Soil Actinomycetes of Vietnam Tropical Forests
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.1.1. Xuan Son National Park
2.1.2. Pu Hoat Nature Reserve
2.1.3. Pu Mat National Park
2.1.4. Kon Chu Rang Nature Reserve
2.1.5. Kon Plong Protected Forest
2.1.6. Kon Ka Kinh National Park
2.2. Research Methods
3. Results
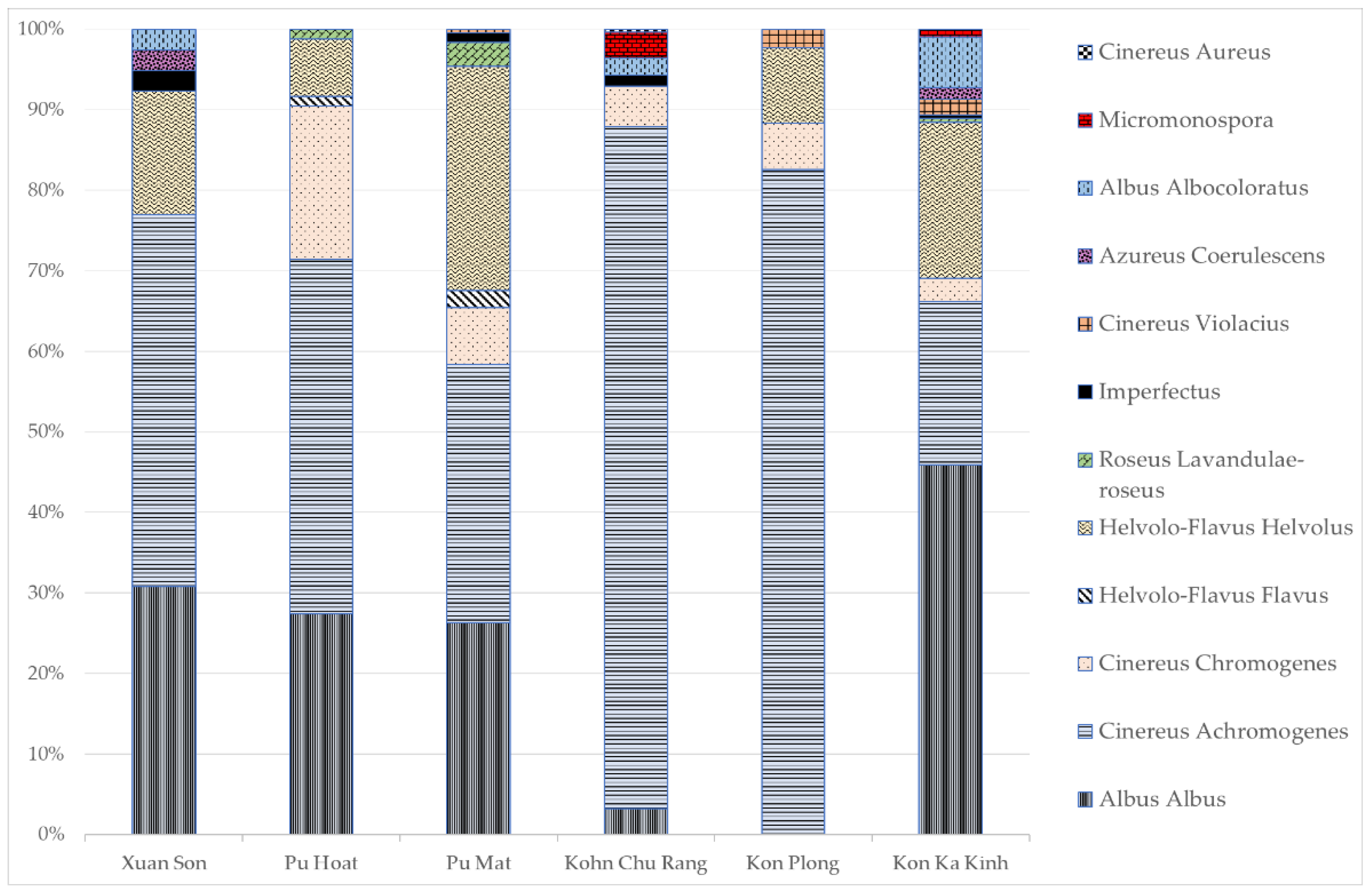
3.1. Determination of the Total Abundance of Actinomycetes, the Length of the Actinomycete Mycelium and the Structure of the Actinomycete Complex in the Studied Samples
3.2. Ecophysiological Properties of Actinomycete Strains Isolated from Tropical Forest Soils
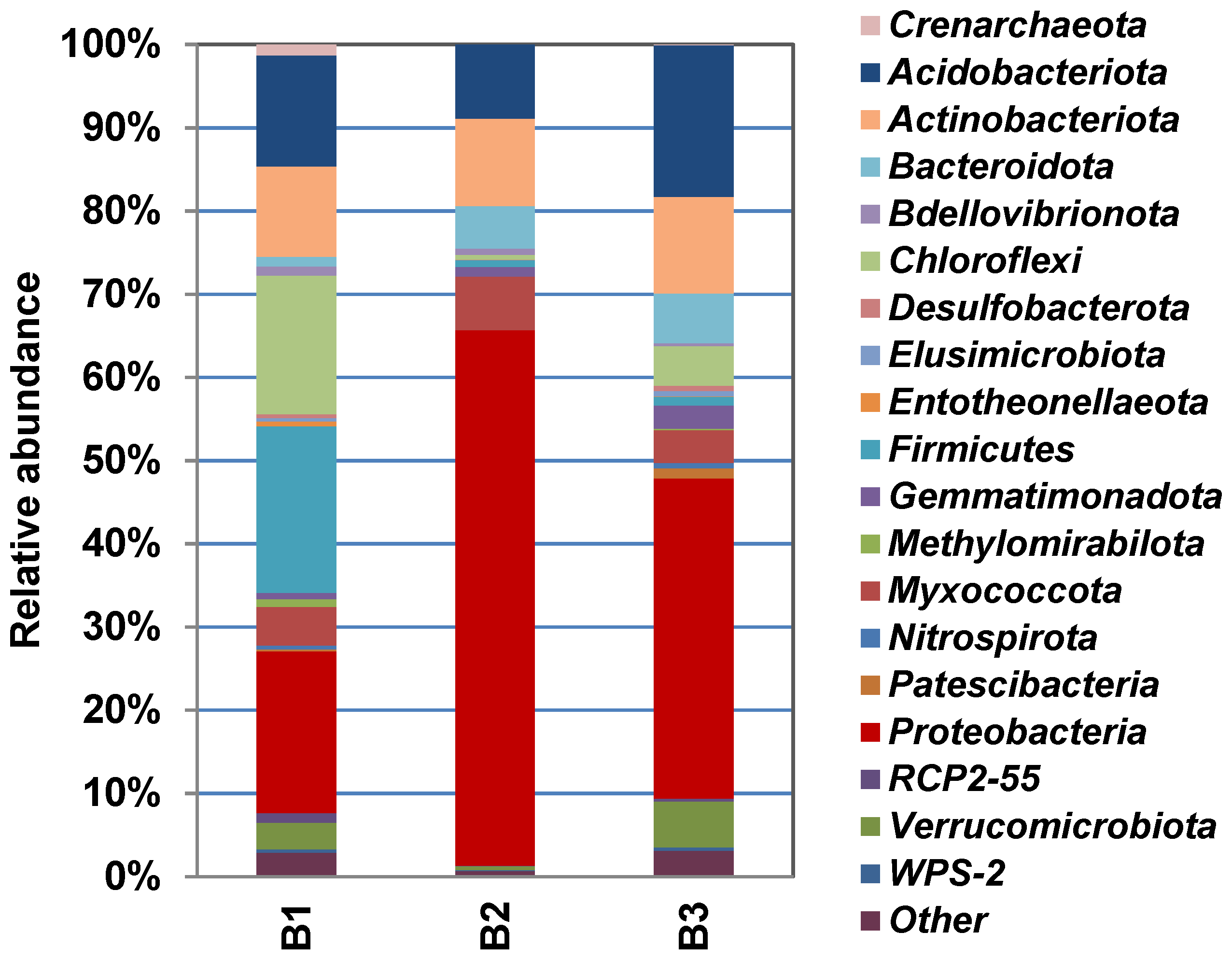
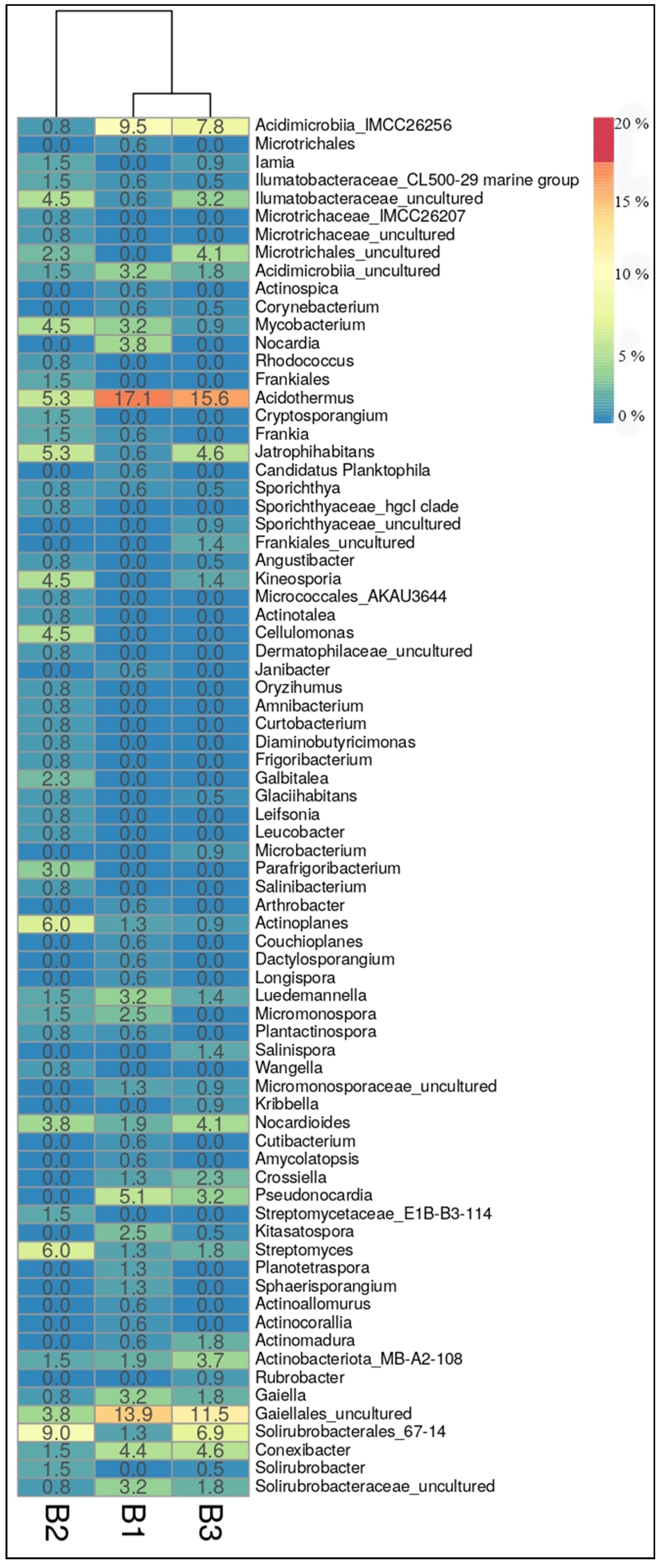
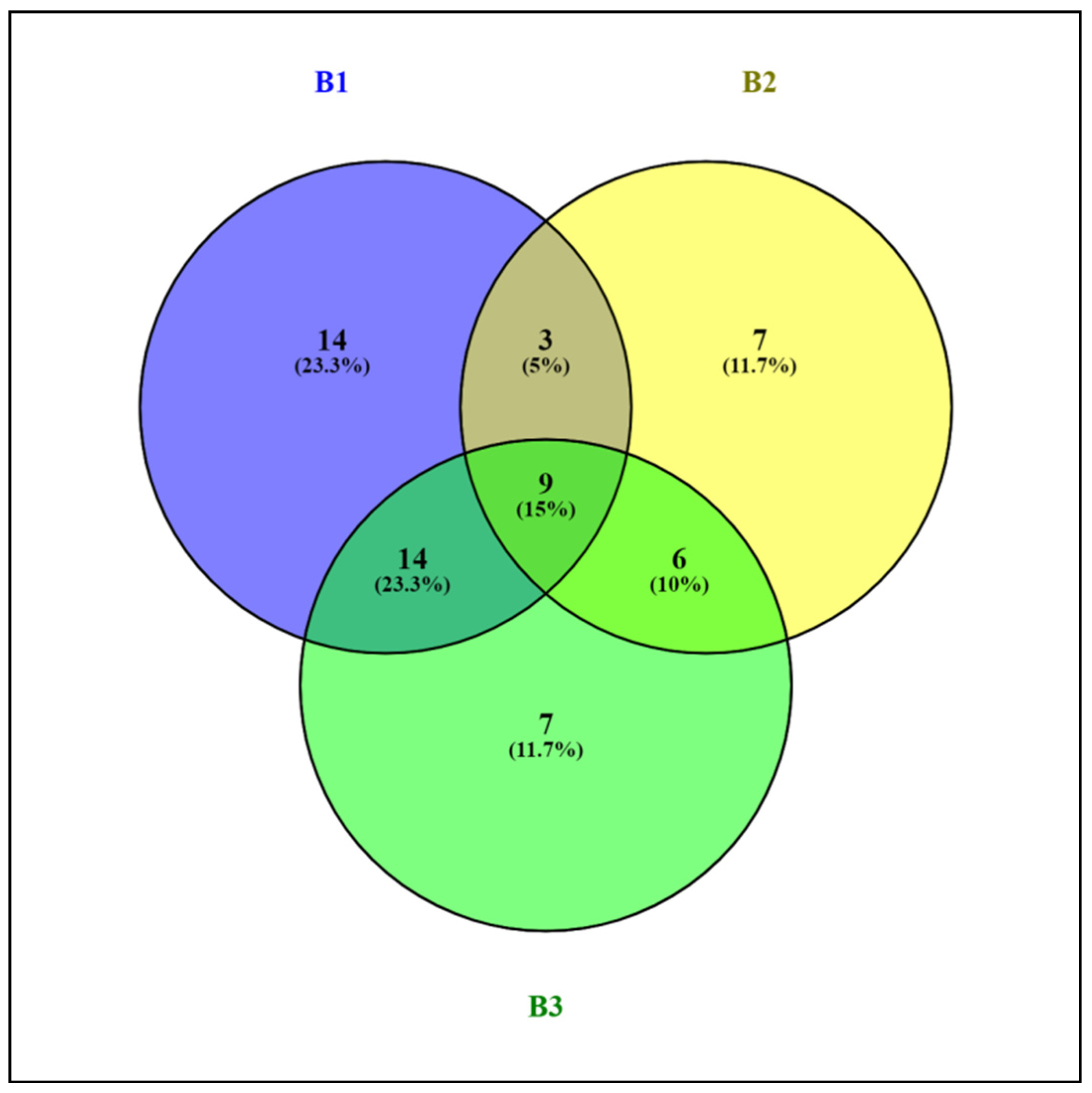
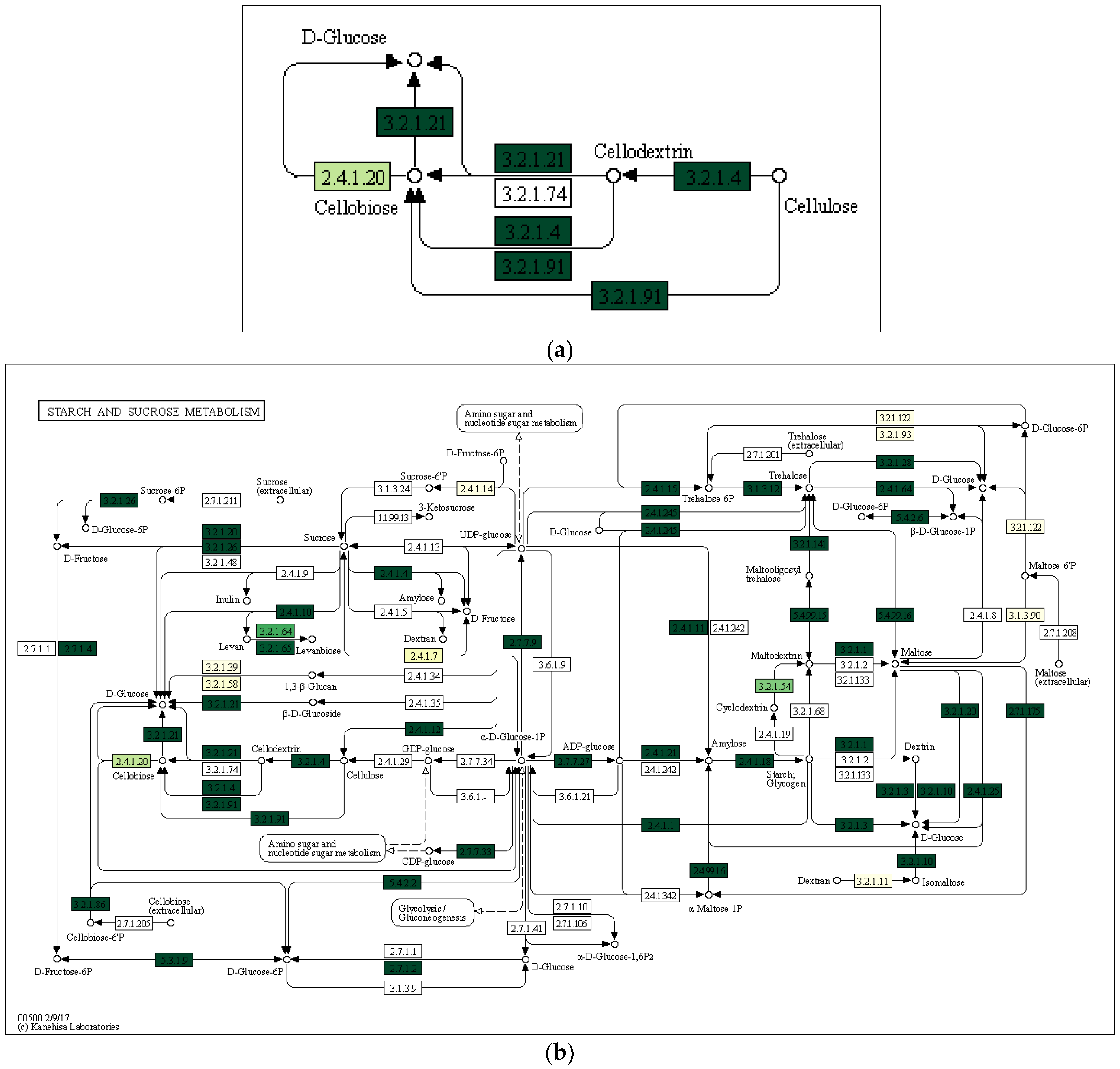
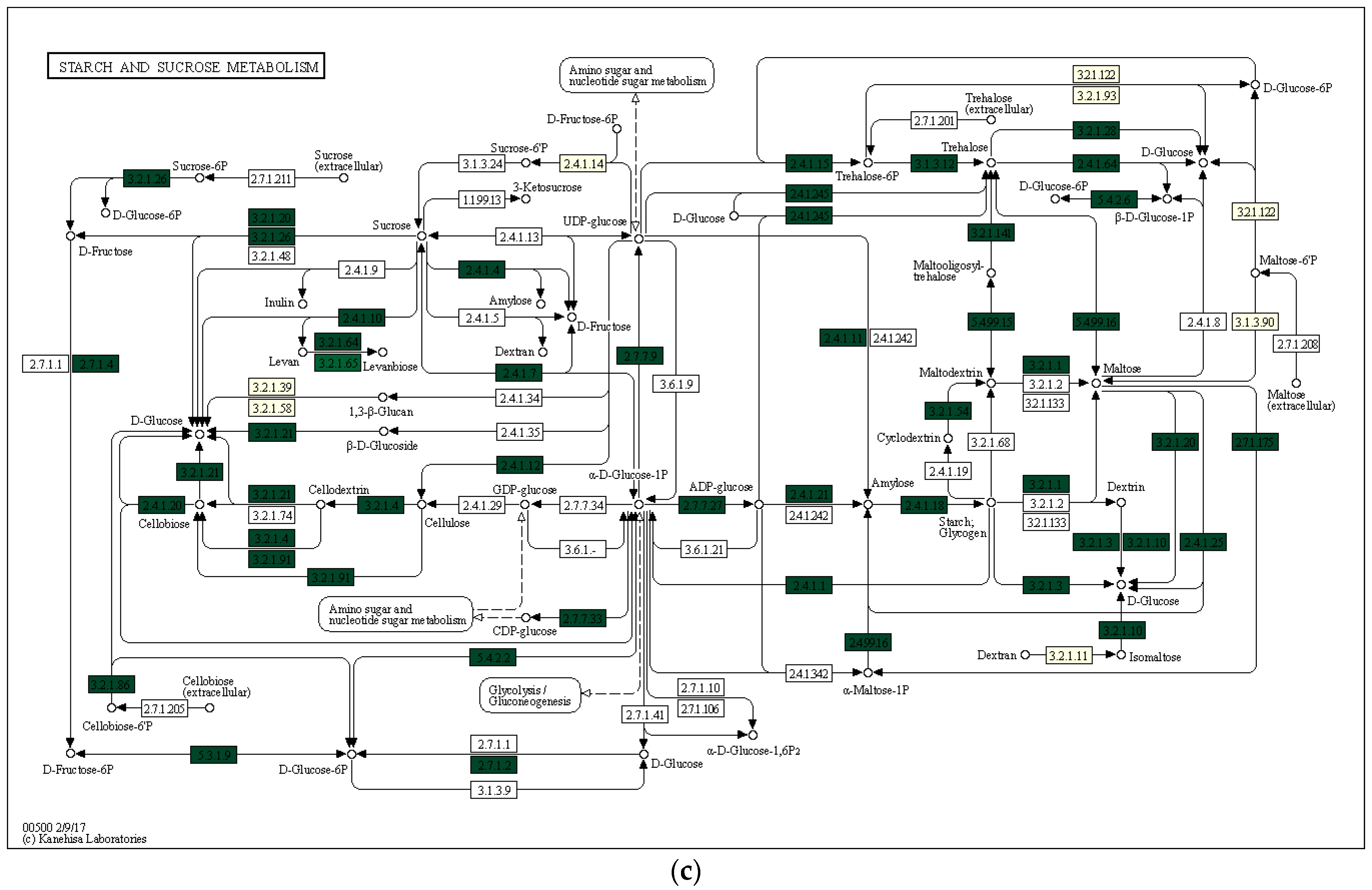
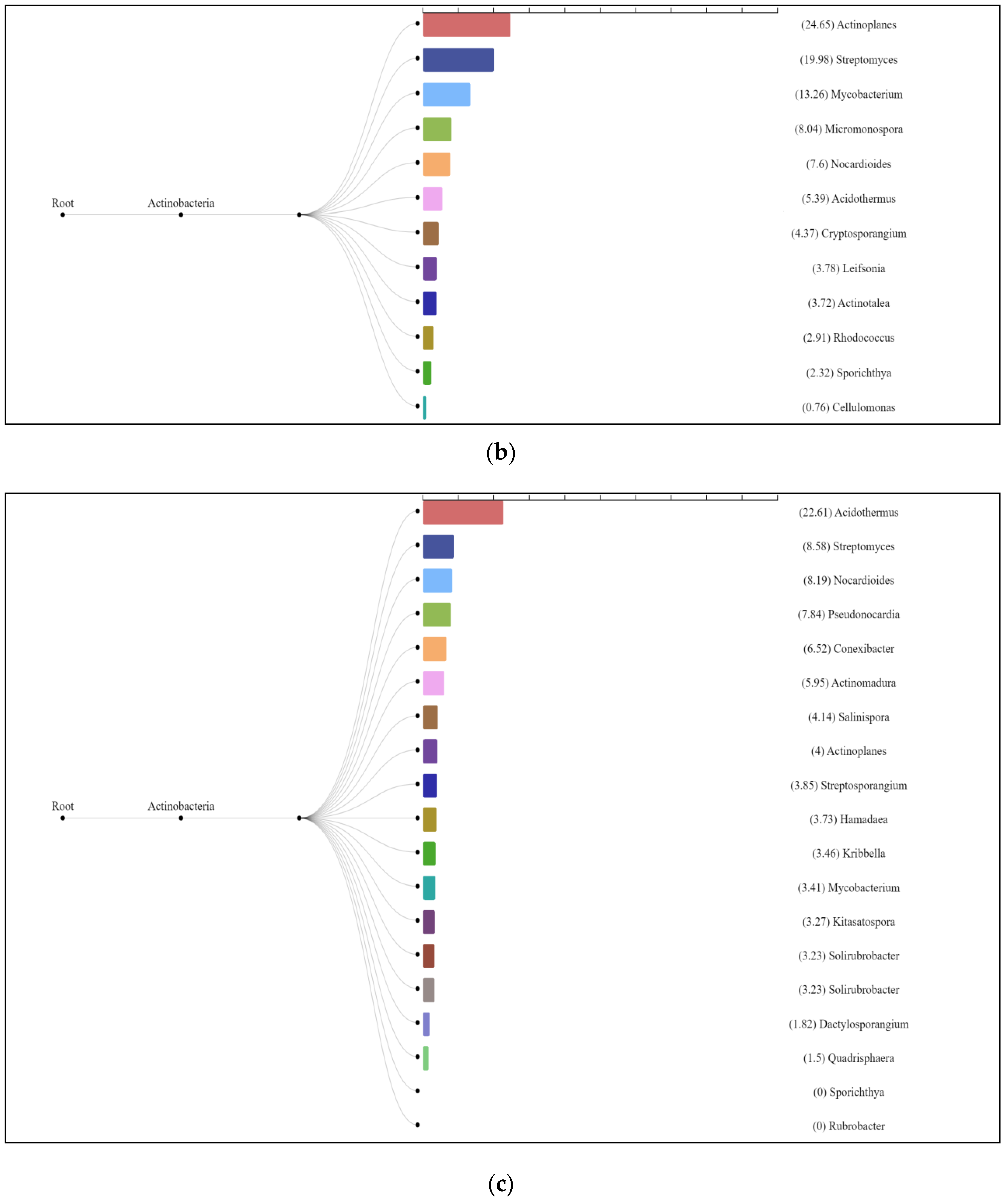
3.3. Characterization of Prokaryotic Community Diversity Using High-Throughput Pyrosequencing of the 16S rRNA Gene
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Laurance, W.F. Reflections on the tropical deforestation crisis. Biol. Conserv. 1999, 91, 109–117. [Google Scholar] [CrossRef]
- Norris, P.R. Class II. Acidimicrobiia class. nov. In Bergey’s Manual of Systematic Bacteriology, 2nd ed.; The Actinobacteria, part B; Goodfellow, M., Kampfer, P., Busse, H.-J., Trujillo, M.E., Suzuki, K.-I., Ludwig, W., Whitman, W.B., Eds.; Springer: New York, NY, USA, 2012; Volume 5. [Google Scholar]
- Stackebrandt, E.; Rainey, F.A.; Ward-Rainey, N.L. Proposal for a new hierarchic classification system, Actinobacteria classis nov. Int. J. Syst. Bacteriol. 1997, 47, 479–491. [Google Scholar] [CrossRef]
- Salam, N.; Jiao, J.-Y.; Zhang, X.-T.; Li, W.-J. Update on the classification of higher ranks in the phylum Actinobacteria. Int. J. Syst. Evol. Microbiol. 2020, 70, 1331–1355. [Google Scholar] [CrossRef]
- Gupta, R.S.; Chen, W.J.; Adeolu, M.; Chai, Y. Molecular signatures for the class Coriobacteriia and its different clades; proposal for division of the class Coriobacteriia into the emended order Coriobacteriales, containing the emended family Coriobacteriaceae and Atopobiaceae fam. nov., and Eggerthellales ord. nov., containing the family Eggerthellaceae fam. nov. Int. J. Syst. Evol. Microbiol. 2013, 63, 3379–3397. [Google Scholar] [PubMed]
- Konig, H. Class III. Coriobacteriia class. nov. In Bergey’s Manual of Systematic Bacteriology, 2nd ed.; The Actinobacteria, part B; Goodfellow, M., Kampfer, P., Busse, H.-J., Trujillo, M.E., Suzuki, K.-I., Ludwig, W., Whitman, W.B., Eds.; Springer: New York, NY, USA, 2012; Volume 5. [Google Scholar]
