Abstract
In Lithuania, the dieback of European ash (Fraxinus excelsior L.), caused by alien ascomycete Hymenoscyphus fraxineus, started in the mid-1990s, resulting in a large-scale decline of F. excelsior and its dominated forest habitats. Nevertheless, the recent inventories show the presence of several hundred hectares of naturally regenerated F. excelsior stands. We used seven naturally regenerated sites and three planted progeny trials of F. excelsior to collect leaves, shoots, roots, and the surrounding soil to study ash-associated fungal communities based on high-throughput sequencing. Results showed that fungal communities associated with F. excelsior in re-emerging stands in post-dieback areas were composed of 1487 fungal taxa. Among these, 60.5% were Ascomycota, 37.5%—Basidiomycota, 1.7%—Zygomycota, and 0.2% were Chytridiomycota. Revealed mycobiota was largely composed of endophytic fungal communities as these were dominated by Cladosporium sp., Fraxinicola fraxini (syn. Venturia fraxini) and Vishniacozyma foliicola. Identified mycobiota also included a range of ash-specific fungal taxa. Hymenoscyphus fraxineus occurred in all stands but was not frequent. Cladosporium sp. showed strongest negative correlation with the presence of H. fraxineus. This ascomycete, given its dominance in leaves, shoots and in the organic soil layer, might be the limiting factor for the infection rate or spread of H. fraxineus. Although fungal communities in asymptomatic and symptomatic samples of F. excelsior differed significantly from each other, the majority of the most frequently found fungal taxa were not host-specific, suggesting that these were negligibly affected by ash dieback. Investigated stands in natural F. excelsior habitats exhibited larger diversity of fungal taxa (especially ash-specific), than progeny trials planted on former grasslands, indicating the importance of natural habitats in F. excelsior restoration programs.
1. Introduction
Since the mid-1990s, a devastating ash dieback epidemic, caused by an ascomycete fungus Hymenoscyphus fraxineus [1] has spread across Europe severely affecting populations of European ash (Fraxinus excelsior L.) [2,3,4]. Other ash species, such as F. angustifolia, were also affected. Reports show that mortality of ash trees vary among different countries due to differences in disease history and disease stage [5]. To date, most European countries have reported the presence of diseased and dead ash trees, as well as incidences of the disease in stands of different age and on various sites [6]. Although H. fraxineus is an invasive pathogen in Europe [7], in its native range in Far East Asia, it is a symptomless colonizer of Manchurian ash (Fraxinus mandshurica Rupr.), which is a close relative of F. excelsior [8]. Younger ash trees affected by ash-dieback may die within few years, while older and larger ones become chronically diseased and susceptible to secondary diseases [9]. This leads to a gradual extinction of F. excelsior from forest ecosystems as the key species. Indeed, the disease has led to deteriorated condition of ash stands and to the increase in sanitary fellings.
The primary infection of H. fraxineus is caused by long-traveling airborne ascospores [10,11,12,13] infecting leaves and petioles, as well as fungal mycelia spreading through the shoot-petiole junction to wood tissues, causing lesions and gradual dieback of tree crowns [14,15,16,17]. Despite necroses around the tree root collar can also be associated with direct infections of H. fraxineus [18,19], leaflets and petioles are considered to be the main entry point for the pathogen [20,21], indicating the importance of foliar mycobiome as a potential biocontrol agent.
Lithuania has one of the longest and most devastating ash dieback disease histories in Europe [22]. Here, ash dieback started in the mid-1990s [23]. Due to subsequent sanitary fellings, the area of ash stands in the country reduced from 50,800 ha in 1995 [24] to 13,013 ha in 2021 [25], or from 2.7 to 0.6% of all forest land. Currently, the epidemic is in its chronic phase, but during the last decade, deterioration of remaining ash stands was slower in comparison to its initial stages (State Forest Service, personal communication).
European ash is of high ecological importance in floodplain and riparian forests, hedgerows, and as a landscape tree [26,27]. As many species are specifically associated with F. excelsior, the loss of ash trees could also result in the loss of their habitat, potentially leading to extinction. However, how many species strictly depend on F. excelsior is obscure largely due to the scarcity of ash-related studies, especially the ones from the pre-dieback era [28]. To date, 463 fungal species are listed among ash-associated mycobiome in the USDA database [29]. In the United Kingdom, 1058 species were identified as being associated with European ash, including 12 species of birds, 55 mammals, 78 vascular plants, 58 bryophytes, and 68 fungi [30].
