Occurrence Prediction of Pine Wilt Disease Based on CA–Markov Model
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Data Acquisition
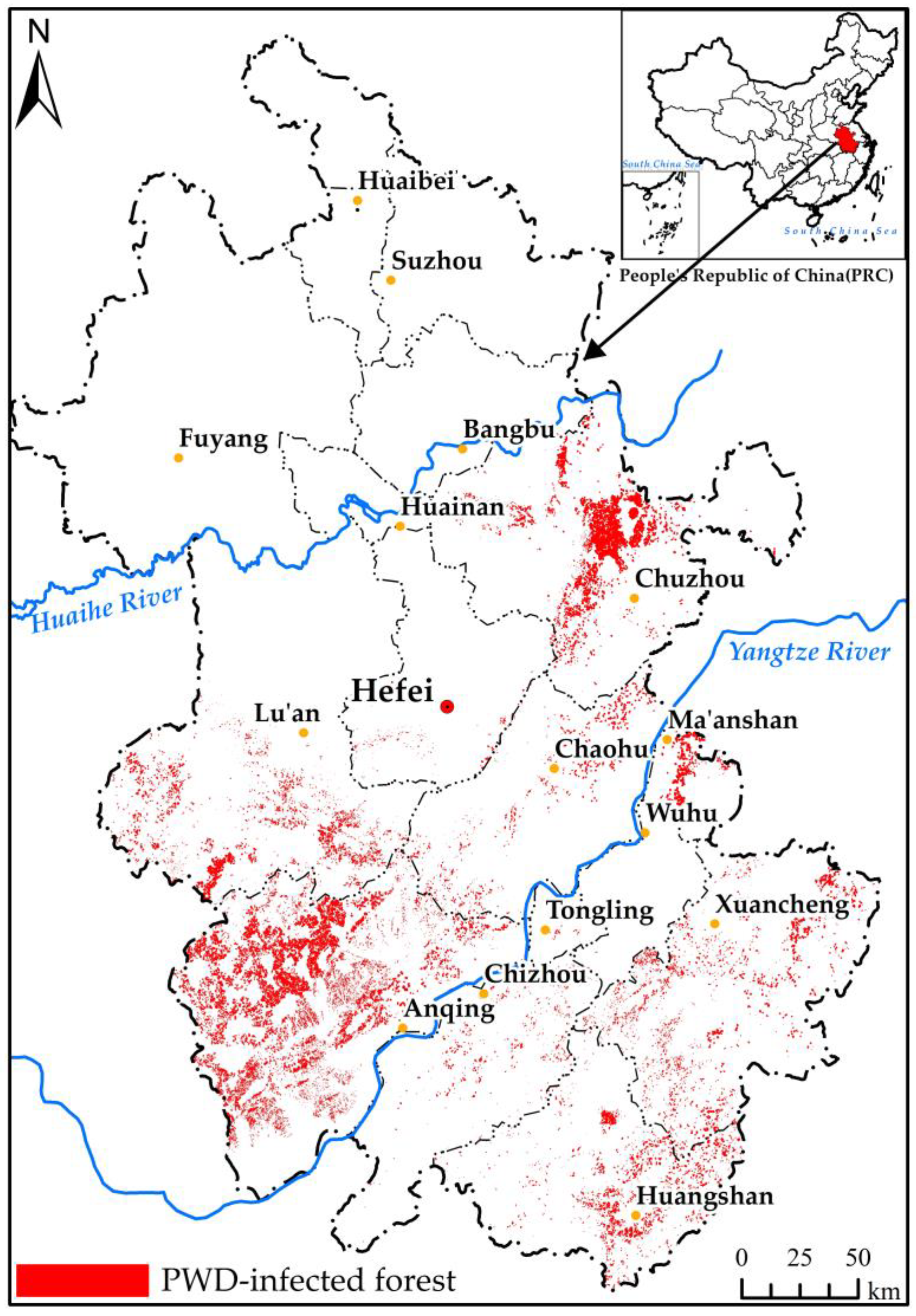
2.2.1. Distribution of Pine Forest and PWD Data
2.2.2. Normalized Vegetation Index (NDVI)
2.2.3. DEM
2.2.4. Meteorological Data
2.2.5. Population Distribution Data
2.2.6. Road Distribution Data
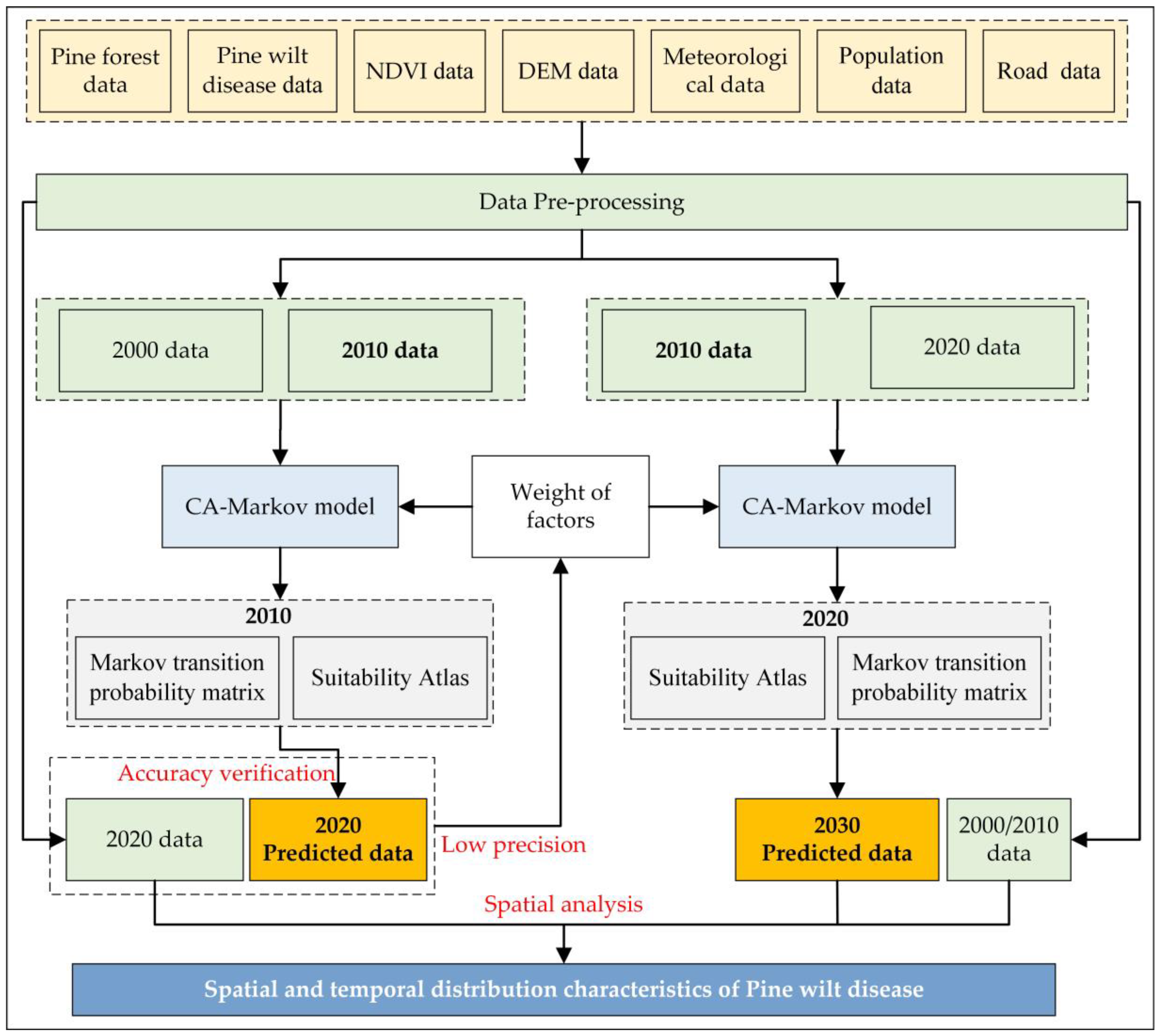
2.3. Methodology
2.3.1. CA–Markov Model
- (1)
- Markov Transition Matrix