- Ludwig, W.; Euzeby, J.; Whitman, W.B. Class IV. Nitriliruptoria class. nov. In Bergey’s Manual of Systematic Bacteriology, 2nd ed.; The Actinobacteria, part B; Goodfellow, M., Kampfer, P., Busse, H.-J., Trujillo, M.E., Suzuki, K.-I., Ludwig, W., Whitman, W.B., Eds.; Springer: New York, NY, USA, 2012; Volume 5, pp. 2000–2001. [Google Scholar]
- Suzuki, K.-I. Class V. Rubrobacteria class. nov. In Bergey’s Manual of Systematic Bacteriology, 2nd ed.; The Actinobacteria, part B; Goodfellow, M., Kampfer, P., Busse, H.-J., Trujillo, M.E., Suzuki, K.-I., Ludwig, W., Whitman, W.B., Eds.; Springer: New York, NY, USA, 2012; Volume 5, pp. 2004–2005. [Google Scholar]
- Suzui, K.-I.; Whitman, W.B. Class VI. Thermoleophilia class. nov. In Bergey’s Manual of Systematic Bacteriology, 2nd ed.; The Actinobacteria, part B; Goodfellow, M., Kampfer, P., Busse, H.-J., Trujillo, M.E., Suzuki, K.-I., Ludwig, W., Whitman, W.B., Eds.; Springer: New York, NY, USA, 2012; Volume 5. [Google Scholar]
- Foesel, B.U.; Geppert, A.; Rohde, M.; Overmann, J. Parviterribacter kavangonensis gen. nov., sp. nov. and Parviterribacter multiflagellatus sp. nov., novel members of Parviterribacteraceae fam. nov. within the order Solirubrobacterales, and emended description of the classes Thermoleophilia and Rubrobacteria and their orders and families. Int. J. Syst. Evol. Microbiol. 2016, 66, 652–665. [Google Scholar] [PubMed]
- Goodfellow, M.; Williams, S.T. Ecology of actinomycetes. Annu. Rev. Microbiol. 1983, 37, 189–216. [Google Scholar] [CrossRef]
- Kochkina, G.A.; Ivanushkina, N.E.; Karasev, S.G.; Gavrish, E.; Gurina, L.V.; Evtushenko, L.I.; Spirina, E.V.; Vorob’eva, E.A.; Gilichinskiĭ, D.A.; Ozerskaia, S.M. Micromycetes and actinobacteria under conditions of many years of natural cryopreservation. Mikrobiologii 2001, 70, 412–420. [Google Scholar]
- Zenova, G.M.; Zvyagintsev, D.G. Diversity of Actinomycetes in Terrestrial Ecosystems; MSU: Moscow, Russia, 2002; p. 132. (In Russian) [Google Scholar]
- Polyanskaya, L.M.; Heidebrecht, V.V.; Stepanov, A.L.; Zvyagintsev, D.G. Distribution of the abundance and biomass of microorganisms according to the profiles of zonal soil types. Eurasian Soil Sci. 1995, 5, 566–572. (In Russian) [Google Scholar]
- Sorokina, L.E. Structure of the Complex of Soil Actinomycetes in Natural and Agroecosystems. Ph.D. Thesis, Moscow State University, Moscow, Russia, 1990. (In Russian). [Google Scholar]
- Guo, X.; Liu, N.; Li, X.; Ding, Y.; Shang, F.; Gao, Y.; Huang, Y. Red soils harbor diverse culturable actinomycetes that are promising sources of novel secondary metabolites. Appl. Environ. Microbiol. 2015, 81, 3086–3103. [Google Scholar] [CrossRef]
- Suksaard, P.; Pathomaree, W.; Duangmal, K. Diversity and plant growth promoting activities of actinomycetes from mangroves. Chiang Mai J. Sci. 2017, 44, 1210–1223. [Google Scholar]
