1. Introduction
The coastal saline-alkali land of the Yellow River Delta suffers frequent natural disasters, soil salinization, and great difficulty in afforestation, which restricts the growth of many tree species [
1,
2,
3]. The selection of salt-tolerant afforestation tree species is a key measure to improving the outcome of afforestation. Willow (
Salix matsudana) is a native arbor species naturally distributed in this area that has a certain tolerance to salt. Selection and breeding of improved willow varieties could greatly help with the construction of shelter forests in the Yellow River Delta.
Early studies on salt resistance in willows have focused on their salt resistance ability and physiological characteristics.
Salix matsudana can grow in a culture medium with a NaCl concentration of 0.1–0.2 g L
−1, whereas the salt stress caused by 0.4 g L
−1 NaCl significantly inhibits its growth [
4]. Salt stress damages the stems and leaves of willow [
5]; reduces its growth increment, biomass, and leaf water content [
6]; and causes ion toxicity, osmotic stress, and secondary oxidative stress to the plant, thereby hindering its photosynthesis, growth, and metabolism [
7]. When osmotic stress occurs, osmotic adjustment substances (soluble sugars and soluble proteins) accumulate in plant cells to maintain a higher water potential [
8,
9]. Osmotic stress and the toxicity of excessive Na
+ will cause the accumulation of reactive oxygen species in plants [
10]. The antioxidant enzyme system (including superoxide dismutase (SOD) and peroxidase (POD)) in plants will be activated [
11], which scavenges peroxide ions and reduces the oxidative damage to the cells [
12,
13].
As molecular biology techniques have advanced, research on the salt tolerance of willows has advanced to the molecular level. Qiao et al. analyzed the salt-stress-responsive proteome of
Salix matsudana [
14]. Zhou et al. discovered significantly different miRNA expressions between salt-sensitive and salt-tolerant
Salix matsudana under various salt stress conditions [
15]. Chen et al. identified candidate genes for salt stress response in
Salix matsudana [
16]. Shan et al. found that the physiological and metabolic processes of plants could be regulated by controlling salt-stress-related genes [
17]. Yang et al. [
18] found that in a high-salt environment, the upregulation of H
+-ATPase gene expression in vacuoles led to cytoplasmic sequestration of Na
+ [
19]. Plants also respond to salt stress by regulating the expression of salt-tolerance-related transcription factors such as WRKY, MYB, and basic helix-loop-helix (BHLH) [
20,
21,
22].
With the gradual maturation of sequencing technology, research based on transcriptome sequencing has expanded from animals to plants. Transcriptomic research in plants has mainly focused on stress resistance, regulation of physiological mechanisms, and nutrient utilization. Studies on stress resistance mainly focus on salt and alkali tolerance, disease resistance, and drought resistance [
23,
24,
25,
26]. Transcriptome sequencing plays an increasingly important role in mining the salt tolerance genes of plant species, such as
Jatropha curcas, citrus, cotton, wheat, and rose [
27,
28,
29,
30,
31,
32]. The application of third-generation single-molecule sequencing technology reduces the difficulty of analyzing the nonparametric transcriptome and allows for the easy sequencing of the complete genome and full-length transcripts, making the in-depth study of transcription mechanisms more convenient. However, there have been few studies on the relationship between the physiological salt-stress-responsive indices and the molecular mechanism of salt tolerance that have used third-generation sequencing technology and have applied the findings to the breeding of salt-tolerant varieties.
The present study examined 16 salt-tolerance-related indices in seedlings of Salix matsudana. On this basis, correlation analysis, grey cluster analysis, principal component analysis, and stepwise regression analysis were performed to screen out the key indices, pick the salt-tolerant and salt-sensitive Salix matsudana clones, and derive the equation that accurately reflected the salinity resistance of Salix matsudana. Transcriptome analysis was carried out on the selected salt-tolerant and salt-sensitive clones. The pathways involved in the salt tolerance of Salix matsudana and their relevant genes were identified on the basis of the differentially expressed genes and the key indices of salt tolerance. Eventually, an identification method of salt tolerance that was stable and efficient in both the physiological and molecular aspects was established for Salix matsudana at the seedling stage. This study might provide a theoretical and technical basis for the selection and breeding of improved varieties that can tolerate the salinity of coastal saline-alkali land.