- (2)
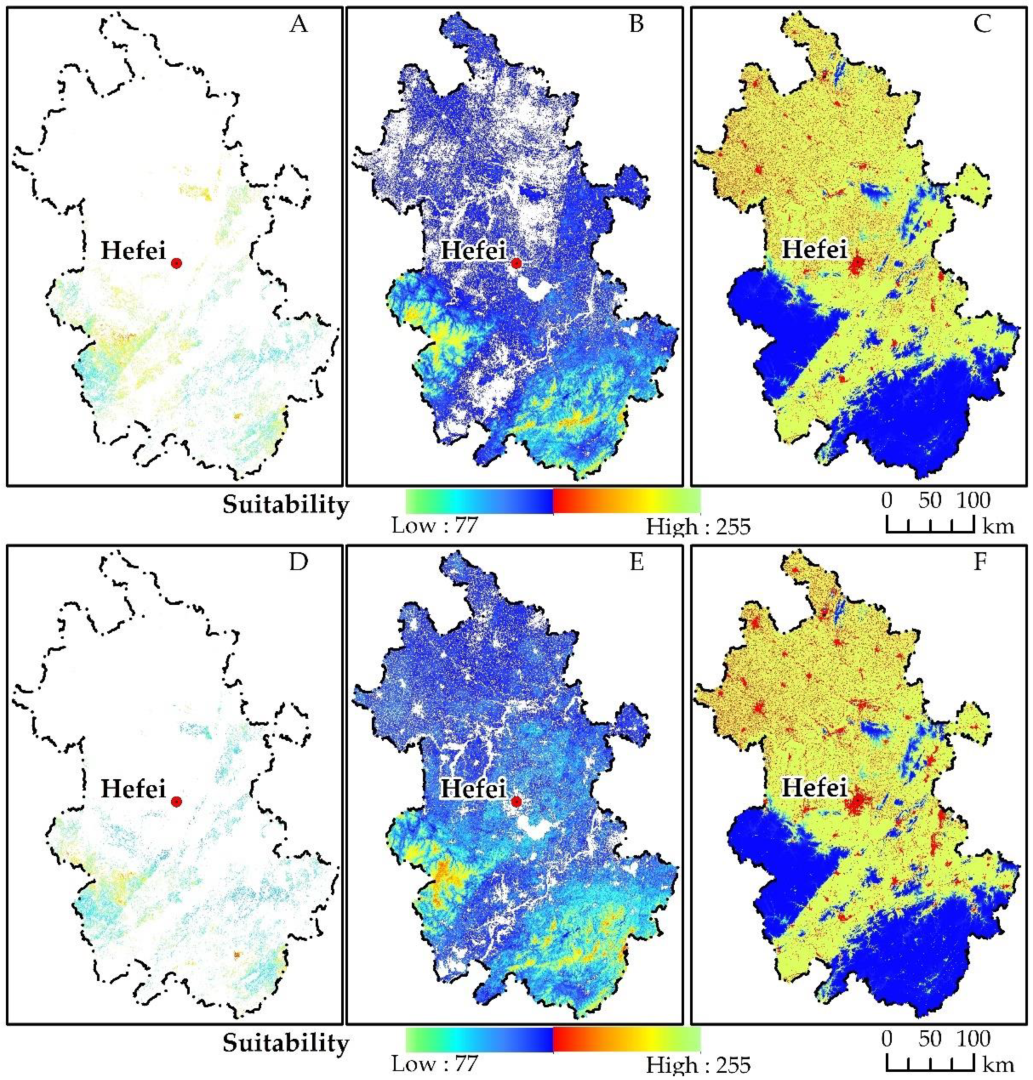
- Designation of suitability Atlas
- (3)
- CA–Markov Prediction
- (4)
- Simulation Accuracy Verification
2.3.2. Directional Distribution
2.3.3. Spatial Autocorrelation Analysis Method
2.3.4. Topographic Analysis
3. Results
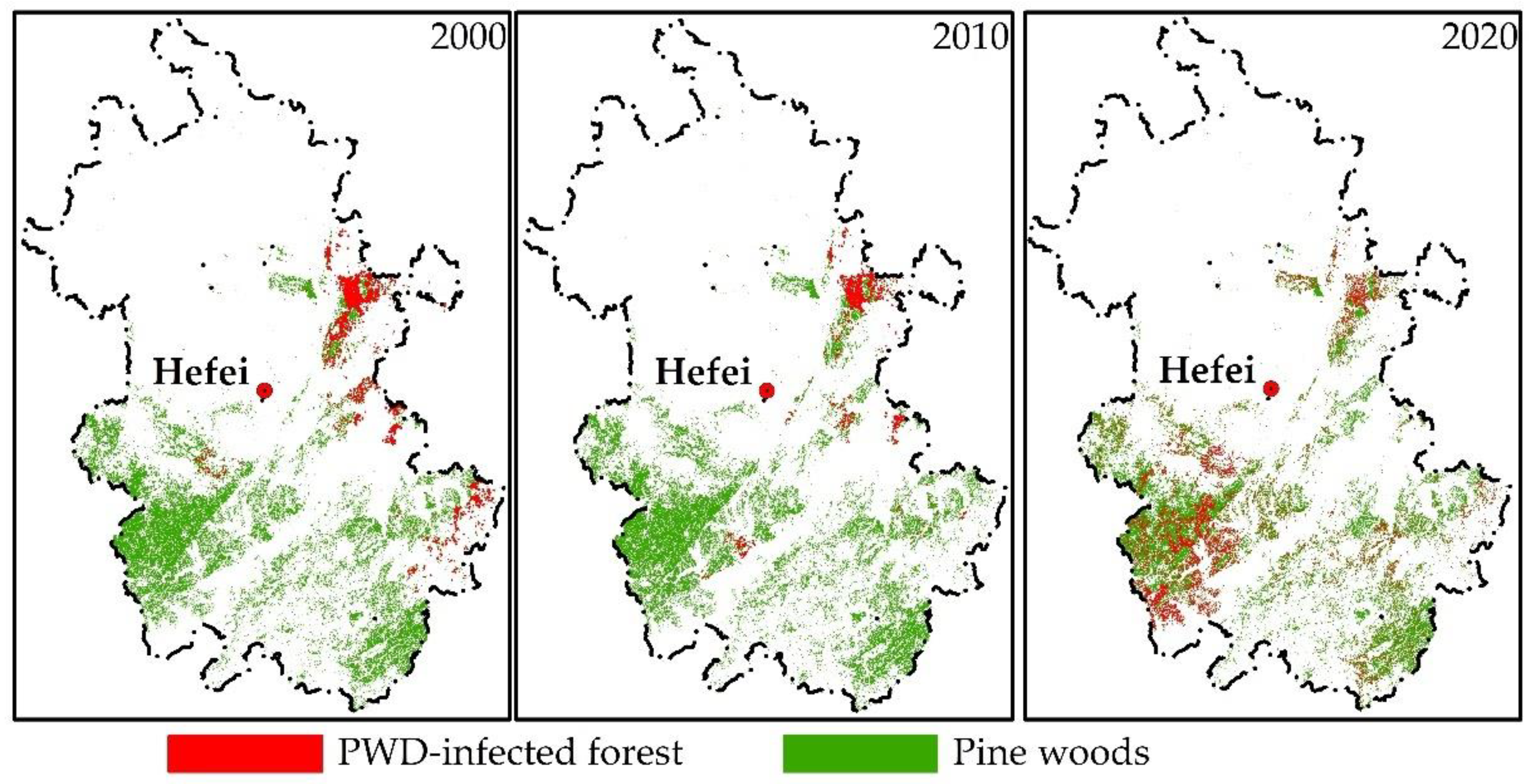
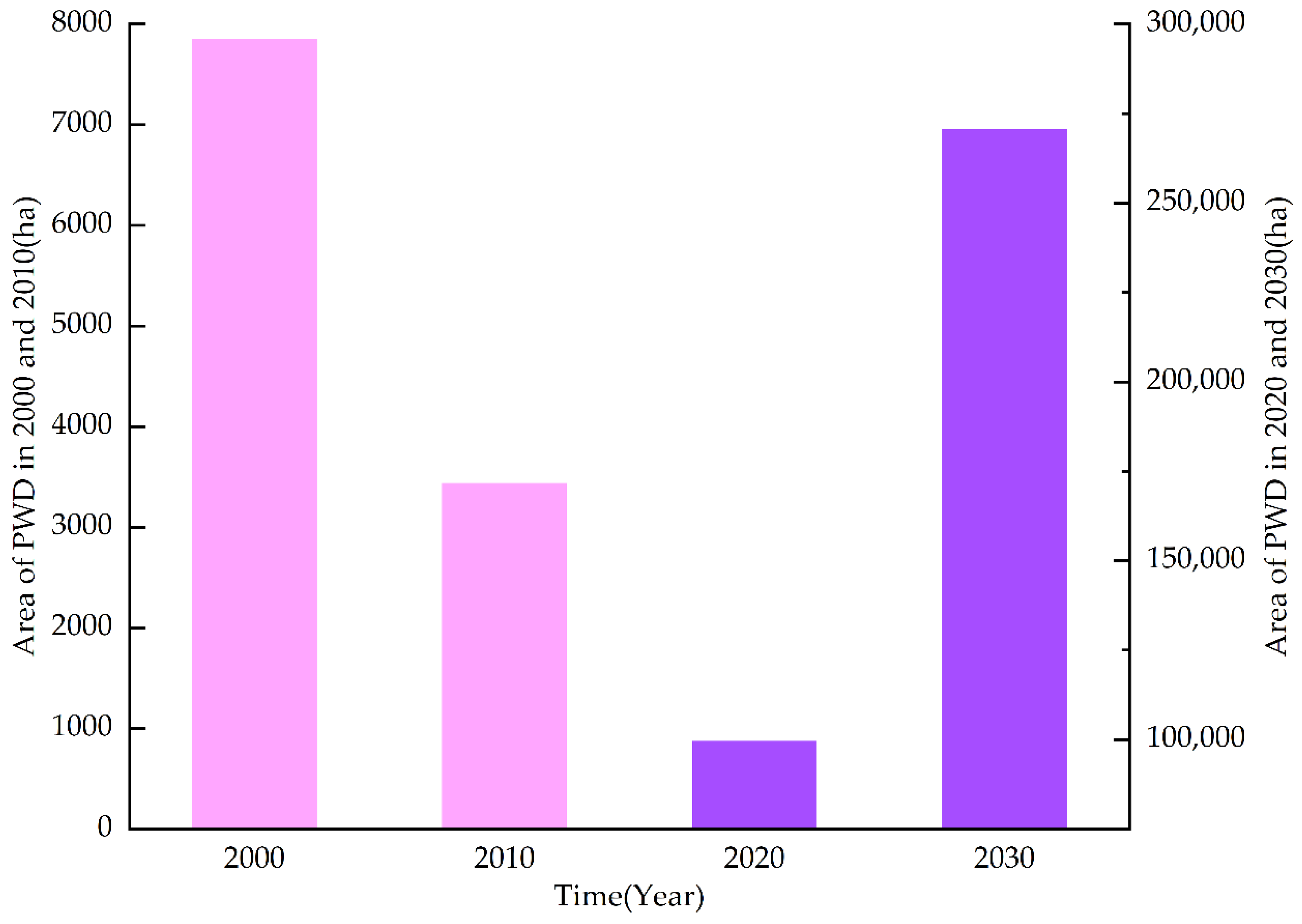
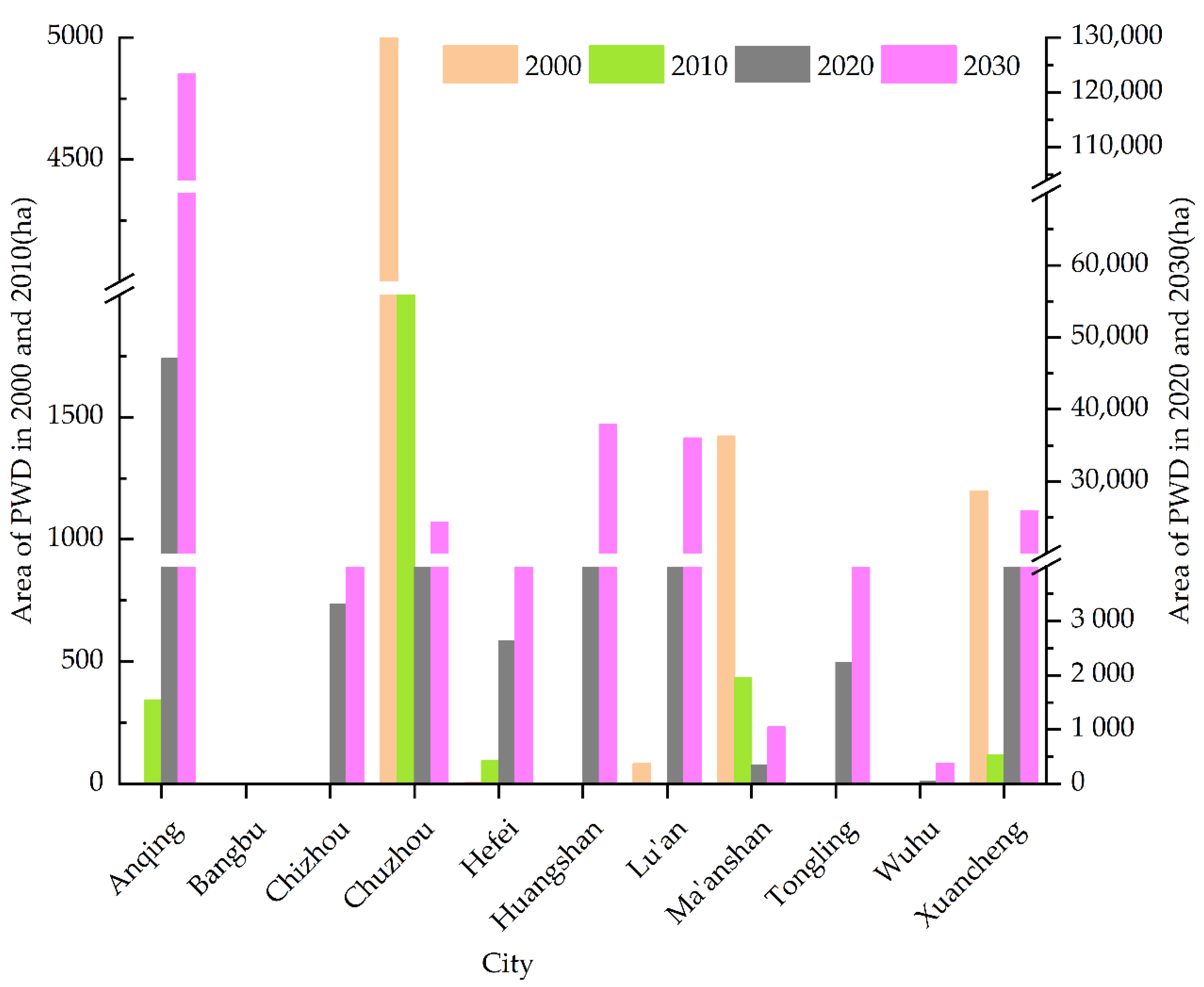
3.1. Spatiotemporal Dynamic Changes in PWD
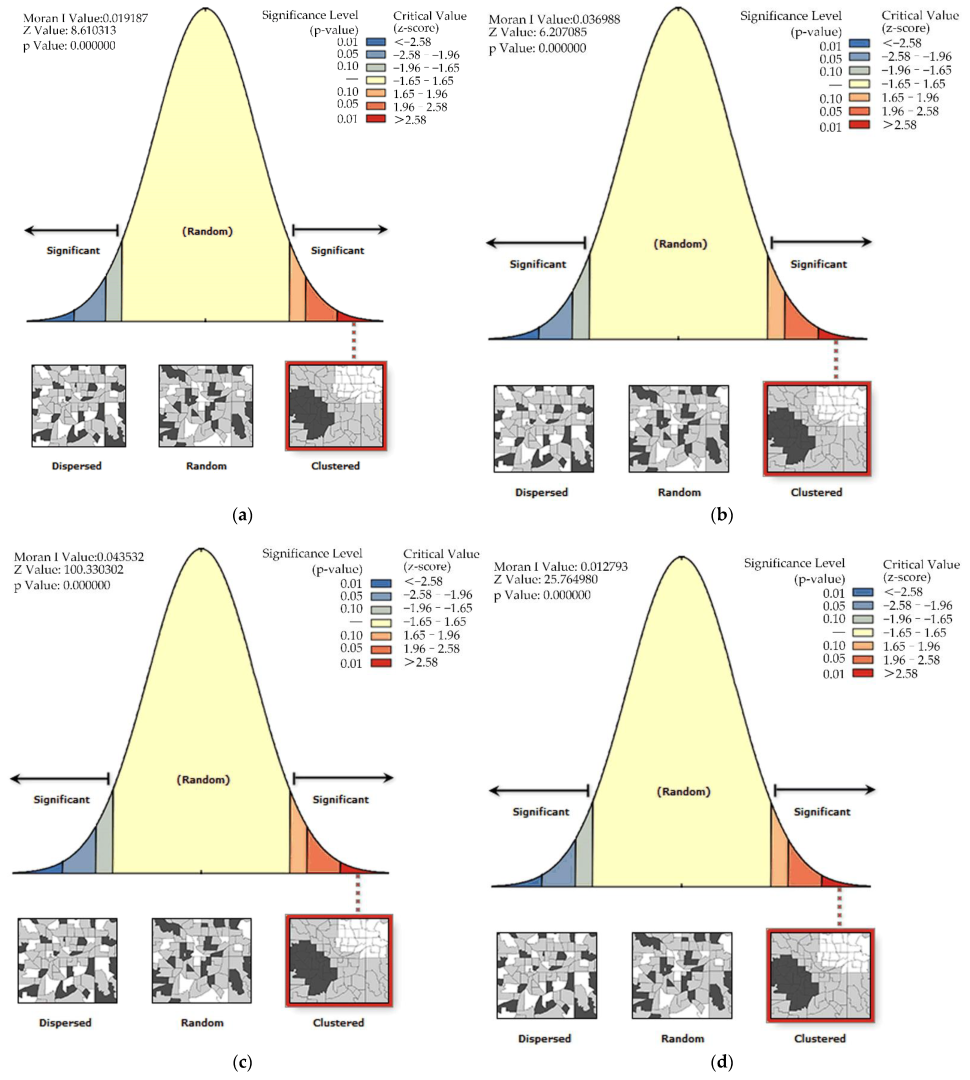