- Qin, S.; Li, J.; Chen, H.H.; Zhao, G.Z.; Zhu, W.Y.; Jiang, C.L.; Li, W.J. Isolation, diversity, and antimicrobial activity of rare actinobacteria from medicinal plants of tropical rain forests in Xishuangbanna, China. Appl. Environ. Microbiol. 2009, 75, 6176–6186. [Google Scholar] [CrossRef] [PubMed]
- Semêdo, L.T.; Linhares, A.A.; Gomes, R.C.; Manfio, G.P.; Alviano, C.S.; Linhares, L.F. Isolation and characterization of actinomycetes from Brazilian tropical soils. Microbiol. Res. 2001, 155, 291–299. [Google Scholar] [CrossRef]
- Lejon, D.P.H.; Chaussod, R.; Ranger, J.; Ranjard, L. Microbial community structure and density under different tree species in an acid forest soil (Morvan, France). Microb. Ecol. 2005, 50, 614–625. [Google Scholar] [CrossRef] [PubMed]
- Hackl, E.; Pfeffer, M.; Donat, C.; Bachmann, G.; Zechmeister-Boltenstern, S. Composition of the microbial communities in the mineral soil under different types of natural forest. Soil Biol. Biochem. 2005, 37, 661–671. [Google Scholar] [CrossRef]
- Van Hop, D.; Sakiyama, Y.; Binh, C.T.T.; Otoguro, M.; Hang, D.T.; Miyadoh, S.; Luong, D.T.; Ando, K. Taxonomic and ecological studies of actinomycetes from Vietnam: Isolation and genus-level diversity. J. Antibiot. 2011, 64, 599. [Google Scholar] [CrossRef][Green Version]
- Dorchenkova, Y.A.; Gracheva, T.A.; Lysak, L.V. Characteristics of the complexes of actinomycetes in the Pu Hoat nature reserve. Eurasian Soil Sci. 2022, 55, 483–487. [Google Scholar] [CrossRef]
- Chernov, T.I.; Zhelezova, A.D.; Tkhakakhova, A.K. Microbiomes of virgin soils of southern Vietnam tropical forests. Microbiology 2019, 88, 489–498. [Google Scholar] [CrossRef]
- Lysak, L.V.; Lapygina, E.V.; Stoletov, G.A.; Gracheva, T.A.; Dorchenkova, Y.A.; Chernov, T.I. Characteristics of microbial communities of soils and plant litter of monsoon tropical forests of national parks in Vietnam. J. Trop. Sci. Technol. 2017, 14, 132–142. [Google Scholar]
- Weber, T.; Charusanti, P.; Musiol-Kroll, E.M.; Jiang, X.; Tong, Y.; Kim, H.U.; Lee, S.Y. Metabolic engineering of antibiotic factories: New tools for antibiotic production in actinomycetes. Trends Biotechnol. 2015, 33, 15–26. [Google Scholar] [CrossRef]
- Shirokikh, I.G.; Shirokikh, A.A.; Ashikhmina, T.Y. Assessing the antagonistic potential and antibiotic resistance of actinomycetes isolated from two zheltozems of southeastern China. Eurasian Soil Sci. 2018, 51, 857–864. [Google Scholar] [CrossRef]
- Eskov, A.K.; Abakumov, E.V.; Tiunov, A.V.; Kuznetsova, O.V.; Dubovikov, D.A.; Prilepsky, N.G.; Antipina, V.A.; Kuznetsov, A.N. 2017. Ageotropic aerial root-traps of nest epiphytes and their role in suspended soils forming. J. Gen. Biol. 2017, 78, 54–68. (In Russian) [Google Scholar]