Mycological studies associated with ash dieback have changed over time, i.e., from identification of causal agent [3,31,32], to the assessment of seasonal patterns of the disease [11,33], fungal diversity, and its seasonal and spatial dynamics in planta [34]. Numerous studies on fungal diversity in and near ash trees covered myciobiota diversity in healthy [35,36,37], as well as diseased [32,34,37,38,39] stands. The number of studies on ash-associated mycobiome (especially endophytes) boosted during several past years. Indeed, the role of the microbiome, as the “second genome” and determinant of plant features, including susceptibility or resistance to pathogens, has become a popular topic in contemporary ecology [40] and is regarded as a possible mean of natural disease control. Endophytes colonize plant’s inter and intracellular space and spend parts of their life cycle or even their whole lives inside plants without causing symptoms [41,42,43]. In some cases, endophytes may contribute to plant growth and increase tolerance to different stress factors, including pathogens [44,45,46]. It is suggested that local biodiversity can play an important role in suppressing the spread of invasive diseases [47,48]. Despite a large number of research studies, the role and dynamics of fungal communities associated with F. excelsior and their effect on ash dieback is not yet fully understood, giving a fragmental picture of ash-associated fungal communities [33].
The rapid spread of H. fraxineus [4] and high mortality rates of infected trees [49] raised a question whether ecosystems dominated by F. excelsior could be restored without artificial measures, such as selection and breeding of disease-resistant ash trees [50,51]. Although the epidemic of ash dieback is ongoing for nearly 30 years, in Lithuanian Forestry Cadastre, there are 1880 ha of F. excelsior dominated stands, which are up to 30 years old, including 1493 ha of stands, which are up to 20 years old [25]. The latter indicates that, in certain small areas, regeneration of F. excelsior is taking place by natural means. Although these stands are affected by ash dieback, i.e., exhibit ash dieback symptoms to various extent, they are in a stable phytosanitary condition and, therefore, are retained by forest managers [24].
The aims of the present study were: (i) to better understand the diversity and composition of fungal communities in asymptomatic and symptomatic functional tissues and adjacent soil of F. excelsior; (ii) to evaluate potential effects of common fungal taxa on the occurrence of ash dieback pathogen H. fraxineus. We hypothesized that fungal communities will differ between asymptomatic and symptomatic samples of respective tissues.
2. Materials and Methods
Study sites and sampling. In total, ten F. excelsior stands were selected in different regions of Lithuania (Figure 1, Table 1). These sites represented native F. excelsior populations of Lithuania [50]. The study sites were selected based on the following criteria: (a) F. excelsior composed at least 40% in a forest stand; (b) stands were formed in the period of the ongoing ash dieback epidemic; (c) stands were of a relatively good and stable phytosanitary condition (mean defoliation < 30%, assessed visually).
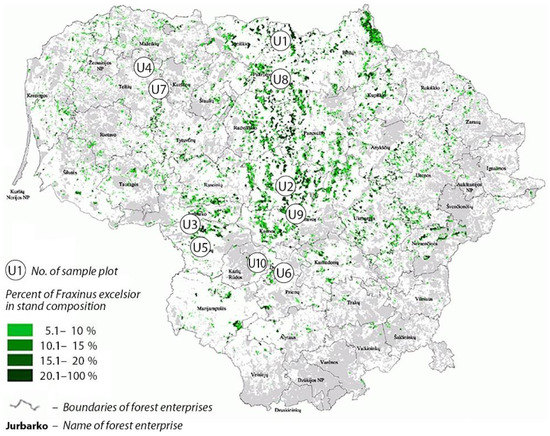
Figure 1.
Map of Lithuania showing the distribution of Fraxinus excelsior forest stands (in green), and the position of sampling sites (U1–U10).

Table 1.
Fraxinus excelsior sites in Lithuania used in the present study.
Prior to the outbreak of ash dieback, U1–U3, U5, U8–U10 sites (Table 1) were comprised of mixed natural forest stands dominated by F. excelsior, which were later subjected to sanitary fellings of various intensity due to ash mortality caused by H. fraxineus. Among all sites of the study, which were formed after the ash dieback outbreak, sites U2 and U3 were of the best phytosanitary conditions with a well-developed main canopy layer comprised almost exclusively of ash. The remaining self-regenerated stands (Table 1) were composed of mixed tree species, putting ash trees in more shaded conditions. Sanitary fellings in self-regenerated sites were not conducted during the last decade (Forest State Enterprise, personal communication). The remaining U4, U6–7 sites (Table 1) were planted as progeny trials of ongoing F. excelsior breeding for resistance programs [50]. Noteworthy, all progeny trials were established on former grasslands but in immediate vicinity to the forest.
At each study site, 12 trees of relatively good phytosanitary conditions, i.e., showing defoliation up to 30%, were selected for sampling. Within each site, all sampled trees were situated at least 50 m from each other. Sampling was carried out in July 2019 by cutting live twigs with leaves from the lower part of the crown (4–12 m above the ground) using a telescopic secateurs. From these, leaf and shoot (ca. 5 cm long and 1–1.5 cm thick) samples were taken from the second-year shoots using hand secateurs. From each respective tree, samples were collected and sorted out, accordingly, to the following sampling categories: (a) asymptomatic leaves; (b) symptomatic leaves showing H. fraxineus necroses; (c) asymptomatic shoots; (d) symptomatic shoots showing H. fraxineus necroses. In symptomatic shoot samples, a borderline between necrotic and healthy tissues was always present. Symptomatic and asymptomatic samples were collected from different twigs. In addition, one lateral root with fine roots per tree was collected. In total, 720 shoot samples (360 symptomless and 360 symptomatic), 720 leaf samples (360 symptomless and 360 symptomatic), and 120 root samples were collected from 120 trees. In addition, in each study site, four random samples of organic and four of mineral soil were collected using soil cores. Each sample was individually packed into plastic bags, transported to the laboratory, and stored at −20 °C until subjected to DNA extraction.
