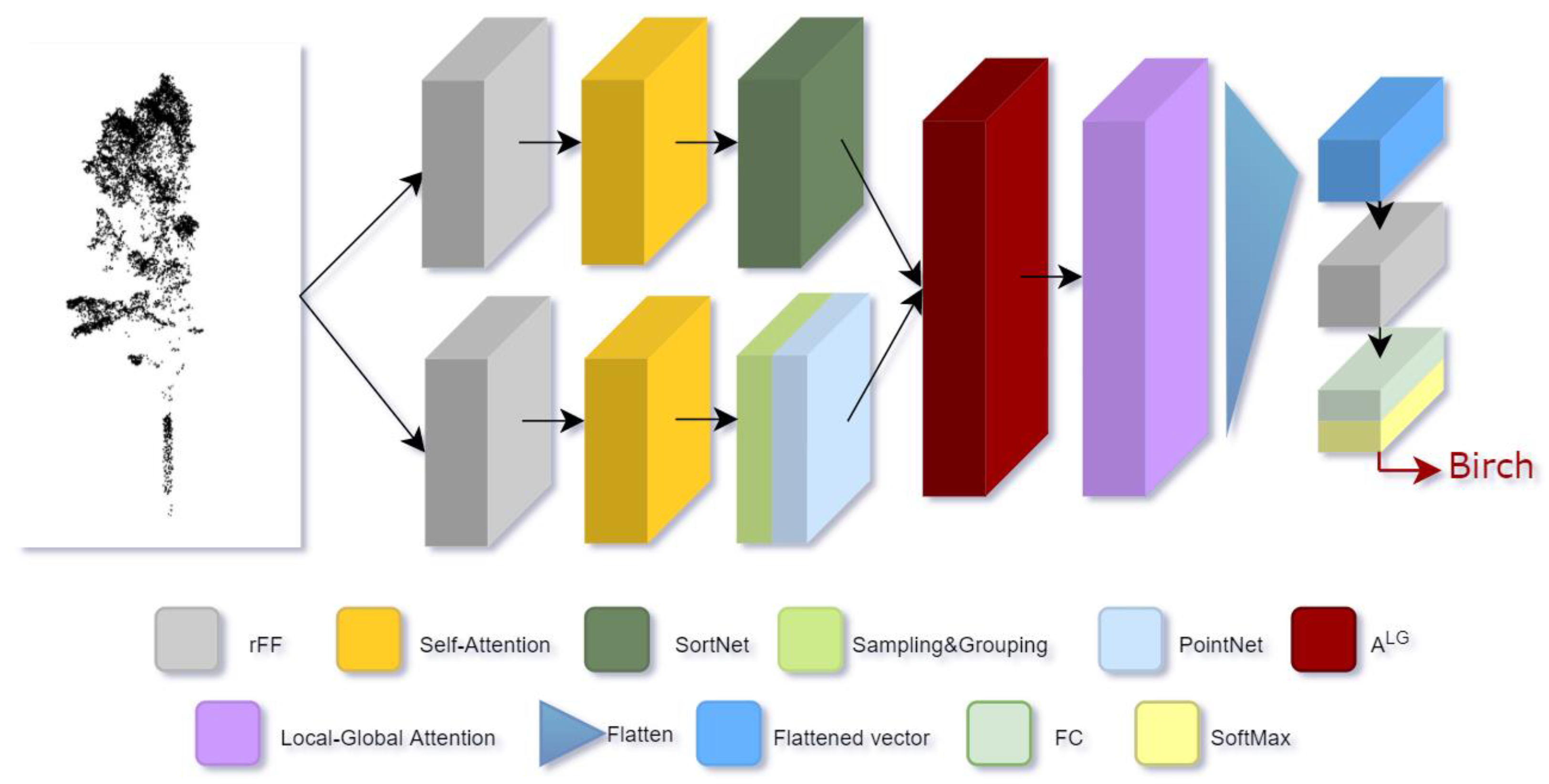
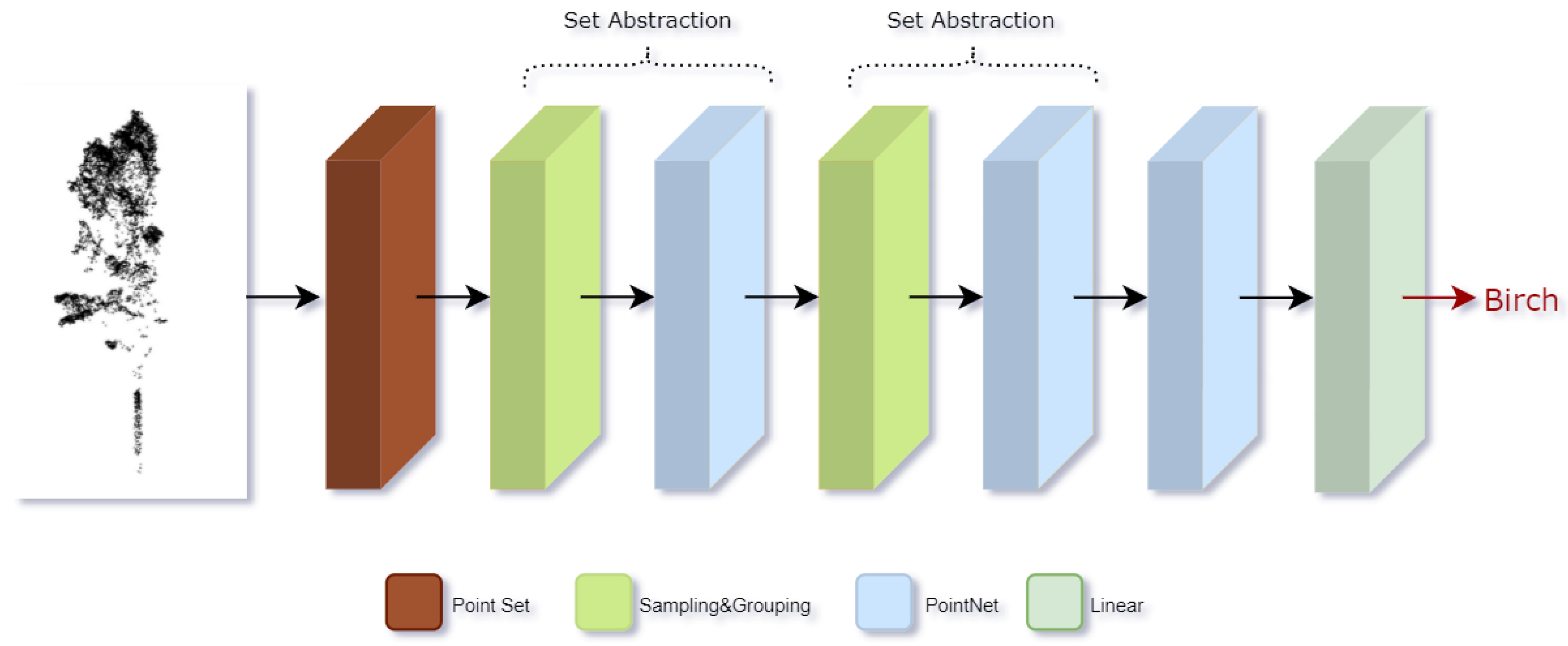
2.3.4. Classifier
Based on Transformer
Based on MLP
Random Forest
Training
2.3.5. Comparison and Evaluation
3. Results
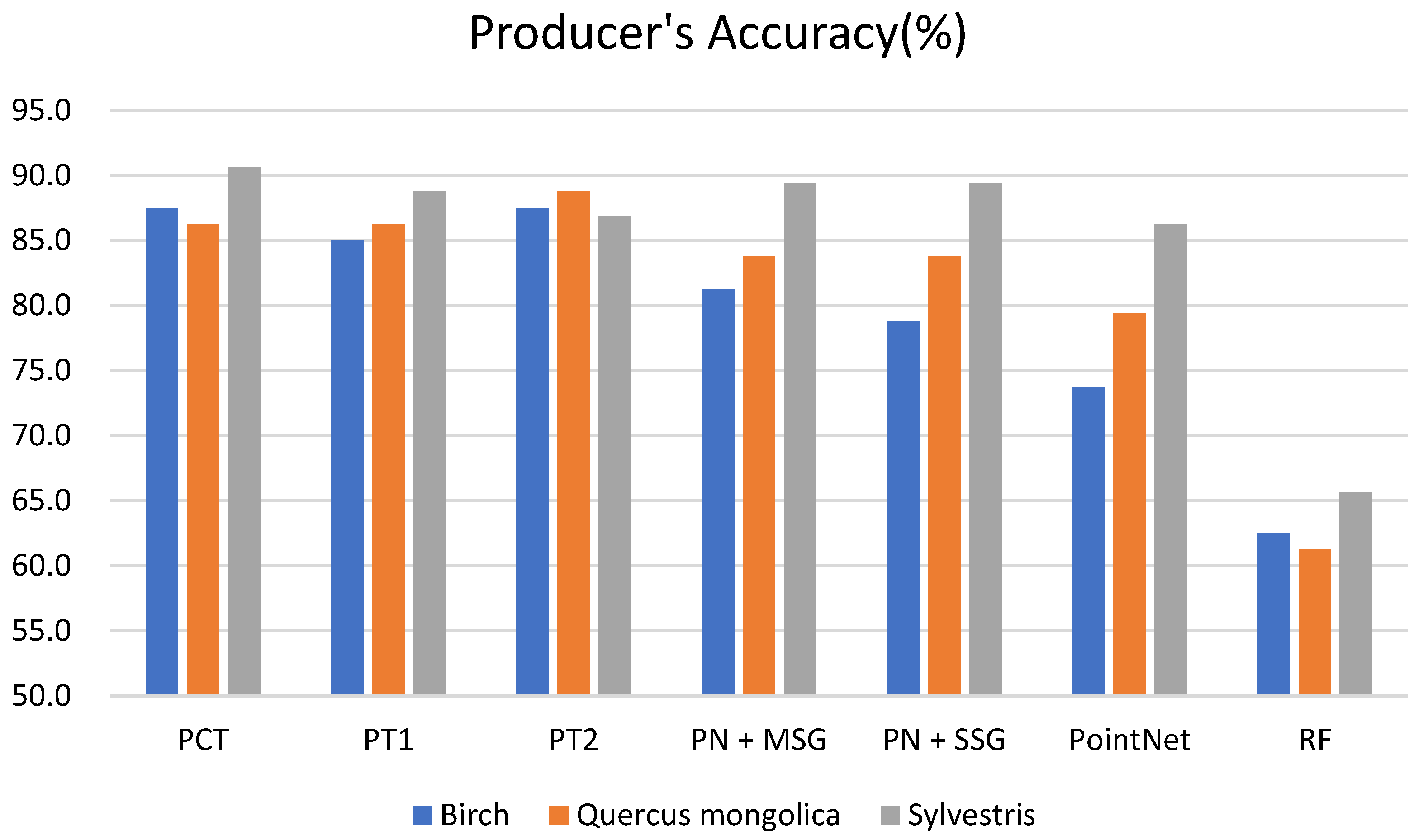
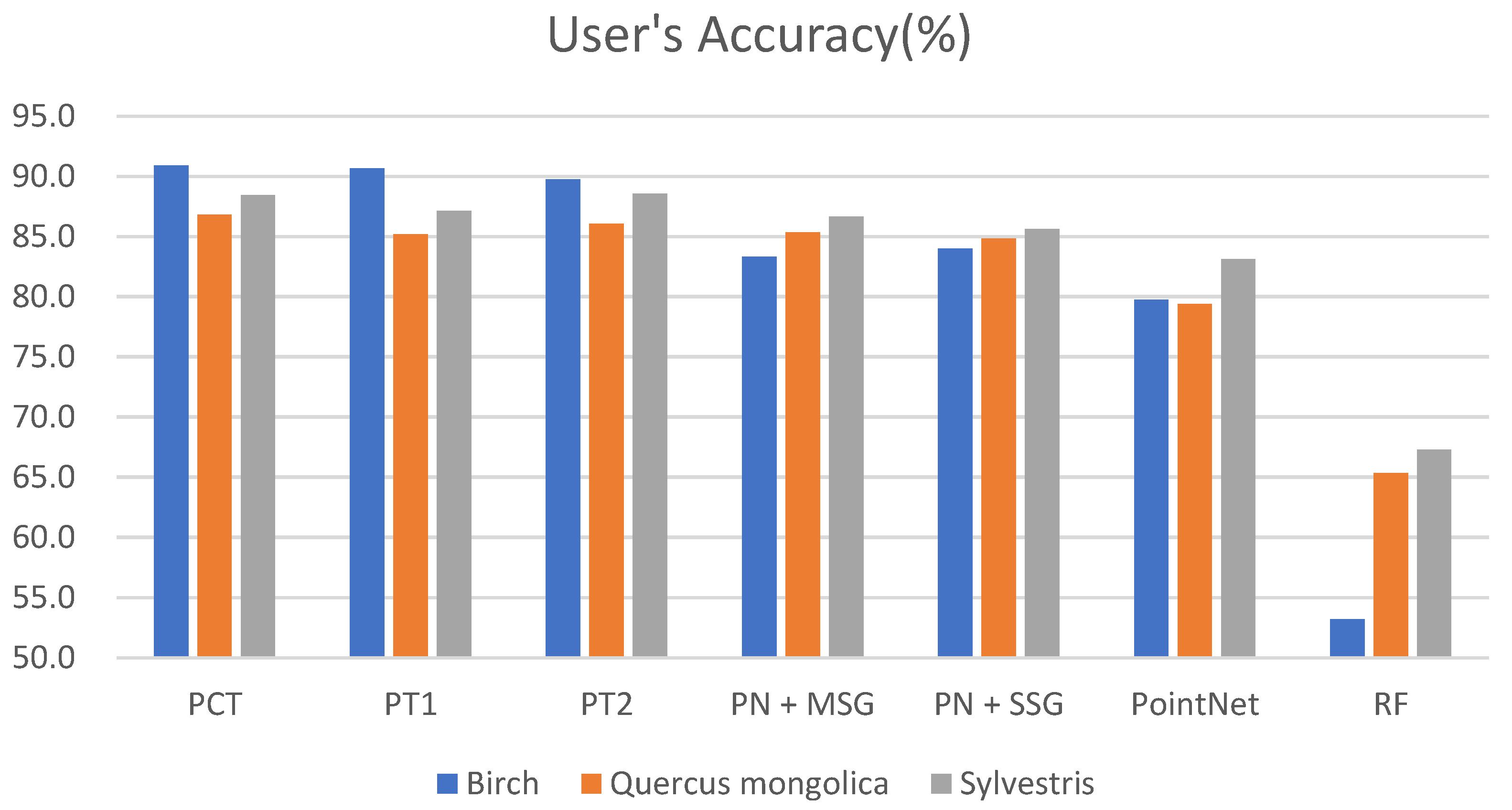
3.1. Classification Results of Different Models
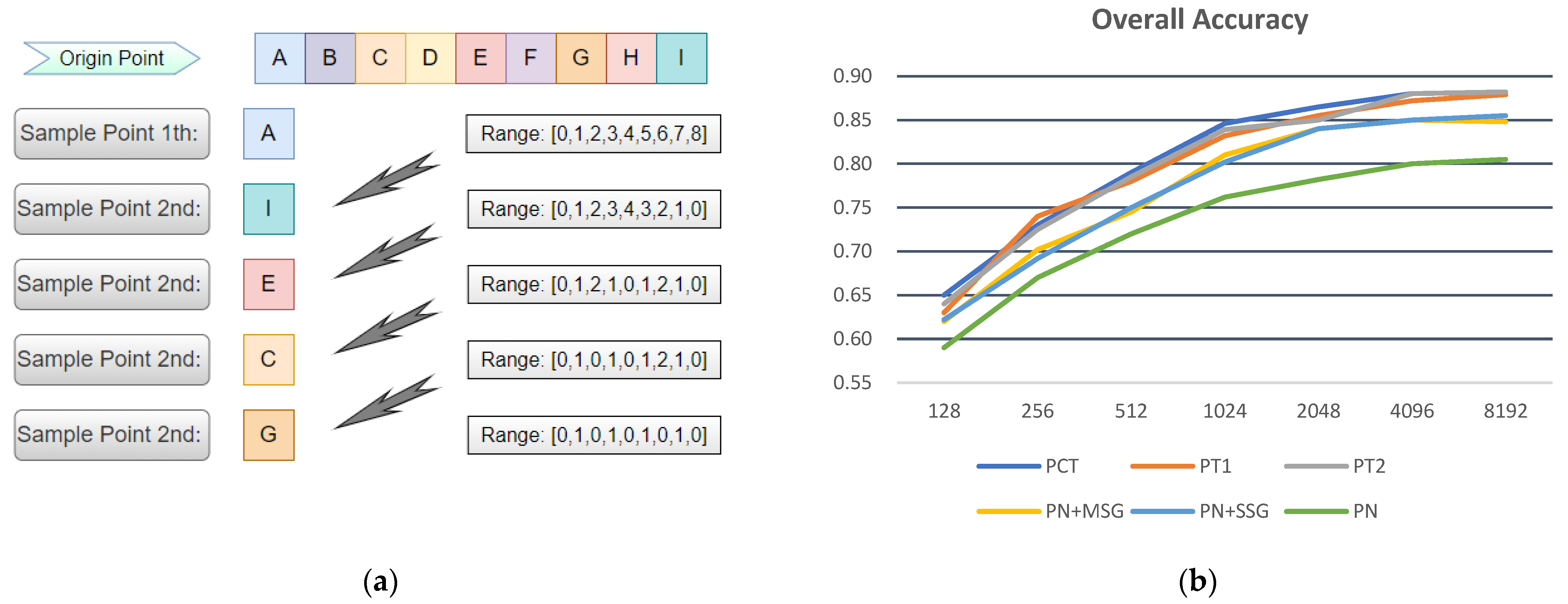
3.2. Classification Results of Different Sample Point Densities
4. Discussion
4.1. Comparison of Different Models
4.2. Comparison of Different Tree Species
4.3. Influence of Sample Point Density on Classification Results
4.4. Comparison with Machine Learning Model
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Jarvis, P.G.; Dewar, R.C. Forests in the Global Carbon Balance: From Stand to Region. In Scaling Physiological Process: Leaf to Globe; Ehleringer, J.R., Field, C.B., Eds.; Academic Press: Cambridge, MA, USA, 1993; pp. 191–221. [Google Scholar]
- McRoberts, R.E.; Cohen, W.B.; Naesset, E. Using remotely sensed data to construct and assess forest attribute maps and related spatial products. Scand. J. For. Res. 2010, 25, 340–367. [Google Scholar] [CrossRef]
- Næsset, E.; Gobakken, T.; Holmgren, J. Laser scanning of forest resources: The Nordic experience. Scand. J. For. Res. 2004, 19, 482–499. [Google Scholar] [CrossRef]
- Lechner, A.M.; Foody, G.M.; Boyd, D.S. Applications in remote sensing to forest ecology and management. One Earth 2020, 2, 405–412. [Google Scholar] [CrossRef]