- Gause, G.F.; Preobrazhenskaya, T.P.; Sveshnikova, M.A.; Terekhova, L.P.; Maksimova, T.S. A Guide for Determination of Actinomycetes; Nauka: Moscow, Russia, 1983; p. 245. (In Russian) [Google Scholar]
- Zvyagintsev, D.G. Methods of Soil Microbiology and Biochemistry; MSU: Moscow, Russia, 1991; pp. 13–18. (In Russian) [Google Scholar]
- Lysak, L.V.; Lihacheva, A.A.; Alferova, I.V. Methods for Isolation and Study of Soil Actinomycetes, Producers of Antibiotics; Max-Press: Moscow, Russia, 2005; pp. 15–16. (In Russian) [Google Scholar]
- Brunk, C.F.; Avaniss-Aghajani, E.; Brunk, C.A. A computer analysis of primer and probe hybridization potential with bacterial small-subunit rRNA sequences. Appl. Environ. Microbiol. 1996, 61, 872–879. [Google Scholar] [CrossRef] [PubMed]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef] [PubMed]
- Nagpal, S.; Haque, M.M.; Singh, R.; Mande, S.S. iVikodak—A Platform and standard workflow for inferring, analyzing, comparing, and visualizing the functional potential of microbial communities. Front. Microbiol. 2019, 9, 3336. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Berry, A.M.; Barabote, R.D.; Normand, P. The Family Acidothermaceae. In The Prokaryotes; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 13–19. [Google Scholar]
- Behrendt, U.; Ulrich, A.; Schumann, P.; Naumann, D.; Suzuki, K.I. Diversity of grass-associated Microbacteriaceae isolated from the phyllosphere and litter layer after mulching the sward; polyphasic characterization of Subtercola pratensis sp. nov., Curtobacterium herbarum sp. nov. and Plantibacter flavus gen. nov., sp. nov. Int. J. Syst. Evol. Microbiol. 2002, 52, 1441–1454. [Google Scholar]
- Jin, L.; Lee, H.-G.; Ko, S.-R.; Ahn, C.-Y.; Oh, H.-M. Jatrophihabitans fulvus sp. nov., an actinobacterium isolated from grass soil. Int. J. Syst. Evol. Microbiol. 2015, 65, 3476–3480. [Google Scholar] [CrossRef]
- Hamid, M.E.; Mahgoub, A.; Babiker, A.J.O.; Babiker, H.A.E.; Holie, M.A.I.; Elhassan, M.M.; Joseph, M.R.P. Isolation and identification of Streptomyces spp. from desert and savanna soils in Sudan. Int. J. Environ. Res. Public. Health 2020, 17, 8749. [Google Scholar] [CrossRef]
- Hashiguchi, Y.; Tezuka, T.; Mouri, Y.; Konishi, K.; Fujita, A.; Hirata, A.; Ohnishi, Y. Regulation of sporangium formation, spore dormancy, and sporangium dehiscence by a hybrid sensor histidine kinase in Actinoplanes missouriensis: Relationship with the global transcriptional regulator TcrA. J. Bacteriol. 2020, 202, e00228-20. [Google Scholar] [CrossRef]
- Kudo, T.; Matsushima, K.; Itoh, T.; Sasaki, J.; Suzuki, K. Description of four new species of the genus Kineosporia: Kineosporia succinea sp. nov., Kineosporia rhizophila sp. nov., Kineosporia mikuniensis sp. nov. and Kineosporia rhamnosa sp. nov., isolated from plant samples, and amended description of the genus Kineosporia. Int. J. Syst. Bacteriol. 1998, 48, 1245–1255. [Google Scholar] [CrossRef]