Preparation of samples and DNA extraction. In the laboratory, collected roots were carefully washed in tap water to remove any of the remaining soil, and fine roots with root tips were separated from coarse roots, which were discarded. Fine roots were cut into ca. 1 cm-long segments and, within each forest stand, pooled together. Soil samples were sieved (mesh size 2 × 2 mm) to remove larger particles and roots and, within each forest stand, pooled together. Leaf and shoot samples were sorted to asymptomatic and symptomatic. In total, ten samples of each roots, mineral soil, organic soil, symptomatic leaves, asymptomatic leaves, symptomatic shoots, and asymptomatic shoots were obtained and used for DNA work. Prior to isolation of DNA, individual leaf, shoot, root, and soil samples were freeze-dried at −60 °C for 24 h. Lyophilized samples were homogenized at 5000 rpm for 60 s using a Precellys 24 tissue homogenizer (Bertin Technologies, Montigny-le-Bretonneux, France). About 50 mg of grounded powder were transferred to 2-mL microcentrifuge tubes, together with two (3 mm in diameter) glass beads, and used for DNA extractions.
The total DNA was isolated from each sample using CTAB protocol [52] by adding 1000 μL of CTAB buffer (0.5 M EDTA pH 8.0, 1 M Tris-HCL pH 8.0, 5 M NaCl, 3% CTAB) and incubating, at 65 °C, for 1 h. Following incubation, the samples were centrifuged at 10,000 rpm for 5 min, and the supernatant was transferred to a new 1.5-mL Eppendorf tube and mixed with an equal volume of chloroform. After centrifugation for 7 min at 10,000 rpm, the supernatant (about 500 μL) was then transferred to a new Eppendorf tube containing an equal volume of 2-propanol that was mixed by vortexing and incubated at room temperature for 30 min, whereupon it was once more centrifuged at 13,000 rpm for 10 min. After centrifugation, the supernatant was discarded, and the remaining pellet was, then, precipitated with 70% ethanol (100 μL) and centrifuged for 5 min at 13,000 rpm. The DNA pellet was dried in airflow for 30 min and, finally, resuspended in 30 μL of sterile milli-Q water. The DNA concentration of each sample was measured using a NanoDrop One spectrophotometer (Thermo Scientific, Rodchester, NY, USA) and, if needed, diluted to 1–10 ng/μL.
PCR amplification and sample preparation for sequencing. The amplification by PCR of the ITS2 rRNA region was done using barcoded primers gITS7 [53] and ITS4 [54]. PCR reactions were performed using the Applied Biosystems 2720 thermal cycler (Applied Biosystems, Foster City, CA, USA) in 50 μL reaction volume using the following final concentrations: 200 nM of dNTPs; 750 nM of MgCl2; 200 nM of each primer; 0.025 nM DreamTaq Green polymerase (5 U/μL) (Thermo Scientific, Waltham, MA, USA); 0.02 ng/μL of template DNA. Sterile milli-Q water was added to make the final volume (50.0 μL) of the reaction. The PCR program started with initial denaturation at 94 °C for 5 min, followed by 30 cycles, each consisting of 94 °C for 30 s, annealing temperature 56 °C for 30 s, and 72 °C for 30 s, with a final extension at 72 °C for 7 min. The PCR products were assessed using gel electrophoresis on 1% agarose gels stained with GelRed (Biotium, Fremont, CA, USA). PCR products were purified using 3 M sodium acetate (pH 5.2) (AppliChem Gmbh, Darmstadt, Germany) and 96% ethanol mixture (1:25). After the quantification of all of the PCR products using a Qubit fluorometer 4.0 (Life Technologies, Stockholm, Sweden), they were pooled in an equimolar mix and sequenced using a PacBio platform and one Sequel SMRT cell at a SciLifeLab facility in Uppsala, Sweden.