- Wulder, M.A.; White, J.C.; Nelson, R.F. Lidar sampling for large-area forest characterization: A review. Remote Sens. Environ. 2012, 121, 196–209. [Google Scholar] [CrossRef]
- Seidel, D.; Ehbrecht, M.; Annighöfer, P. From tree to stand-level structural complexity-which properties make a forest stand complex? Agric. For. Meteorol. 2019, 2781, 07699. [Google Scholar] [CrossRef]
- Abd Rahman, M.Z.; Gorte, B.G.H.; Bucksch, A.K. A new method for individual tree delineation and undergrowth removal from high resolution airborne lidar. In Proceedings of the ISPRS Workshop Laserscanning 2009, Paris, France, 1–2 September 2009; Volume XXXVIII. Part 3/W8. [Google Scholar]
- Qi, W.; Dubayah, R.O. Combining Tandem-X InSAR and simulated GEDI lidar observations for forest structure mapping. Remote Sens. Environ. 2016, 187, 253–266. [Google Scholar] [CrossRef]
- Brandtberg, T.; Warner, T.A. Detection and analysis of individual leaf-Off tree crowns in small footprint, high sampling density LIDAR data from the eastern deciduous forest in North America. Remote Sens. Environ. 2003, 85, 290–303. [Google Scholar] [CrossRef]
- Cao, J.; Leng, W.; Liu, K. Object-based mangrove species classification using unmanned aerial vehicle hyperspectral images and digital surface models. Remote Sens. 2018, 10, 89. [Google Scholar] [CrossRef]
- Li, J.; Hu, B.; Noland, T.L. Classification of tree species based on structural features derived from high density LiDAR data. Agric. For. Meteorol. 2013, 171, 104–114. [Google Scholar] [CrossRef]