- Reddy, T.V.K.; Mahmood, S.; Paris, L.; Reddy, Y.H.K.; Wellington, E.M.H.; Idris, M.M. Streptomyces hyderabadensis sp. nov., an actinomycete isolated from soil. Int. J. Syst. Evol. Microbiol. 2011, 61, 76–80. [Google Scholar] [CrossRef] [PubMed]
- Taddei, A.; Rodríguez, M.J.; Márquez-Vilchez, E.; Castelli, C. Isolation and identification of Streptomyces spp. from Venezuelan soils: Morphological and biochemical studies. Microbiol. Res. 2006, 161, 222–231. [Google Scholar] [CrossRef] [PubMed]
- Yamano, Y.; Sugimoto, S.; Matsunami, K. Complete genome sequence of Actinoplanes sp. strain L3-i22, isolated from soil in Japan. Microbiol. Resour. Announc. 2021, 10, e0079821. [Google Scholar] [CrossRef] [PubMed]
- Poomthongdee, N.; Duangmal, K.; Pathomaree, W. Acidophilic actinomycetes from rhizosphere soil: Diversity and properties beneficial to plants. J. Antibiot. 2015, 68, 106–114. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Chen, X.; Cao, Y.; Ren, Z. Diversity of Cultivable Actinomycetes in Tropical Rainy Forest of Xishuangbanna, China. Open J. Soil Sci. 2013, 3, 9–14. [Google Scholar] [CrossRef]
- Mohanta, Y.K. Isolation of cellulose-degrading actinomycetes and evaluation of their cellulolytic potential. Bioeng. Biosci. 2014, 2, 1–5. [Google Scholar] [CrossRef]
- Jones, B.; Kleij Wilhelmus, A.; Van Solingen, P.; Weyler, W. Cellulase Producing Actinomycetes, Cellulase Produced there from and Method of Producing Same. EP 1408108 B1, 11 August 2004. [Google Scholar]
- Tamreihao, K.; Salam, N.; Ningthoujam, D.S. Use of acidophilic or acidotolerant actinobacteria for sustainable agricultural production in acidic soils. In Extremophiles in Eurasian Ecosystems: Ecology, Diversity, and Applications; Egamberdieva, D., Birkeland, N.K., Panosyan, H., Li, W.J., Eds.; Springer: Singapore, 2018; pp. 453–464. [Google Scholar]
- Tanaka, Y.; Omura, S. Agroactive compounds of microbial origin. Annu. Rev. Microbiol. 1993, 47, 57–87. [Google Scholar] [CrossRef]
- Doumbou, C.L.; Hamby Salove, M.K.; Crawford, D.L.; Beaulieu, C. Actinomycetes, promising tools to control plant diseases and to promote plant growth. Phytoprotection 2001, 82, 85–102. [Google Scholar] [CrossRef]



| Protected Area | Sampling Location (Altitude, m) | Soil Type (WRB) |
|---|---|---|
| Xuan Son National Park | Karst valley, broadleaf forest (580 m) | Calcisols |
| Mount Ten, broadleaf forest (1200 m) | Ferralsols | |
| Pu Hoat Nature Reserve | Broadleaf forest in river valley (845 m) | Fluvisols |
| Suspended soils | ||
| Tall-stemmed coniferous forest (1370 m) | Ferralsols | |
| Pu Mat National Park | Tall-stemmed forest in river valley (200 m) | Ferralsols |
| Kon Chu Rang Nature Reserve | Broadleaf forest (1000 m) | Ferralsols |