2. Materials and Methods
2.1. Experimental Materials
The research objects included a total of five willow clones: the improved salt-tolerant varieties Luliu 2 and Luliu 6, which were registered by the Shandong Academy of Forestry Sciences, as well as three unapproved clones with good traits (preliminarily named Jinan 1, Jinan 2, and Binzhou 1) in the experimental forest of Jinan, Shandong Province. The widely popular Willow No. 172 was used as the control. We numbered Luliu 2, Luliu 6, Jinan 1, Jinan 2, Binzhou 1, and Willow No. 172 as Sm2, Sm6, SmA, SmB, SmC, and Sm172 for this paper, respectively. The branches used in this study were collected from the experimental forest of Xicang Village, Changqing District, Jinan City, Shandong Province.
2.2. Experimental Methods
(1) Pot experiment: In April 2017, potted seedlings were cultivated with sieved soil. The soil was taken from the Forestry Experimental Station of Shandong Agricultural University. Before loading, it was screened to remove impurities and homogenized. The soil is sandy loam with a pH of 7.05 ± 0.03, a total nitrogen content of 1.27 ± 0.14 g·kg−1, the hydrolyzable nitrogen content of 94.36 ± 13.89 mg·kg−1, the available phosphorus content of 32.01 ± 4.10 mg·kg−1, the available potassium content of 56.72 ± 7.83 mg·kg−1, and the organic matter content of 16.27 ± 2.39 g·kg−1. The cuttings of Salix matsudana clones Sm6 and Sm172 were selected and planted into greenhouse flowerpots at the Forestry Experimental Station of Shandong Agricultural University. The upper diameter of the flowerpots was 30 cm, the lower diameter was 20 cm, and the height was 25 cm. The weight of the soil in each pot was 10 kg. After planting, each cutting protruded approximately 2 cm above the soil surface. Regular watering management was conducted during the study. After the cuttage seedlings survived, the weak seedlings were eliminated in July 2017, while the seedlings that grew well and had no obvious diseases and pests were selected for experiments. The greenhouse was equipped with a water curtain, fan, and other temperature control equipment. During the experiment, the day and night temperatures in the greenhouse were controlled at (25 ± 2) °C and (20 ± 2) °C respectively, and the relative humidity was 65–70%.
Soil salinity was determined by the gravimetric method. A soil salinity gradient (0.1%, 0.3%, 0.5%, and 0.7%) was prepared, and deionized water was used as the blank control. First, the mass of NaCl needed for each stress level was calculated. NaCl solutions with the corresponding concentrations were prepared in deionized water and added three times in equal amounts (irrigation once every 7 days). A tray was placed below each pot to prevent the loss of salt. The physiological and biochemical indices were measured 15 days after the last addition of the salt.