3.2. Spatial Autocorrelation Characteristics of PWD
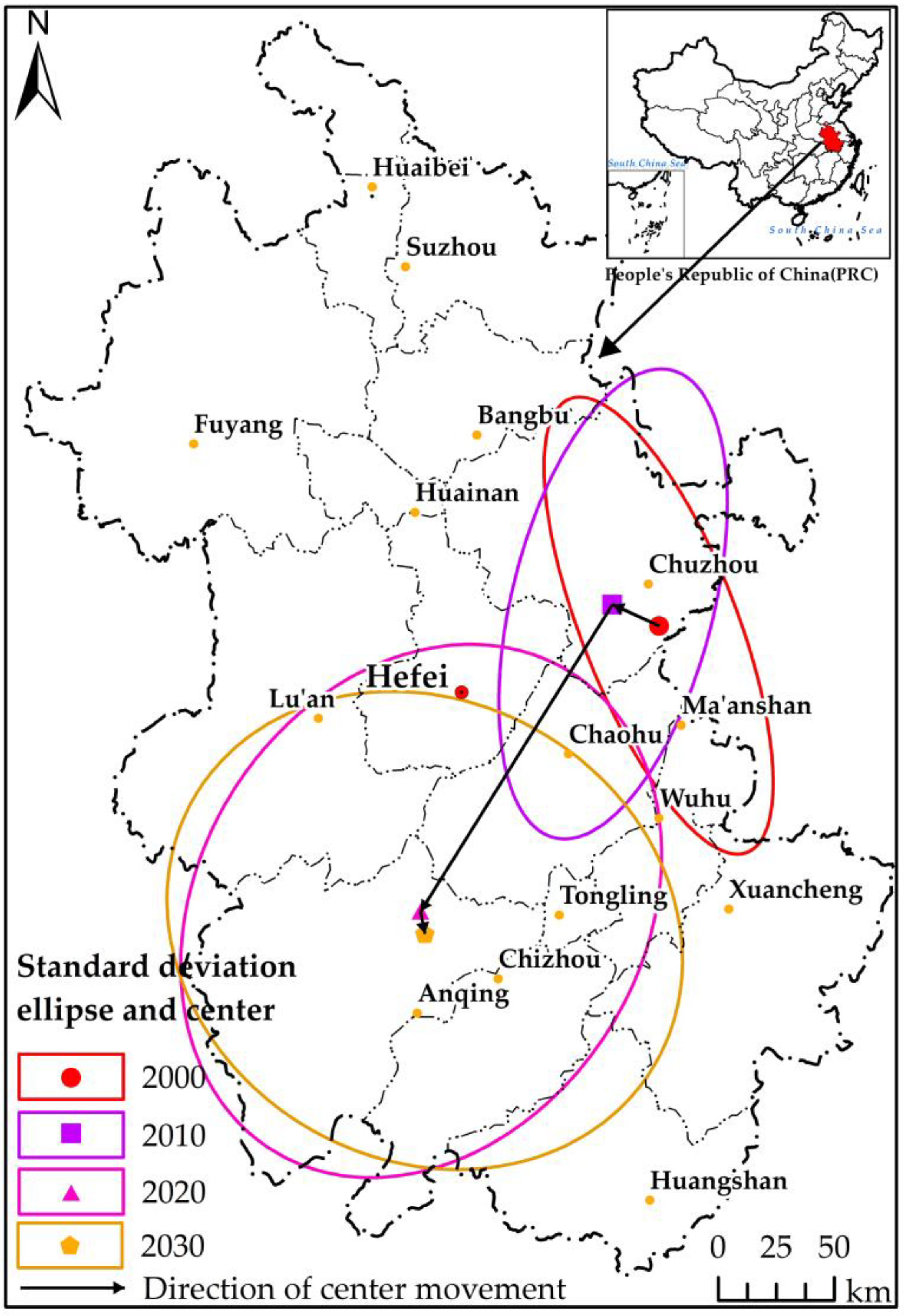
3.3. Characteristics of Directional Distribution and Center Movement of PWD
3.4. Distribution Characteristics of PWD According to Topographic Conditions
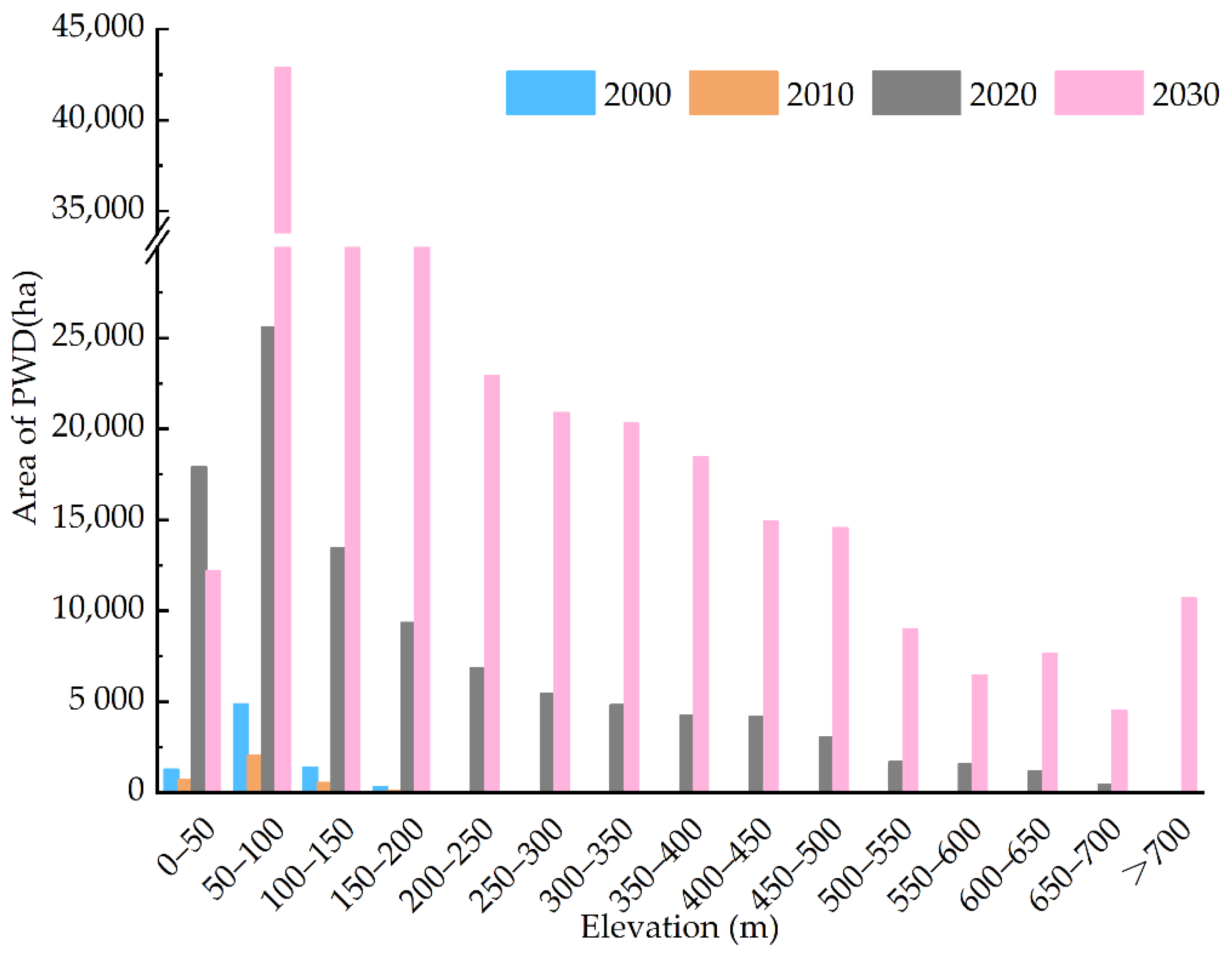
3.4.1. PWD Distribution along Elevation
3.4.2. PWD Distribution along Slope Gradient
3.4.3. Distribution of PWD along Aspect
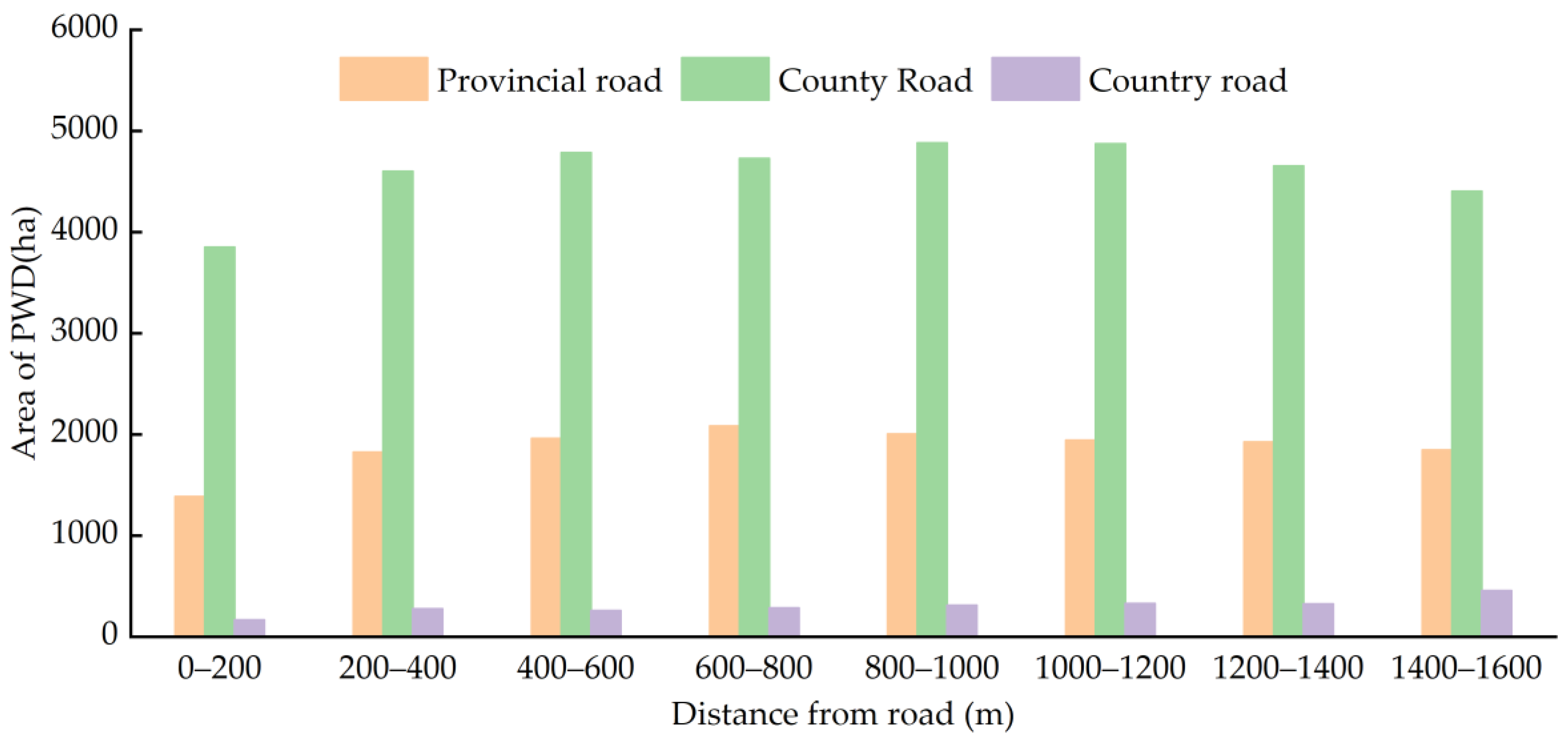
3.5. Relationship between the Occurrence Area and Road of PWD
4. Discussion
4.1. Research Contributions
4.2. Limitations and Prospects
4.3. Countermeasures and Suggestions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Olsson, P.; Heliasz, M.; Jin, H.; Eklundh, L. Mapping the reduction in gross primary productivity in subarctic birch forests due to insect outbreaks. Biogeosciences 2017, 14, 1703–1719. [Google Scholar] [CrossRef]
- Bergdahl, D.R. Impact of pinewood nematode in North America: Present and future. J. Nematol. 1988, 20, 260–265. [Google Scholar] [PubMed]
- Li, Y.; Zhang, X. Analysis on the trend of invasion and expansion of Bursaphelenchus xylophilus. For. Pest Dis. 2018, 37, 1–4. [Google Scholar]
- State Forestry and Grassland Administration. 2022 Announcement of Pine Wood Nematode Epidemic Area. Available online: http://www.gzhs.gov.cn/zfbm/hsxlyj_5721086/zcwj_5721053/202205/t20220511_73990365.html (accessed on 29 July 2022).