| Coniferous forest (1050 m) | Ferralsols | |
| Broadleaf forest (1025 m) | Ferralsols | |
| Suspended soils | ||
| Mixed forest (995 m) | Ferralsols | |
| Kon Plong protected forest | Tall-stemmed mixed forest (1400 m) | Ferralsols |
| Kon Ka Kinh National Park | Eastern part of the park, forest on slope (700 m) | Cambisols |
| Suspended soils | ||
| Forest on ridge (900 m) | Ferralsols | |
| Southwestern part of the park, tall-stemmed forest in valley (1000 m) | Ferralsols (Gleyic) | |
| Southwestern part of the park, mid-mountain tall-stemmed forest (1500 m) | Ferralsols | |
| Southwestern part of the park, low-stemmed forest (860 m) | Fluvisols | |
| Southwestern part of the park, mixed forest (1160 m) | Cambisols |
| Protected Area | Ecotope | Sample | Abundance of Actinomycetes, CFU/g | Length of Actinomycete Mycelium, m/g |
|---|---|---|---|---|
| Xuan Son National Park | Karst valley, broadleaf forest, dark brown ferrallitic soil | Litter | 1.0 × 108 | 1000 |
| Soil (0–20 cm) | 0.5 × 106 | 300 | ||
| Mount Ten, broadleaf forest, mountainous dark brown ferrallitic soil | Litter | 2.0 × 105 | 400 | |
| Soil (0–20 cm) | 0.6 × 105 | 800 | ||
| Pu Hoat Nature Reserve | River valley, broadleaf forest, alluvial brown soil | Litter | 9.2 × 106 | 800 |
| Soil (0–5 cm) | 9.3 × 105 | 178 | ||
| Soil (5–20 cm) | 3.1 × 105 | 406 | ||
| River valley, broadleaf forest, suspended soil | Soil from epiphyte baskets | 6.0 × 105 | 580 | |
| Ridge slope, tall-stemmed forest, mountainous red-yellow humus–ferrallitic soil | Litter | 1.1 × 105 | 800 | |
| Soil (0– 5 cm) | 2.0 × 105 | 200 | ||
| Soil (5–20 cm) | 2.0 × 104 | 406 | ||
| Pu Mat National Park | River valley, tall-stemmed forest, red-yellow humus–ferrallitic soil | Litter | 1.4 × 105 | 820 |
| Soil (0–5 cm) | 8.5 × 105 | 240 | ||
| Soil (5–20 cm) | 1.1 × 106 | 380 | ||
| Kon Chu Rang Nature Reserve | Broadleaf forest, mountainous red-yellow humus–ferrallitic soil | Litter | 2.0 × 104 | 200 |
| Soil (0–5 cm) | 2.0 × 104 | 280 | ||
| Soil (5–20 cm) | 4.1 × 105 | 360 | ||
| Broadleaf forest, suspended soil | Soil from epiphyte baskets | 2.1 × 105 | 680 | |
| Coniferous forest, red-yellow humus–ferrallitic soil | Litter | 0.7 × 106 | 100 | |
| Soil (0–20 cm) | 1.6 × 106 | 400 | ||
| Mixed forest, mountainous red-yellow humus–ferrallitic soil | Litter | 0.7 × 105 | 800 | |
| Soil (0–20 cm) | 1.5 × 105 | 300 | ||
| Kon Plong | Tall-stemmed mixed forest, mountainous red-yellow humus–ferrallitic soil | Litter | 3.9 × 106 | 400 |
| Soil (0–20 cm) | 1.8 × 106 | 240 | ||
| Kon Ka Kinh National Park | Eastern part of the park, tall-stemmed forest on a slope, brown tropical thin sandy loam | Litter | 6.1 × 106 | 126 |
| Soil (0–5 cm) | 5.2 × 106 | 170 | ||