Bioinformatics. The sequences generated were subjected to quality control and clustering in the SCATA NGS sequencing pipeline at http://scata.mykopat.slu.se (accessed on 25 June 2021). Quality filtering included the removal of short sequences (<200 bp), sequences with low read quality, primer dimers, and homopolymers, which were collapsed to three base pairs (bp) before clustering. Sequences that were missing a tag or primer were excluded. The primer and sample tags were then removed from the sequence, but information on the sequence association with the sample was stored as meta-data. The sequences were then clustered into different taxa using single-linkage clustering based on 98% similarity. The most common genotype (real read) for each cluster was used to represent each taxon. For clusters containing two sequences, a consensus sequence was produced. Fungal taxa were taxonomically identified using the GenBank (NCBI) and UNITE databases and the Blastn algorithm. The criteria used for identification were: sequence coverage > 80%; similarity to taxon level 98–100%, similarity to genus level 94–97%. Sequences not matching these criteria were considered unidentified and were given unique names. Representative sequences of all fungal non-singletons as the Targeted Locus Study project have been deposited in GenBank under accession number KFVX00000000.
Statistical analyses. To evaluate the relationship between the sampling intensity and the number of fungal taxa in F. excelsior samples (leaves, shoots, roots, and the soil), rarefaction curves were generated using Analytical Rarefaction v.1.3 (http://www.uga.edu/strata/software/index.html) (accessed on 19 October 2021). The diversity and species richness of fungal communities was evaluated, calculating Shannon diversity index and qualitative Sørensen similarity index. Nonparametric Mann–Whitney test in Minitab v.19.2 (Minitab® Inc., Pennsylvania State University, State College, PA, USA) was used to evaluate if the Shannon diversity index differed significantly between asymptomatic and symptomatic samples of leaves and shoots, as well as between organic and mineral soil. Nonparametric chi-square test was used to analyze differences in the richness of fungal taxa between different stands and types of samples (symptomatic vs. asymptomatic). Non-parametric Kruskall–Wallis test, combined with Dunn’s pairwise analysis and Bonferoni correction, was used to evaluate differences in diversity of fungal communities among different sites and types of samples using the XLSTAT v. 2021 statistical software package (Addinsoft Inc., Damrémont, France).
Pearson’s correlation between frequencies of sequences of different taxa were computed using CORR procedure of the SAS software (SAS® Analytics Pro 9.4, 2020, San Francisco, CA, USA). To obtain normal distribution of residuals and variance homogeneity, the SQRT transformation was applied for the sequence frequency data. Assumptions of normal distribution of residuals and variance homogeneity were tested with GLM and UNIVARIATE procedures in SAS (SAS® Analytics Pro 9.4, 2020). Nonmetric multidimensional scaling (NMDS), using Bray–Curtis distance, was accomplished in Canoco 5 (v5.12) program package [55]. Permutational multivariate analysis of variance (PERMANOVA) with the Bray–Curtis distance metric was used to assess the significance of community similarity. This analysis was done using the adonis2 function from the vegan package in R version 4.1.2 (available at https://www.r-project.org/) (accessed on 19 October 2021) using 999 permutations.
3. Results
High-throughput sequencing resulted in 586,427 reads. After quality filtering, 236,532 (40.3%) high-quality reads were retained. Clustering analysis at 98% similarity showed the presence of 1487 non-singleton fungal taxa (Tables S1–S4). Among the detected fungi, 60.5% were Ascomycota, 37.5%—Basidiomycota, 1.7%—Zygomycota, and 0.2% were Chytridiomycota (Table 2). In total, 266 (17.9%) taxa were identified to taxon level, 262 (17.6%) to genus level, and 913 (61.4%) remained unidentified. Analysis showed that there were 936 fungal taxa in leaves, followed by 846 taxa in shoots, 304 in root, and 239 in the soil (Table 3). Most frequently detected fungi in leaves, shoots, roots, and the rhizosphere soil are in Table 4, Table 5, Table 6 and Table 7, respectively.

Table 2.
Distribution of sequences and taxa, by phylum, in different types of samples of Fraxinus excelsior.

Table 3.
Generated high-quality sequences and detected diversity of fungal taxa in samples of different types in trials and natural populations of Fraxinus excelsior in Lithuania.

Table 4.
Occurrence and relative abundance (%) of the 40 most common fungal taxa (shown as a proportion of all high-quality fungal sequences) in asymptomatic and symptomatic leaf samples in ten populations of Fraxinus excelsior in Lithuania.

Table 5.
Occurrence and relative abundance (%) of the 40 most common fungal taxa (shown as a proportion of all high-quality fungal sequences) in asymptomatic and symptomatic shoot samples in ten populations of Fraxinus excelsior in Lithuania.

Table 6.
Occurrence and relative abundance (%) of the 40 most common fungal taxa (shown as a proportion of all high-quality fungal sequences) in root samples from ten populations of Fraxinus excelsior in Lithuania.

Table 7.
Occurrence and relative abundance (%) of the 40 most common fungal taxa (shown as a proportion of all high-quality fungal sequences) in organic and mineral soil samples from ten populations of Fraxinus excelsior in Lithuania.
Rarefaction analysis of leaf, shoot, root, and the soil samples revealed that rarefaction curves did not reach the asymptote (Figure 2).