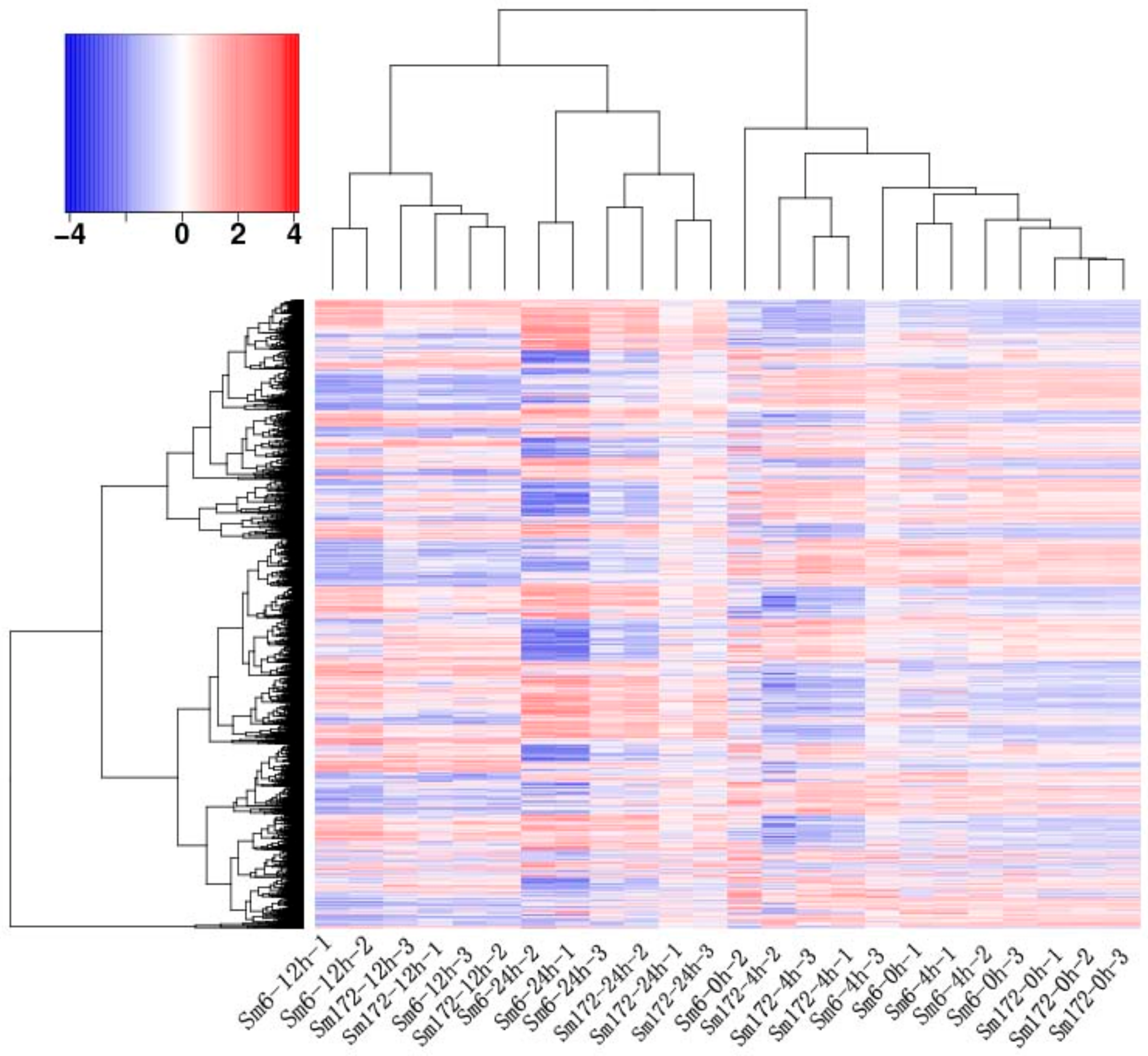
(2) Transcriptome analysis: Transcriptome sequencing was performed using a combination of second-generation and third-generation sequencing technologies. Normally growing Sm6 and Sm172 adult plants (one each) were selected. Several branches with similar growth were cut at a length of approximately 15 cm. The cuttings were placed in beakers filled with deionized water and cultivated in a constant-temperature light incubator. After the branches took root and grew new leaves, various groups of branches were soaked in 200 mmol·L−1 NaCl solution for 0, 4, 12, and 24 h. The leaves were collected from all groups immediately after the soaking treatment. During sampling, leaves were collected from 3 plants in each group and mixed. Immediately after collection, the samples were placed in liquid nitrogen and then stored in a −80 °C freezer. The samples were saved for procedures such as RNA extraction.