- Gent, D.H.; Schwartz, H.F. Validation of potato early blight disease forecast models for Colorado using various sources of meteorological data. Plant Dis. 2003, 87, 78–84. [Google Scholar] [CrossRef] [PubMed]
- Roubal, C.; Regis, S.; Nicot, P.C. Field models for the prediction of leaf infection and latent period of Fusicladium oleagineum on olive based on rain, temperature and relative humidity. Plant Pathol. 2013, 62, 657–666. [Google Scholar] [CrossRef]
- Kumar, A.; Agrawal, R.; Chattopadhyay, C. Weather based forecast models for diseases in mustard crop. Mausam 2013, 64, 663–670. [Google Scholar] [CrossRef]
- Valdez-Torres, J.B.; Soto-Landeros, F.; Osuna-Enciso, T.; Baez-Sanudo, M.A. Phenological prediction models for white corn (Zea mays L.) and fall armyworm (Spodoptera frugiperda J.E. Smith). Agrociencia 2012, 46, 399–410. [Google Scholar]
- Schwartz, M.W. Potential effects of global climate change on the biodiversity of plants. For. Chron. 1992, 68, 462–471. [Google Scholar] [CrossRef]
- Laderach, P.; Ramirez-Villegas, J.; Navarro-Racines, C.; Zelaya, C.; Martinez-Valle, A.; Jarvis, A. Climate change adaptation of coffee production in space and time. Clim. Chang. 2017, 141, 47–62. [Google Scholar] [CrossRef]
- Dukes, J.S.; Pontius, J.; Orwig, D.; Garnas, J.R.; Rodgers, V.L.; Brazee, N.; Cooke, B.; Theoharides, K.A.; Stange, E.E.; Harrington, R.; et al. Responses of insect pests, pathogens, and invasive plant species to climate change in the forests of northeastern North America: What can we predict? Can. J. For. Res. 2009, 39, 231–248. [Google Scholar] [CrossRef]
- Brown, N.; Vanguelova, E.; Parnell, S.; Broadmeadow, S.; Denman, S. Predisposition of forests to biotic disturbance: Predicting the distribution of Acute Oak Decline using environmental factors. For. Ecol. Manag. 2018, 407, 145–154. [Google Scholar] [CrossRef]
- Giliba, R.A.; Mpinga, I.H.; Ndimuligo, S.A.; Mpanda, M.M. Changing climate patterns risk the spread of Varroa destructor infestation of African honey bees in Tanzania. Ecol. Process. 2020, 9, 48. [Google Scholar] [CrossRef]
- Guru-Pirasanna-Pandi, L.G.; Choudhary, J.S.; Chemura, A.; Basana-Gowda, G.; Annamalai, M.; Patil, N.; Adak, T.; Rath, P.C. Predicting the brown planthopper, Nilaparvata lugens (Stal) (Hemiptera: Delphacidae) potential distribution under climatic change scenarios in India. Curr. Sci. India 2021, 121, 1600–1609. [Google Scholar] [CrossRef]
- Bosso, L.; Di Febbraro, M.; Cristinzio, G.; Zoina, A.; Russo, D. Shedding light on the effects of climate change on the potential distribution of Xylella fastidiosa in the Mediterranean basin. Biol. Invasions 2016, 18, 1759–1768. [Google Scholar] [CrossRef]
- Bosso, L.; Russo, D.; Di Febbraro, M.; Cristinzio, G.; Zoina, A. Potential distribution of Xylella fastidiosa in Italy: A maximum entropy model. Phytopathol. Mediterr. 2016, 55, 62–72. [Google Scholar] [CrossRef]
- Poutsma, J.; Loomans, A.; Aukema, B.; Heijerman, T. Predicting the potential geographical distribution of the harlequin ladybird, Harmonia axyridis, using the CLIMEX model. BioControl 2008, 53, 103–125. [Google Scholar] [CrossRef]
- Phillips, S.J.; Anderson, R.P.; Schapire, R.E. Maximum entropy modeling of species geographic distributions. Ecol. Model. 2006, 190, 231–259. [Google Scholar] [CrossRef]
- Peterson, A.T.; Papes, M.; Eaton, M. Transferability and model evaluation in ecological niche modeling: A comparison of GARP and Maxent. Ecography 2007, 30, 550–560. [Google Scholar] [CrossRef]
- Beaumont, L.J.; Hughes, L.; Poulsen, M. Predicting species distributions: Use of climatic parameters in BIOCLIM and its impact on predictions of species’ current and future distributions. Ecol. Model. 2005, 186, 250–269. [Google Scholar] [CrossRef]
- Kelly, M.; Meentemeyer, R.K. Landscape dynamics of the spread of sudden oak death. Photogramm. Eng. Remote Sens. 2002, 68, 1001–1010. [Google Scholar]
- Volpi, I.; Guidotti, D.; Mammini, M.; Marchi, S. Predicting symptoms of downy mildew, powdery mildew, and gray mold diseases of grapevine through machine learning. Ital. J. Agrometeorol. 2021, 2, 57–69. [Google Scholar] [CrossRef]
- BenDor, T.K.; Metcalf, S.S.; Fontenot, L.E.; Sangunett, B.; Hannon, B. Modeling the spread of the emerald ash borer. Ecol. Model. 2006, 197, 221–236. [Google Scholar] [CrossRef]
- Howell, B.E.; Burns, K.S.; Kearns, H.S. Biological Evaluation of a Model for Predicting Presence of White Pine Blister Rust in Colorado Based on Climatic Variables and Susceptible White Pine Species Distribution: USDA Forest Service, Rocky Mountain Region, Renewable Resources. 2006. Available online: http://www.fs.usda.gov/Internet/FSE_DOCUMENTS/fsbdev3_039457.pdf (accessed on 20 September 2022).