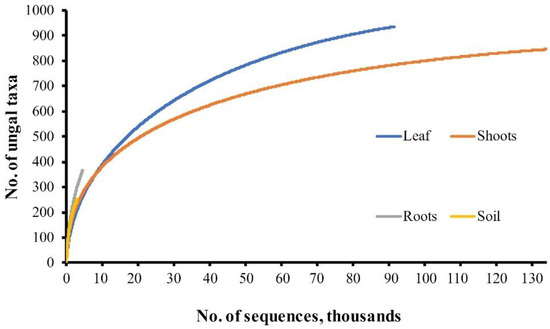
Figure 2.
Rarefaction curves showing the relationship between the cumulative number of fungal taxa and the sequencing intensity.
There were 388 fungal taxa, which were found in a single site out of ten sampled; by contrast, 105 fungal taxa were found in all ten sites. As many different fungal taxa were detected at different sites, Sørensen’s qualitative similarity index of fungal communities among different study sites was moderate (0.49–0.63).
Among fungal taxa that were found in leaf samples, 279 (29.8%) were exclusively found in asymptomatic leaves, as well as 205 (21.9%) in symptomatic leaves, while 452 (48.3%) fungal taxa were common to both categories of leaf samples (Figure 3).
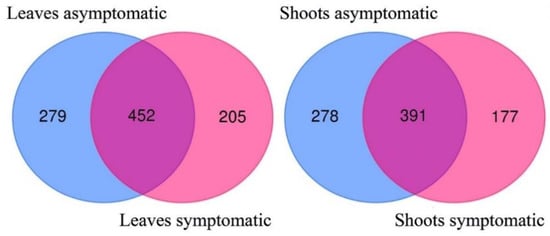
Figure 3.
Venn diagrams showing the diversity and overlap of fungal taxa in asymptomatic and symptomatic samples of leaves and shoots of Fraxinus excelsior.
The most frequently detected fungal taxa were Cladosporium sp. 5239_0, Fraxinicola fraxini (=Venturia fraxini), and basidiomycetous yeast Vishniacozyma foliicola, found in all ten sites and in all sample types. Together, the three most abundant fungal taxa included 32.5% of all high-quality sequences. These taxa, together with another frequently detected fungi such as Elsinoe heveae, Lecania naegelii, or Phyllactinia fraxini, were dominating in fungal communities of analyzed F. excelsior samples in shoots and leaves (Table 4, Table 5, Table 6 and Table 7).
Nonmetric multidimensional scaling of fungal communities demonstrated that fungal communities in shoots were largely separated and differed significantly between symptomatic and asymptomatic samples (p < 0.002), yet they intermixed between asymptomatic and symptomatic samples in leaves (p > 0.095) (Figure 4). Fungal communities in mineral and organic soil samples were largely overlapping, but statistical analysis showed that these differed significantly from each other (p < 0.041) (Figure 4).
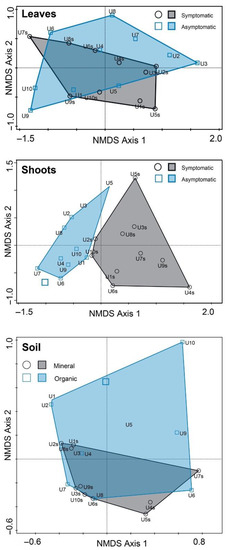
Figure 4.
Ordination diagram based on nonmetric multidimensional scaling of fungal communities in leaf, shoot, root, and soil samples. Each point in the diagram represents a respective site of F. excelsior.
The most frequent fungi found in soil samples were Trechispora invisitata (0.65% of all reads, Table 6), Malassezia restricta, and Cladosporium sp., which were found in the soil of all study sites. The causal agent of ash dieback H. fraxineus was consistently found in all sites (Table 4 and Table 5), but it was relatively unabundant (0.89% of all high-quality reads). The lowest abundance of H. fraxineus was in the site U2, which exhibited the lowest ash defoliation of all investigated sites. H. fraxineus was most frequent in shoots (1.05% of reads), followed by roots (0.86% of reads) and leaf (0.69% of reads) samples. On few occasions, H. fraxineus was also present in the soil, comprising 0.25% of all sequences found in soil samples. Among leaf and shoot samples, 94.2% of all H. fraxineus sequences were detected in symptomatic tissues. Among leaves and shoots, the detected frequency of H. fraxineus did not correlate significantly with the presence of other fungal taxa, except for a positive correlation between the abundance of H. fraxineus and Ramularia vizellae on leaves (r = 0.576, p = 0.008), a positive correlation between the abundance of H. fraxineus and Pleosporales sp. on shoots (r = 0.562, p = 0.010), and a negative correlation between the abundance of H. fraxineus and Cladosporium sp. on shoots (r = −0.548, p = 0.012). Significantly positive correlation between the abundance of H. fraxineus and Ph. fraxini (r = 0.622, p < 0.001) was detected in sites U2 and U3 in asymptomatic leaves. In root samples, significant correlations between the abundance of H. fraxineus were found with the abundance of Knufia sp. (r = 0.782, p = 0.001), Malassezia restricta (r = 0.736, p = 0.001), Sphaerulina rhabdoclinis (r = 0.656, p = 0.002), Vishniacozyma follicola (r = 0.641, p = 0.002), Fraxinicola fraxini (r = 0.639, p = 0.002), Lecania naegelii (r = 0.614, p = 0.004), and Phyllactina fraxini (r = 0.602, p = 0.005). The overall species richness was significantly higher in asymptomatic vs. symptomatic leaves and shoots (chi-square tests, p < 0.001). In a total of over 10 sites, 731 taxa were found in asymptomatic leaves, while 657 taxa were found in symptomatic leaves (Table 2). The Shannon diversity index was also higher in asymptomatic leaves than in symptomatic leaves (3.59 vs 3.01, Table 3), but the difference was