2.3. Determination of Salt Tolerance Indices and Data Analysis
We grouped the indices related to the salt tolerance of
Salix matsudana into four groups, salt injury indices, physiological and biochemical indices, biomass indices, and photosynthetic indices (
Table 1), and used them to establish the standard equation of salt tolerance index. The root fresh weight (RFW), shoot fresh weight (SFW), relative leaf water content (RWC), relative seedling height (RH), and relative diameter (RD) were determined according to the method developed by Chen et al. [
33]. The chlorophyll content was measured following Wang et al. [
34]. Using a portable photosynthesis system (CIRAS-2), the net photosynthetic rate (Pn), stomatal conductance (GS), and intercellular CO
2 concentration (Ci) were measured between 9:00 a.m. and 11:00 a.m. at a saturated light intensity of 1200 μmol·m
−2s
−1 and a leaf chamber temperature of 25 °C. The relative conductivity (RC) and malondialdehyde (MDA) content were determined as described by Qin et al. [
35]. The activities of peroxidase (POD) and superoxide dismutase (SOD) were determined following Li et al., and the contents of soluble sugar (SS) and starch (SST) were determined according to Wang et al. [
36,
37].
All statistical analyses were conducted using Statistical Product Service Solutions for Windows 22.0 (SPSS, Chicago, IL, USA).
2.4. Sequencing Method and Analysis
Leaf RNA was extracted, and a cDNA library was constructed. Second-generation sequencing was performed on the Illumina HiSeq high-throughput sequencing platform. The third-generation full-length transcriptome sequencing was performed by Anoroad Gene Technology Co., Ltd. (Beijing, China) with the PacBio Sequel sequencer. Sequencing quality was assessed using FastQC. Functional annotation was performed with Trinotate. GO classification was performed with BLAST2GO. COG classification was performed using eggNOG. Metabolic pathway analysis was performed using the KEGG. Differential expression analysis was conducted with DESeq. The GO classification of the differentially expressed Unigene and Pathway enrichment analysis were performed using GOseq.
2.5. Verification of the Differentially Expressed Genes
RNAs were extracted from Sm6 and Sm172 that had received salt treatment for 0, 4, 12, or 24 h. The RNAs were reverse-transcribed, and 12 randomly selected salt-tolerance-related genes of Salix matsudana were subjected to real-time fluorescence quantitative verification. The polymerase chain reaction (PCR) system (20 μL) was set up as follows: SYBR Green 10 μL, forward primer (10 mM) 1 μL, reverse primer (10 mM) 1 μL, cDNA 2 μL, and ddH2O to a total volume of 20 μL. The PCR conditions were as follows: predenaturation at 95 °C for 30 s and 40 cycles of denaturation at 95 °C for 5 s and optimal annealing temperature 60 °C for 30 s.
4. Discussion
This study analyzed the chosen indices using a combination of the analytic hierarchy process and principal component analysis, which was more reasonable than directly doing principal component analysis on all indices. SOD, POD, chlorophyll, and net photosynthetic rate may be used as key indices to identify the salt tolerance of
Salix matsudana at the seedling stage [
38,
39,
40]. Upon salt stress, the antioxidant enzymes such as SOD and POD were activated in all clones of
Salix matsudana, which led to the elimination of the toxicity of free radicals. SOD prevents the formation of free radicals and the damage to cells mainly by regulating the content of O
2−. It is the first line of defense against cell damage [
39,
41]. POD plays an important role in protecting cells from H
2O
2 stress [
42]. In response to salt stress, photosynthesis is reduced in
Salix matsudana. The accumulation of photosynthetic products is inhibited, which hinders the growth and development of plants and even causes plant death [
43]. As an important photosynthetic pigment, chlorophyll plays an important role in absorption, transmission, and transformation of light energy [
3,
44,
45]. Therefore, the change in chlorophyll content is an important index of plant growth [
46]. Salt stress leads to ion toxicity, hinders the absorption of Mg
2+ by plants, blocks the synthesis of chlorophyll, reduces chlorophyll content, and thus seriously hinders photosynthesis [
47,
48,
49,
50,
51]. Such findings are consistent with the research results of Gong et al. [
52,
53].