- Kim, S.; McGaughey, R.J.; Andersen, H.E. Tree species differentiation using intensity data derived from leaf-on and leaf-off airborne laser scanner data. Remote Sens. Environ. 2009, 113, 1575–1586. [Google Scholar] [CrossRef]
- Shoot, C.; Andersen, H.E.; Moskal, L.M. Classifying forest type in the national forest inventory context with airborne hyperspectral and lidar data. Remote Sens. 2021, 13, 1863. [Google Scholar] [CrossRef]
- Alonzo, M.; Bookhagen, B.; Roberts, D.A. Urban tree species mapping using hyperspectral and lidar data fusion. Remote Sens. Environ. 2014, 148, 70–83. [Google Scholar] [CrossRef]
- Sun, Y.; Huang, J.; Ao, Z. Deep learning approaches for the mapping of tree species diversity in a tropical wetland using airborne LiDAR and high-spatial-resolution remote sensing images. Forests 2019, 10, 1047. [Google Scholar] [CrossRef]
- Mizoguchi, T.; Ishii, A.; Nakamura, H. Individual tree species classification based on terrestrial laser scanning using curvature estimation and convolutional neural network. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2019, XLII-2/W13, 1077–1082. [Google Scholar] [CrossRef]
- Xi, Z.; Hopkinson, C.; Rood, S.B. See the forest and the trees: Effective machine and deep learning algorithms for wood filtering and tree species classification from terrestrial laser scanning. ISPRS J. Photogramm. Remote Sens. 2020, 168, 1–16. [Google Scholar] [CrossRef]