insignificant (p > 0.05). Overall, 669 taxa were found in asymptomatic shoots, while 568—in symptomatic ones (Table 2). Contrary to this, the Shannon diversity index was higher in symptomatic than in asymptomatic shoots (4.15 vs. 3.49, Table 2), but the difference was also insignificant (p > 0.05). The Shannon diversity index did not differ significantly between the organic and mineral soil (p > 0.05). Correlations of abundance of H. fraxineus with the Shannon diversity index were insignificant both in asymptotic leaves (r = −0.588, p = 0.073) and in symptomatic leaves (r = −0.169, p = 0.640). No significant correlations were also found for symptomatic (r = −0.034, p = 0.926) and asymptomatic (r = −0.118, p = 0.745) shoot samples.
4. Discussion
Since the outbreak of ash dieback in Lithuania in mid-1990s, silviculture in F. excelsior-dominated forest ecosystems was shifted towards the cultivation of other tree species or left for self-regeneration [22]. Cultivation of F. excelsior stands was generally abandoned. The only notable exception from this was a small number of dedicated F. excelsior progeny trials and clonal archives, created to look for a viable solution to restore F. excelsior stands using ash material resistant to H. fraxineus. Such trials, in combination with disease resistance and due to self-thinning and sanitary thinning, resulted in healthier stands in comparison to natural ash stands, but with ambiguous results [50].
In addition, a small number of young, naturally regenerated F. excelsior stands, which were often mixed with other tree species, emerged on sites of former ash stands deteriorated by ash dieback during the first decade of this still ongoing epidemic. Although F. excelsior trees in these stands are not free of H. fraxineus infections, the sanitary conditions of investigated stands were stable and generally satisfactory for the last five years (State Forest Service, personal communication). Phytosanitary conditions of investigated stands U2 and U3 were good, as these included only a limited number of diseased ash trees and with a crown defoliation under 30 %.
It should be noted that all F. excelsior stands (and investigated trees) selected for this study were of young age, modest diameter, and modest height, usually not exceeding 15 m. The young ash trees are generally considered to be more susceptible to ash dieback than the older ones [56,57].
Due to our sampling approach, the composition of mycobiome species was determined, in shoots and leaves, of the lower part of the crowns of investigated trees. The results of the present study should be interpreted with caution, as H. fraxineus infection in the tops of F. excelsior trees were reported to be typical [58]. On the other hand, ash-associated mycobiome is usually most species-rich in the lower part of the crown, i.e., in a more shaded part of the F. excelsior crowns [35,36]. Noteworthy, the observable defoliation of crown tops was absent among ash trees selected for sampling and was not typical in assessed stands overall.
In general, our study revealed a high diversity of fungal taxa in both leaf and shoot samples, which was in agreement with earlier findings of culture-independent studies [34,39,59]. Species composition was clearly dominated by Ascomycota, which included representatives of endophytic and other plant colonizing fungi [42]. Species accumulation curves (Figure 2) showed that the species saturation was not reached, indicating that a potentially higher diversity of fungal taxa could be detected by broader sampling and sequencing effort. Indeed, a higher sampling intensity of leaves and shoots resulted in much higher species richness in leaf and shoot samples compared to those detected in root and soil samples.
The frequency of H. fraxineus appearance among detected fungal taxa was rather low in all sampling categories. As the ash dieback epidemic was ongoing for years, the possibility should not be excluded that, due to the competition for the same ecological niche, H. fraxineus was gradually replaced by opportunistic pathogens and/or secondary colonizers. This fact is in contradiction with some important studies, where H. fraxineus was among the most abundant species [60] in leaf mycobiome. However, a relatively small proportion of H. fraxineus among leaf colonizing fungi is also reported by Cross et al. [34], Agan et al. [61], and Agostinelli et al. [39], where the abundance of H. fraxineus biomass in colonized leaf substrate was significantly increased only towards the second half or the end of the vegetation season.