| Soil (5–20 cm) | 6.1 × 106 | n.d. | ||
| Eastern part of the park, tall-stemmed forest, suspended soil | Soil from epiphyte baskets | 2.1 × 106 | n.d. | |
| Eastern part of the park, tall-stemmed forest on the ridge, mountainous red-yellow humus–ferrallitic soil | Litter | 4.0 × 105 | 101 | |
| Soil (0–5 cm) | 5.3 × 104 | 50 | ||
| Soil (5–20 cm) | 2.0 × 104 | 75 | ||
| Valley tall-stemmed forest, hydromorphic dark humus–ferrallitic soil | Litter | 3.5 × 106 | 155 | |
| Soil (0–20 cm) | 1.9 × 106 | 160 | ||
| Mid-mountain tall-stemmed forest, mountainous red-yellow humus–ferrallitic soil | Litter | 6.9 × 106 | 165 | |
| Soil (0–20 cm) | 0.6 × 105 | 160 | ||
| Southwestern part of the park, low-stemmed forest, alluvial brown sandy loamy soil | Litter | 3.3 × 105 | 332 | |
| Soil (0–5 cm) | 1.4 × 105 | 252 | ||
| Soil (5–20 cm) | 8.7 × 104 | 503 | ||
| Southwestern part of the park, mixed forest, brown tropical thin clay soil | Litter | 3.3 × 105 | 40 | |
| Soil (0–5 cm) | 8.0 × 104 | n.d. | ||
| Soil (5–20 cm) | 2.0 × 105 | n.d. |
| Library Name | Number of Reads | Number of OTUs | Goods Coverage, % | Chao1 Index | Simpson Index | Shannon Diversity Index |
|---|---|---|---|---|---|---|
| Alluvial brown soil (B1) | 1471 | 1196 | 30 | 6086.2 | 0.02 | 6.96 |
| Litter (B2) | 1286 | 1214 | 10 | 17191.6 | 0.01 | 7.07 |
| Soil from epiphyte baskets (B3) | 1887 | 1569 | 28 | 8526.4 | 0.01 | 7.27 |
| Class | Order | Family | Content, % |
|---|---|---|---|
| Actinomycetia | Frankiales | Acidothermaceae | 2 |
| Frankiaceae | 0.1 | ||
| Sporichthyaceae | 0.05 | ||
| Corynebacteriales | Mycobacteriaceae | 0.5 | |
| Nocardiaceae | 0.2 | ||
| Corynebacteriaceae | 0.01 | ||
| Micromonosporales | Micromonosporaceae | 0.7 | |
| Streptomycetales | Streptomycetaceae | 0.6 | |
| Streptosporangiales | Streptosporangiaceae | 0.2 | |
| Thermomonosporaceae | 0.2 | ||
| Pseudonocardiales | Pseudonocardiaceae | 0.2 | |
| Micrococcales | Micrococcaceae | 0.1 | |
| Intrasporangiaceae | 0.07 | ||
| Propionibacteriales | Nocardioidaceae | 0.04 | |
| Propionibacteriaceae | 0.03 | ||
| Catenulisporales | Actinospicaceae | 0.01 | |
| Thermoleophilia | Solirubrobacterales | Solirubrobacteraceae | 0.02 |
| TM146 | 0.9 | ||
| YNPFFP1 | 0.2 | ||
| Elev-16S-1332 | 0.03 | ||
| Gaiellales | Gaiellaceae | 0.03 | |
| uncultured | 0.8 | ||
| Acidimicrobiia | Acidimicrobiales | Acidimicrobiaceae | 0.09 |
| uncultured | 1 |
| Class | Order | Family | Content, % |
|---|---|---|---|
| Actinomycetia | Frankiales | Acidothermaceae | 0.2 |
| Frankiaceae | 0.4 | ||
| Cryptosporangiaceae | 0.09 | ||
| Sporichthyaceae | 0.05 | ||
| Kineosporiales | Kineosporiaceae | 0.6 | |
| Corynebacteriales | Mycobacteriaceae | 0.5 | |
| Nocardiaceae | 0.05 | ||
| Micromonosporales | Micromonosporaceae | 0.9 | |