- Liu, M.; Han, Z.; Chen, Y. Tree species classification of LiDAR data based on 3D deep learning. Measurement 2021, 177, 109301. [Google Scholar] [CrossRef]
- Qi, C.R.; Su, H.; Mo, K. Pointnet: Deep learning on point sets for 3d classification and segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 652–660. [Google Scholar]
- Qi, C.R.; Yi, L.; Su, H. Pointnet++: Deep hierarchical feature learning on point sets in a metric space. Adv. Neural Inf. Process. Syst. 2017, 30, 5105–5114. [Google Scholar]
- Vaswani, A.; Shazeer, N.; Parmar, N. Attention is all you need. Adv. Neural Inf. Process. Syst. 2017, 30, 1–15. [Google Scholar]
- Pang, Y.; Wang, W.; Tay, F.E.; Liu, W. Masked autoencoders for point cloud self-supervised learning. arXiv 2022, arXiv:2203.06604. [Google Scholar]
- Guo, M.H.; Cai, J.X.; Liu, Z.N. Pct: Point cloud transformer. Comput. Vis. Media 2021, 7, 187–199. [Google Scholar] [CrossRef]
- Zhang, W.; Qi, J.; Wan, P. An easy-to-use airborne LiDAR data filtering method based on cloth simulation. Remote Sens. 2016, 8, 501. [Google Scholar] [CrossRef]
- Lu, X.; Guo, Q.; Li, W. A bottom-up approach to segment individual deciduous trees using leaf-off lidar point cloud data. ISPRS J. Photogramm. Remote Sens. 2014, 94, 1–12. [Google Scholar] [CrossRef]
- Véga, C.; Hamrouni, A.; el Mokhtari, S. PTrees: A point-based approach to forest tree extraction from lidar data. Int. J. Appl. Earth Obs. Geoinf. 2014, 33, 98–108. [Google Scholar] [CrossRef]