Under salt stress, reactive oxygen species activate a variety of
MAPK signaling cascades, inducing specific cellular responses. This study found that in
Salix matsudana exposed to salt stress, the most strongly expressed
MAPK signaling pathway was the mitogen-activated protein kinase (
MPK). This study showed that H
2O
2 phosphorylated mitogen-activated protein kinase kinase 4/5 (
MKK4/5) by activating
ANP1 upstream of the
MPK cascade, thereby phosphorylating
MPK3. This result is consistent with the findings of Kim et al. [
54] and Kovtun et al. [
55]. H
2O
2 also directly activated
MPK3 by inducing the formation of nucleoside diphosphate kinase 2 (
NDPK2), in line with the findings of Moon et al. [
56]. The serine-threonine protein kinase
OXI1 (oxidative-signal-inducible 1) was also activated by peroxide and responded through the downstream
MPK3. In the process of cell death caused by reactive oxygen species through the above three pathways,
MPK3 is significantly upregulated, and the level of reactive oxygen species (such as H
2O
2) is increased [
57], which eventually leads to cell death. When
Salix matsudana is exposed to low salt stress or short-term salt stress, the upregulation of
MPK3 and the increase in H
2O
2 lead to elevated activities of SOD and POD in the antioxidant system, which allows the resistance to the damage of reactive oxygen species and maintenance of normal physiological function of cells. Therefore, improving the activity of antioxidant enzymes in plants and increasing the level of plant antioxidant metabolism are important ways to enhance the salt tolerance of plants. Alleviation of the harm of salt stress to
Salix matsudana could be achieved by inhibiting the expression of the MPK3-related gene
Sm6-f3p60-931, which suppressed the activity of
MPK3 and inhibited the production of reactive oxygen species. This biological process played a certain positive role in the salt stress resistance of
Salix matsudana and provided evidence that SOD and POD are key indices of salt tolerance in
Salix matsudana.
In addition, this study showed that
MPK3 of the
MAPK pathway cascade was significantly upregulated under salt stress, which reduced the function of
SPEECHLESS (
SPCH) and inhibited stomatal development. The basic helix-loop-helix (
bHLH)-type transcription factors that regulate stomatal development include three members:
SPCH,
MUTE, and
FAMA. These proteins are involved in the regulation of the initiation of the M cell lineage, the formation of guard mother cells, and the symmetrical division of guard mother cells [
58,
59].
SPCH has a region between the
bHLH domain and the C-terminal region that is rich in the acidic amino acids serine and threonine. It mediates the direct phosphorylation of
SPCH by
MPK3 [
60,
61,
62], which hinders stomatal development and directly hinders photosynthesis. Therefore, physiological study of
Salix matsudana showed that, with more concentrated or longer salt treatment, the net photosynthetic rate was reduced in each
Salix matsudana clone. This finding provides evidence that the net photosynthetic rate is a key index of salt tolerance in
Salix matsudana.
Moreover, genes with large differences in expression levels are involved in the synthesis of
NCED. According to the metabolic pathway map of carotenoid biosynthesis, carotenoid biosynthesis is related to ABA synthesis [
54,
63]. ABA plays a central role in plant responses to stressful environments [
64,
65]. In the process of ABA synthesis, zeaxanthin epoxidase catalyzes the conversion of carotene to violaxanthin.
NCED further catalyzes the conversion of 9-cis-flavin to xanthotoxin, which further leads to the production of abscisic aldehyde. Abscisic aldehyde is eventually converted into ABA under the action of an amine oxidase. These findings are consistent with the results of Li et al. [
23].
NCED is a member of the carotenoid cleavage dioxygenase family. It is the most important rate-limiting enzyme in the process of ABA biosynthesis in higher plants and belongs to a class of key genes involved in the regulation of ABA biosynthesis [
66,
67].
NCED expression level is positively correlated with ABA content in plants [
68,
69,
70]. Therefore, the goal of plant physiological self-regulation might be achieved through the control of the expression of
NCED synthesis-related genes such as
Sm172-f2p28-2386,
Sm172-f2p30-2392,
Sm172-f2p39-2386, and
Sm6-f8p60-2372, which further control the expression of
NCED and regulate the content of ABA in plants. In future research on the salt tolerance of
Salix matsudana, ABA might become a key index.
These data provide a scientific basis for future research on the mechanism of Salix matsudana salt tolerance, deliver a simpler method for the identification and breeding of salt-tolerant Salix matsudana varieties, and can guide the selection of salt-tolerant tree species in the coastal saline-alkali land of the Yellow River Delta.