Our study indicates some impact of H. fraxineus on total diversity of fungal communities: on 8 out of 10 sites studied, the Shannon diversity index was higher in asymptomatic leaves than in symptomatic ones (H = 3.59 and H = 3.01 on asymptomatic and symptomatic, respectively, Table 2). Meanwhile, the opposite tendency was observed for shoot samples (H = 3.49 and H = 4.15 on asymptomatic and symptomatic samples, respectively (Table 2).
The impact of H. fraxineus on composition of fungal taxa and distribution of ash-colonizing mycobiome was among primary objectives of many recent studies [58]. However, the extent of such impact is difficult to evaluate, mainly due to the lack of knowledge of F. excelsior-associated fungal diversity prior to the outbreak of ash dieback disease. The phenomena of the replacement of autochthonous leaf decomposer Hymenoscyphus albidus by alien pathogenic H. fraxineus is described in several studies, e.g., [51,62]. In our case, H. albidus was not detected. The presence or absence of H. albidus among dieback-affected European ash mycobiome was often in the focus, as it occupies similar ecological niche as H. fraxineus. Besides, H. albidus can be considered as a reference species, and its abundance may indicate potential changes in fungal communities associated with F. excelsior. Although H. albidus was often reported to be absent in recent studies [20,39,60], its abundance in ash leaves was never exactly clear. In recent studies on ash dieback, where H. albidus was actually found, it appeared in low numbers [62,63,64]. Czech studies on ash-dieback [63,64] imply insignificant impact of H. fraxineus to the prevalence of H. albidus. Its low colonization frequency appears to be typical, even before the outbreak of ash dieback in Europe [65,66].
Ash-specific Fraxinicola fraxini was found to be absent or rare in some Fennoscandian stands of F. excelsior [39,67]. In the present study, F. fraxini was the key component of European ash leaf-colonizing mycobiome in both asymptomatic and symptomatic leaf samples, consistently found in all locations and in all samples, and its appearance was higher in symptomatic leaves in the majority of investigated sites. By contrast, the abundance of F. fraxini in symptomatic leaves in sites U2 and U3, which were characterized by a low defoliation, was lower than it was in asymptomatic leaves. This is in agreement with other studies, where possible F. fraxini association with premature ash defoliation was discussed, regardless that this ascomycete is often considered as a weak leaf pathogen [61,62]. As an early leaf colonizer and the species able to coexist with H. fraxineus in colonized necrotic petioles of European ash, F. fraxini was sometimes considered as a possible antagonist to H. fraxineus [60]. We did not detect a significant correlation between the frequency of these two fungal species, which can be due to the fact that F. fraxini and H. fraxineus colonize different parts of ash foliage [60].
Cryptosphaeria eunomia, which specifically colonizes and decomposes dead F. excelsior twigs, was among the most frequently detected fungi in symptomatic shoot samples, but on different sites, it was found very inconsistently. In ash progeny trials, this species was found very seldom, indicating its strong dependence on natural ash habitats.
Although Alternaria alternata (as well as the other Alternaria spp.) was not among the most abundant fungal taxa, it requires special attention. For about two decades, the cosmopolitan necrotroph A. alternata was considered as one of the most frequently found and identified species associated with symptomatic leaves, shoots, and stems of European ash [38,68,69], and it was considered as the species which may contribute to the worsening phytosanitary conditions of dieback-affected F. excelsior stands [32,70]. It is noteworthy that this particular species appeared among frequently found taxa in other studies, based on the culture-dependent fungal identification approach. However, in our studies and similar high-throughput sequencing-based studies, A. alternata appeared in much lower proportions [39,59], suggesting that, due to fast growth rate, it can be overrepresented in culture-dependent studies, but it probably has a limited role among ash-associated mycobiome. One of such possible roles is an extensive leaf colonization towards the end of the vegetation season in the tops of the crowns, causing premature defoliation as it was suggested by Scholtysik et al. [36].
Similarly to A. alternata, another set of fungal taxa, traditionally regarded among the most prevalent in European ash [32,36,69], namely Aspergillus sp., Epicoccum nigrum, Giberella avenacea, Phoma sp., and Phomopsis sp., were identified in low abundances. These findings are in line with other high-throughput sequencing studies [34,39,59], but they strongly contradict the results of culture-based studies [38,59,60,69]. Evidently, each of the aforementioned methods have their advantages and shortcomings, suggesting that the use of both methods can provide a valuable complementary information.
Interestingly, a parasitic ascomycete Botryosphaeria stevensii (anamorph state is Diplodia mutila), a known pathogen of F. excelsior [69], was missing in all samples of this study. By contrast, B. stevensii was reported in necrotic shoots [32,70], leaves [59,60], and seeds [59] of F. excelsior.
Another member of the genus Diplodia, which is associated with F. excelsior, namely Diplodia fraxini, was a frequently and consistently found fungal taxon in symptomatic apical parts of investigated ash. It is a frequently detected fungal taxon in necrotic tissues of diseased F. excelsior [60,71]. Albeit we did not find significant statistical evidence of an antagonistic relationship between H. fraxineus and D. fraxini, based on relative scarcity of the former and abundance of the latter, the possibility should not be excluded that the spread of H. fraxineus in European ash could be limited by other ash-association pathogens, such as D. fraxini.