- Shendryk, I.; Broich, M.; Tulbure, M.G. Bottom-up delineation of individual trees from full-waveform airborne laser scans in a structurally complex eucalypt forest. Remote Sens. Environ. 2016, 173, 69–83. [Google Scholar] [CrossRef]
- Wu, Z.; Song, S.; Khosla, A. 3d shapenets: A deep representation for volumetric shapes. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 1912–1920. [Google Scholar]
- Zhao, H.; Jiang, L.; Jia, J. Point transformer. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, BC, Canada, 11–17 October 2021; pp. 16259–16268. [Google Scholar]
- Engel, N.; Belagiannis, V.; Dietmayer, K. Point transformer. IEEE Access 2021, 9, 134826–134840. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Belgiu, M.; Drăguţ, L. Random forest in remote sensing: A review of applications and future directions. ISPRS J. Photogramm. Remote Sens. 2016, 114, 24–31. [Google Scholar] [CrossRef]
- Liaw, A.; Wiener, M. Classification and Regression by randomForest. R News 2002, 2, 18–22. [Google Scholar]
- Brede, B.; Lau, A.; Bartholomeus, H.M. Comparing RIEGL RiCOPTER UAV LiDAR derived canopy height and DBH with terrestrial LiDAR. Sensors 2017, 17, 2371. [Google Scholar] [CrossRef]
- Liu, L.; Coops, N.C.; Aven, N.W. Mapping urban tree species using integrated airborne hyperspectral and LiDAR remote sensing data. Remote Sens. Environ. 2017, 200, 170–182. [Google Scholar] [CrossRef]