| Streptomycetales | Streptomycetaceae | 0.7 | |
| Micrococcales | Microbacteriaceae | 0.9 | |
| Cellulomonadaceae | 0.4 | ||
| Promicromonosporaceae | 0.1 | ||
| Propionibacteriales | Nocardioidaceae | 0.2 | |
| Thermoleophilia | Solirubrobacterales | Solirubrobacteraceae | 0.05 |
| Conexibacteraceae | 0.1 | ||
| Patulibacteraceae | 0.05 | ||
| TM146 | 0.2 | ||
| uncultured | 0.05 | ||
| Elev-16S-1332 | 0.3 | ||
| Gaiellales | Gaiellaceae | 0.05 | |
| Acidimicrobiia | Acidimicrobiales | Acidimicrobiaceae | 0.4 |
| Iamiaceae | 0.1 | ||
| uncultured | 0.2 | ||
| AcidimicrobialesIncertae Sedis | 0.07 |
| Class | Order | Family | Content, % |
|---|---|---|---|
| Actinomycetia | Frankiales | Acidothermaceae | 3 |
| Frankiaceae | 0.7 | ||
| uncultured | 0.2 | ||
| Sporichthyaceae | 0.1 | ||
| Streptosporangiales | Thermomonosporaceae | 0.5 | |
| Streptosporangiaceae | 0.08 | ||
| Kineosporiales | Kineosporiaceae | 0.2 | |
| Corynebacteriales | Mycobacteriaceae | 0.04 | |
| Corynebacteriaceae | 0.04 | ||
| Micromonosporales | Micromonosporaceae | 0.5 | |
| Pseudonocardiales | Pseudonocardiaceae | 0.4 | |
| Streptomycetales | Streptomycetaceae | 0.3 | |
| Micrococcales | Microbacteriaceae | 0.05 | |
| Propionibacteriales | Nocardioidaceae | 0.5 | |
| Thermoleophilia | Solirubrobacterales | Solirubrobacteraceae | 0.09 |
| Patulibacteraceae | 0.1 | ||
| TM146 | 0.5 | ||
| YNPFFP1 | 0.1 | ||
| Gsoil-1167 | 0.02 | ||
| Elev-16S-1332 | 0.7 | ||
| Gaiellales | Gaiellaceae | 0.2 | |
| uncultured | 1 | ||
| Acidimicrobiia | Acidimicrobiales | Acidimicrobiaceae | 0.3 |
| Iamiaceae | 0.09 | ||
| uncultured | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dorchenkova, Y.A.; Gracheva, T.A.; Babich, T.L.; Sokolova, D.S.; Alexandrova, A.V.; Pham, G.T.H.; Lysak, L.V.; Golovchenko, A.V.; Manucharova, N.A. Soil Actinomycetes of Vietnam Tropical Forests. Forests 2022, 13, 1863. https://doi.org/10.3390/f13111863
Dorchenkova YA, Gracheva TA, Babich TL, Sokolova DS, Alexandrova AV, Pham GTH, Lysak LV, Golovchenko AV, Manucharova NA. Soil Actinomycetes of Vietnam Tropical Forests. Forests. 2022; 13(11):1863. https://doi.org/10.3390/f13111863
Chicago/Turabian StyleDorchenkova, Yuliya A., Tatiana A. Gracheva, Tamara L. Babich, Diyana Sh. Sokolova, Alina V. Alexandrova, Giang T. H. Pham, Lyudmila V. Lysak, Alla V. Golovchenko, and Natalia A. Manucharova. 2022. "Soil Actinomycetes of Vietnam Tropical Forests" Forests 13, no. 11: 1863. https://doi.org/10.3390/f13111863
APA StyleDorchenkova, Y. A., Gracheva, T. A., Babich, T. L., Sokolova, D. S., Alexandrova, A. V., Pham, G. T. H., Lysak, L. V., Golovchenko, A. V., & Manucharova, N. A. (2022). Soil Actinomycetes of Vietnam Tropical Forests. Forests, 13(11), 1863. https://doi.org/10.3390/f13111863