- Wang, S.H.; Dai, J.G.; Zhao, Q.Z.; Cui, M.N. Application of grey systems in predicting the degree of cotton spider mite infestations. Grey Syst. Theory Appl. 2017, 7, 353–364. [Google Scholar] [CrossRef]
- Aparecido, L.; Rolim, G.D.; De Moraes, J.; Costa, C.; de Souza, P.S. Machine learning algorithms for forecasting the incidence of Coffea arabica pests and diseases. Int. J. Biometeorol. 2020, 64, 671–688. [Google Scholar] [CrossRef]
- Fabre, F.; Pierre, J.S.; Dedryver, C.A.; Plantegenest, M. Barley yellow dwarf disease risk assessment based on Bayesian modelling of aphid population dynamics. Ecol. Model. 2006, 193, 457–466. [Google Scholar] [CrossRef]
- Hanks, E.M.; Hooten, M.B.; Baker, F.A. Reconciling multiple data sources to improve accuracy of large-scale prediction of forest disease incidence. Ecol. Appl. 2011, 21, 1173–1188. [Google Scholar] [CrossRef] [PubMed]
- Iordache, M.; Mantas, V.; Baltazar, E.; Pauly, K.; Lewyckyj, N. A machine learning approach to detecting pine wilt disease using airborne spectral imagery. Remote Sens. 2020, 12, 2280. [Google Scholar] [CrossRef]
- Wu, W.; Zhang, Z.; Zheng, L.; Han, C.; Wang, X.; Xu, J.; Wang, X. Research Progress on the Early Monitoring of Pine Wilt Disease Using Hyperspectral Techniques. Sensors 2020, 20, 3729. [Google Scholar] [CrossRef]
- Lee, D.; Choi, W.I.; Nam, Y.; Park, Y. Predicting potential occurrence of pine wilt disease based on environmental factors in South Korea using machine learning algorithms. Ecol. Inform. 2021, 64, 101378. [Google Scholar] [CrossRef]
- Han, Y.; Wang, Y.; Xiang, Y.; Ye, J. Prediction of potential distribution of Bursaphelenchus xylophilus in China based on Maxent ecological niche model. J. Nanjing For. Univ. (Nat. Sci. Ed.) 2015, 39, 6–10. [Google Scholar] [CrossRef]
- Li, M.Y.; Liu, M.L.; Liu, M.; Ju, Y.W. Prediction of Pine Wilt Disease in Jiangsu Province Based on Web Dataset and GIS. In Web Information Systems and Mining; Springer: Berlin/Heidelberg, Germany, 2010; Volume 6318, pp. 146–153. [Google Scholar]
- Ju, Y.; Li, M.; Wu, W. Predictive Methods of Pine Wilt Disease in Jiangsu Province. Sci. Silvae Sin. 2010, 46, 91–96. [Google Scholar]
- He, S.; Wen, J.; Luo, Y.; Zong, S.; Zhao, Y.; Han, J. The predicted geographical distribution of Bursaphelenchus xylophilus in China under climate warming. Chin. J. Appl. Entomol. 2012, 49, 236–243. [Google Scholar]
- Zhao, J.; Han, X.; Shi, J. Potential distribution of Bursaphelenchus xylophilus in China due to adaptation cold conditions. J. Biosaf. 2017, 26, 191–198. [Google Scholar]
- Gruffudd, H.R.; Jenkins, T.A.R.; Evans, H.F. Using an evapo-transpiration model (ETpN) to predict the risk and expression of symptoms of pine wilt disease (PWD) across Europe. Biol. Invasions 2016, 18, 2823–2840. [Google Scholar] [CrossRef]
- An, H.; Lee, S.; Cho, S.J. The Effects of Climate Change on Pine Wilt Disease in South Korea: Challenges and Prospects. Forests 2019, 10, 486. [Google Scholar] [CrossRef]
- Hirata, A.; Nakamura, K.; Nakao, K.; Kominami, Y.; Tanaka, N.; Ohashi, H.; Takano, K.T.; Takeuchi, W.; Matsui, T. Potential distribution of pine wilt disease under future climate change scenarios. PLoS ONE 2017, 12, e0182837. [Google Scholar] [CrossRef]
- de la Fuente, B.; Saura, S.; Beck, P. Predicting the spread of an invasive tree pest: The pine wood nematode in Southern Europe. J. Appl. Ecol. 2018, 55, 2374–2385. [Google Scholar] [CrossRef]
- de la Fuente, B.; Saura, S. Long-Term Projections of the Natural Expansion of the Pine Wood Nematode in the Iberian Peninsula. Forests 2021, 12, 849. [Google Scholar] [CrossRef]
- Hao, Z.; Fang, G.; Huang, W.; Ye, H.; Zhang, B.; Li, X. Risk Prediction and Variable Analysis of Pine Wilt Disease by a Maximum Entropy Model. Forests 2022, 13, 342. [Google Scholar] [CrossRef]
- Tang, X.; Yuan, Y.; Li, X.; Zhang, J. Maximum Entropy Modeling to Predict the Impact of Climate Change on Pine Wilt Disease in China. Front. Plant Sci. 2021, 12, 652500. [Google Scholar] [CrossRef]
- Nguyen, T.V.; Park, Y.; Jeoung, C.; Choi, W.; Kim, Y.; Jung, I.; Shigesada, N.; Kawasaki, K.; Takasu, F.; Chon, T. Spatially explicit model applied to pine wilt disease dispersal based on host plant infestation. Ecol. Model. 2017, 353, 54–62. [Google Scholar] [CrossRef]
- Takasu, F. Individual-based modeling of the spread of pine wilt disease: Vector beetle dispersal and the Allee effect. Popul. Ecol. 2009, 51, 399–409. [Google Scholar] [CrossRef]
- Robinet, C.; Van Opstal, N.; Baker, R.; Roques, A. Applying a spread model to identify the entry points from which the pine wood nematode, the vector of pine wilt disease, would spread most rapidly across Europe. Biol. Invasions 2011, 13, 2981–2995. [Google Scholar] [CrossRef]
- Yoshimura, A.; Kawasaki, K.; Takasu, F.; Togashi, K.; Futai, K.; Shigesada, N. Modeling the spread of pine wilt disease caused by nematodes with pine sawyers as vector. Ecology 1999, 80, 1691–1702. [Google Scholar] [CrossRef]
- Choi, W.I.; Song, H.J.; Kim, D.S.; Lee, D.; Lee, C.; Nam, Y.; Kim, J.; Park, Y. Dispersal Patterns of Pine Wilt Disease in the Early Stage of Its Invasion in South Korea. Forests 2017, 8, 411. [Google Scholar] [CrossRef]
- Aslam, A.; Ozair, M.; Hussain, T.; Awan, A.U.; Tasneem, F.; Shah, N.A. Transmission and epidemiological trends of pine wilt disease: Findings from sensitivity to optimality. Results Phys. 2021, 26, 104443. [Google Scholar] [CrossRef]
- Shi, X.; Song, G. Analysis of the Mathematical Model for the Spread of Pine Wilt Disease. J. Appl. Math. 2013, 2013, 184054. [Google Scholar] [CrossRef]
- Dimitrijevic, D.D.; Bacic, J. Mathematical analysis of dynamic spread of Pine Wilt disease. Commun. Agric. Appl. Biol. Sci. 2013, 78, 389–399. [Google Scholar]
- Kobayashi, F.; Yamane, A.; Ikeda, T. The Japanese pine sawyer beetle as the vector of pine wilt disease. Annu. Rev. Entomol. 1984, 29, 115–135. [Google Scholar] [CrossRef]
- Deng, R.; Guo, F. Occurrence regularity and control strategy of pine wood nematode in Shangyou County. J. Green Sci. Technol. 2019, 21, 204–205. [Google Scholar]
- Jing, Y.; Zhang, F.; Zhang, Y. Change and prediction of the land use /cover in Ebinur Lake Wetland Nature Reserve based on CA-Markov model. Chin. J. Appl. Ecol. 2016, 27, 3649–3658. [Google Scholar] [CrossRef]