- Sothe, C.; De Almeida, C.M.; Schimalski, M.B. Comparative performance of convolutional neural network, weighted and conventional support vector machine and random forest for classifying tree species using hyperspectral and photogrammetric data. GIScience Remote Sens. 2020, 57, 369–394. [Google Scholar] [CrossRef]
- Hartling, S.; Sagan, V.; Sidike, P. Urban tree species classification using a WorldView-2/3 and LiDAR data fusion approach and deep learning. Sensors 2019, 19, 1284. [Google Scholar] [CrossRef] [PubMed]


| Instrument Parameters | Zenmuse L1 Settings |
|---|---|
| Flying height | 35 m |
| Flying speed | 3 m/s |
| Point cloud density | 1772 points/m2 |
| Side overlap | 65% |
| Course angle | 28° |
| Echo mode | Triple |
| Sample rate | 160 khz |
| Scan mode | Repeat |
| Quantity | Tree Species | Number of Each Species | |
|---|---|---|---|
| Birch | 320 | ||
| Training Set | 1600 | Quercus mongolica | 640 |
| Sylvestris | 640 | ||
| Birch | 80 | ||
| Test Set | 400 | Quercus mongolica | 160 |
| Sylvestris | 160 |
| Model | Predicted Class | True Class | Total | ||
|---|---|---|---|---|---|
| Birch | Quercus mongolica | Sylvestris | |||
| Birch | 70 | 5 | 2 | 77 | |
| PCT | Quercus mongolica | 8 | 138 | 13 | 159 |
| Sylvestris | 2 | 17 | 145 | 164 | |
| Birch | 68 | 5 | 2 | 75 | |
| PT1 | Quercus mongolica | 8 | 138 | 16 | 162 |
| Sylvestris | 4 | 17 | 142 | 163 | |
| Birch | 70 | 4 | 4 | 78 | |
| PT2 | Quercus mongolica | 6 | 142 | 17 | 165 |
| Sylvestris | 4 | 14 | 139 | 157 | |