- Zhou, C.; Ou, Y.; Ma, T.; Tan, B. Theoretical Perspectives of CA-based Geographical System Modeling. Prog. Geogr. 2009, 28, 833–838. [Google Scholar]
- Xin, F.; Wang, X.; Yang, Y.J. Deriving suitability factors for CA-Markov land use simulation model based on local historical data. J. Environ. Manag. 2017, 206, 10–19. [Google Scholar]
- Jossart, J.; Theuerkauf, S.J.; Wickliffe, L.C.; Morris, J.A., Jr. Applications of Spatial Autocorrelation Analyses for Marine Aquaculture Siting. Front. Mar. Sci. 2020, 6, 806. [Google Scholar] [CrossRef]
- Rangel, T.; Diniz-Filho, J.; Bini, L.M.; Rangel, T.; Diniz-Filho, J.A.F.; Bini, L.M. Towards an integrated computational tool for spatial analysis in macroecology and biogeography. Glob. Ecol. Biogeogr. 2006, 15, 321–327. [Google Scholar] [CrossRef]
- Hao, Z.; Huang, J.; Zhou, Y.; Fang, G. Spatiotemporal Pattern of Pine Wilt Disease in the Yangtze River Basin. Forests 2021, 12, 731. [Google Scholar] [CrossRef]
- Zhang, X. Major Biological Disaster of Forest in China; China Forestry Press: Beijing, China, 2003. [Google Scholar]
- Li, Y. Landscape Patterns on Influence of Population Density and Genetic diversity of Monochamus alternatus Hope (Coleoptera: Cerambycidae). Master’s Thesis, Fujian Agriculture and Forestry University, Fuzhou, China, 2020. [Google Scholar] [CrossRef]
- Xiao, G. Forest Insects in China, 2nd ed.; China Forestry Press: Beijing, China, 1992. [Google Scholar]
- Tobler, W.R. A computer movie simulating urban growth in the Detroit region. Econ. Geogr. 1970, 46, 234–240. [Google Scholar] [CrossRef]
- Zhang, X.; Zhou, J.; Li, M. Analysis on spatial and temporal changes of regional habitat quality based on the spatial pattern reconstruction of land use. Acta Geogr. Sin. 2020, 75, 160–178. [Google Scholar]
- Zhao, J.; Zhang, H.; Qiao, Z.; Zhang, Z.; Hou, G. Dynamic simulation of land cover change in Xianghai wetland based on CA-Markov model. J. Nat. Resour. 2009, 24, 2178–2186. [Google Scholar]
- Cui, L.; Zhao, Y.H.; Liu, J.C.; Wang, H.Y.; Han, L.; Li, J.; Sun, Z.H. Vegetation Coverage Prediction for the Qinling Mountains Using the CA-Markov Model. ISPRS Int. J. Geo-Inf. 2021, 10, 679. [Google Scholar] [CrossRef]
- Rimal, B.; Zhang, L.F.; Keshtkar, H.; Wang, N.; Lin, Y. Monitoring and Modeling of Spatiotemporal Urban Expansion and Land-Use/Land-Cover Change Using Integrated Markov Chain Cellular Automata Model. ISPRS Int. J. Geo-Inf. 2017, 6, 288. [Google Scholar] [CrossRef]
- Mondal, M.S.; Sharma, N.; Kappas, M.; Garg, P.K. Modeling of spatio-temporal dynamics of land use and land cover in a part of Brahmaputra River basin using Geoinformatic techniques. Geocarto Int. 2013, 28, 632–656. [Google Scholar] [CrossRef]
- Luo, Z.; Hu, X.; Wei, B.; Cao, P.; Cao, S.; Du, X. Urban Landscape Pattern Evolution and Prediction Based on Multi-Criteria CA-Markov Model:Take Shanghang County as an Example. Econ. Geogr. 2020, 40, 58–66. [Google Scholar] [CrossRef]
- Lu, X.; Huang, J.; Li, X.; Fang, G.; Liu, D. The interaction of environmental factors increases the risk of spatiotemporal transmission of pine wilt disease. Ecol. Indic. 2021, 133, 108394. [Google Scholar] [CrossRef]


| Time | 2010 | Time | 2020 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| States of Pine Forest | Healthy Pine Forest | Infected Pine Forest | Non-Pine Forest | States of Pine Forest | Healthy Pine Forest | Infected Pine Forest | Non-Pine Forest | ||
| 2000 | healthy pine forest | 0.8472 | 0.1528 | 0 | 2010 | healthy pine forest | 0.7682 | 0.2318 | 0 |
| infected pine forest | 0 | 0 | 1 | infected pine forest | 0 | 0 | 1 | ||
| non-pine forest | 0.065 | 0.0125 | 0.9225 | non-pine forest | 0.0439 | 0.0202 | 0.9359 | ||
| Limiting Factors | Influencing Factors | Functional Relationships | Weights |
|---|---|---|---|
| Building up | NDVI | Diminishing—J Shape | 0.205 |
| / | Average wind speed | Diminishing—Sigmoidal | 0.203 |
| / | Solar radiation intensity | Diminishing—Sigmoidal | 0.104 |
| / | Population density | Diminishing—J Shape | 0.073 |
| / | Average relative humidity | Diminishing—J Shape | 0.052 |
| / | Average rainfall | Diminishing—Sigmoidal | 0.034 |
| / | Maximum temperature | Diminishing—Sigmoidal | 0.032 |
| / | DEM | Diminishing—Sigmoidal | 0.007 |
| / | Distance from the road | Diminishing—Sigmoidal | 0.29 |
| Years | Moran’s I | Z-Score | p Value |
|---|---|---|---|
| 2000 | 0.019187 | 8.610313 | 0.000000 |
| 2010 | 0.036988 | 6.207085 | 0.000000 |
| 2020 | 0.043532 | 100.330302 | 0.000000 |
| 2030 | 0.012793 | 25.764980 | 0.000000 |
| Year | Distribution Area of PWD on Different Grade Slope/ha | |||||
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | Total | |
| 2000 | 7066 | 665 | 126 | 7857 | ||
| 2010 | 3190 | 206 | 43 | 3440 | ||
| 2020 | 64,695 | 26,884 | 7385 | 640 | 0.41 | 99,605 |
| 2030 | 127,888 | 103,165 | 35,824 | 3509 | 245 | 270,632 |
| Total | 202,839 | 130,920 | 43,378 | 4149 | 245 | 381,532 |
| Year | Distribution Area of PWD on Different Grade Aspect/ha | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| North | Northeast | East | Southeast | South | Southwest | West | Northwest | Total | |
| 2000 | 1229 | 864 | 618 | 481 | 841 | 1094 | 1500 | 1231 | 7857 |
| 2010 | 595 | 427 | 211 | 285 | 344 | 456 | 526 | 597 | 3440 |
| 2020 | 17,718 | 12,083 | 8145 | 7349 | 10,879 | 13,677 | 14,761 | 14,993 | 99,605 |
| 2030 | 32,116 | 22,449 | 16,676 | 28,875 | 40,058 | 42,158 | 45,328 | 42,972 | 270,632 |
| Total | 51,658 | 35,822 | 25,649 | 36,990 | 52,122 | 57,384 | 62,115 | 59,792 | 381,533 |
| Road Type | 2020 Correlation | 2030 Correlation |
|---|---|---|
| Provincial road | 0.491 | 0.626 |
| County Road | 0.392 | 0.270 |
| Country road | 0.896 ** | 0.950 ** |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, D.; Zhang, X. Occurrence Prediction of Pine Wilt Disease Based on CA–Markov Model. Forests 2022, 13, 1736. https://doi.org/10.3390/f13101736
Liu D, Zhang X. Occurrence Prediction of Pine Wilt Disease Based on CA–Markov Model. Forests. 2022; 13(10):1736. https://doi.org/10.3390/f13101736
Chicago/Turabian StyleLiu, Deqing, and Xiaoli Zhang. 2022. "Occurrence Prediction of Pine Wilt Disease Based on CA–Markov Model" Forests 13, no. 10: 1736. https://doi.org/10.3390/f13101736
APA StyleLiu, D., & Zhang, X. (2022). Occurrence Prediction of Pine Wilt Disease Based on CA–Markov Model. Forests, 13(10), 1736. https://doi.org/10.3390/f13101736