| Birch | 65 | 9 | 4 | 78 | |
| PN + MSG | Quercus mongolica | 10 | 134 | 13 | 157 |
| Sylvestris | 5 | 17 | 143 | 165 | |
| Birch | 63 | 10 | 2 | 75 | |
| PN + SSG | Quercus mongolica | 9 | 134 | 15 | 158 |
| Sylvestris | 8 | 16 | 143 | 167 | |
| Birch | 59 | 11 | 4 | 74 | |
| PointNet | Quercus mongolica | 15 | 127 | 18 | 160 |
| Sylvestris | 6 | 22 | 138 | 166 | |
| Birch | 50 | 25 | 19 | 94 | |
| RF | Quercus mongolica | 16 | 98 | 36 | 150 |
| Sylvestris | 14 | 37 | 105 | 156 | |
| Model | Overall Accuracy % | Kappa Coefficient |
|---|---|---|
| PCT | 88.3 | 0.82 |
| PT1 | 87.3 | 0.80 |
| PT2 | 87.8 | 0.81 |
| PN + MSG | 85.5 | 0.77 |
| PN + SSG | 85.0 | 0.76 |
| PointNet | 80.5 | 0.70 |
| RF | 63.3 | 0.43 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, P.; Yuan, X.; Li, D. Classification of Individual Tree Species Using UAV LiDAR Based on Transformer. Forests 2023, 14, 484. https://doi.org/10.3390/f14030484
Sun P, Yuan X, Li D. Classification of Individual Tree Species Using UAV LiDAR Based on Transformer. Forests. 2023; 14(3):484. https://doi.org/10.3390/f14030484
Chicago/Turabian StyleSun, Peng, Xuguang Yuan, and Dan Li. 2023. "Classification of Individual Tree Species Using UAV LiDAR Based on Transformer" Forests 14, no. 3: 484. https://doi.org/10.3390/f14030484
APA StyleSun, P., Yuan, X., & Li, D. (2023). Classification of Individual Tree Species Using UAV LiDAR Based on Transformer. Forests, 14(3), 484. https://doi.org/10.3390/f14030484