The very first culture-independent studies of endophytic communities within plants [72] revealed that these communities vary a lot within a single plant species, as well as over relatively short distances, sensitively reacting to the key substrate and various other resources. Considering the abovementioned factors and tendencies, we can conclude that the mycobiome diversity between seven self-regenerated stands and three ash progeny trials assessed in the present study was remarkably similar, sharing 85 taxa among the 100 most frequently found ones. Apparently, the site (self-regenerated forest stands vs. planted and carefully thinned progeny trials) did not have a pronounced impact on qualitative species richness, although in general numbers of sequence reads among samples collected in progeny trials, these were significantly lower. Hereby, H. fraxineus was several times less abundant in samples from ash progeny trials, yet the actual phytosanitary conditions in trials were slightly worse than those of self-regenerated stands. From the 100 most frequently detected fungal taxa in leaf, shoot, and root samples, 85 of them were omnipresented at both types of sites (≥ 5 reads). Ascomycetes Cosmospora obscura, Nigrograna mycophile, Paraphaeosphaeria sp. 5239_60, Pirozynskiella laurisilvatica, Pleosporales sp. 5239_32, Prosthemium sp. 5239_69, Sclerostagonospora sp. 5239_35, basidiomycete Peniophora sp., and several unidentified genera were identified, almost exclusively, on self-regenerated sites, indicating the dependence of these fungi on natural habitats. Among typical ash-related fungal taxa [33,36,39], Cryptosphaeria eunomia, Didymella sp., Dioszegia sp., D. fraxini, Knufia sp., Ph. fraxini, and Sphaerulina azalea also clearly favored typical conditions of self-regenerated sites. It is suggested that sun-exposed parts of F. excelsior crowns (such conditions were more pronounced in progeny trials vs. self-regenerated sites) may be more susceptible to H. fraxineus infections due to lower diversity of endophytic mycobiome, in comparison to the parts of the crowns situated under more shady, moister conditions and (possibly) higher density of fungal spores [58]. This also corresponds well with the observation of spaciously planted and thinned from unwanted vegetation F. excelsior clonal archives in Lithuania, where ash clones in close vicinity to neighborhood forest canopy (20 m or less) were of significantly better phytosanitary conditions than trees constantly sunlit during daytime (unpublished data).
In the present study, a high abundance of biotrophic powdery mildew Ph. fraxini proved it to be a keystone species in foliage of F. excelsior. Its positive correlation in generally healthy sites with H. fraxineus is in accordance with [34] and [61], where strong connections between these two fungal species were revealed in leaves of investigated European ash, suggesting Ph. fraxini as an interesting investigation prospect. The possibility should not be excluded that leaf colonization by Ph. fraxini could predispose overwise healthy tissues to infections by H. fraxineus, thereby allowing it to more easily overcome tree defense.
The diversity of fungal taxa in soil and root samples was surprisingly low (Figure 2), repeatedly showing that a higher diversity of fungal taxa could be detected using a larger sample size and deeper sequencing. Following the general classification of soil fungi to three main functional groups—symbiotrophs, saprotrophs, and pathogens [73]—fungal communities in the soil were dominated by saprobic species, and diversity was greatly influenced through vegetation properties of the sites, especially leaf litter. Most abundant and consistently found species in the soil were Cladosporium sp., Vishniacozyma foliicola, Fraxinocola fraxini, and Didymella sp. pointing out these fungal taxa as important decomposers of F. excelsior apical parts on forest floor. The absence of Glomeromycota, among identified species in assessed soil and root samples, is surprising, as F. excelsior is known to be a host of arbuscular mycorrhizal fungi [74]. Evidently, to reveal arbuscular mycorrhiza fungi related to the European ash, a more extensive sampling effort is required. Such evidence is in accordance with the study of [75], where only a small proportion of root tips of Fraxinus were found to be colonized by just a few species of arbuscular mycorrhiza fungi.
Occurrence of H. fraxineus in soil samples was very low (only 0.9 reads per sample), in comparison to quantities of ascomata of this species on the forest floor reported by [58], indicating that the propagule pressure of this pathogenic species in investigated stands was lower than expected, regardless of the fact what we excluded ground litter from our study.
Noteworthy, Armillaria spp., known as a secondary pathogen of ash-dieback-affected F. excelsior, was detected very seldom in three sites of naturally regenerated forest.
5. Conclusions
Functional tissues (leaves, shoots, and roots) and adjacent soil of F. excelsior were associated with species-rich communities of fungi. Although the ash dieback pathogen H. fraxineus was relatively rare in all sites and substrates (leaves, shoots, roots, and the soil), in leaves and shoots, there were profound differences in the richness of fungal taxa between symptomatic and asymptomatic samples, suggesting that the state of infection influenced overall fungal diversity. The abundance of several fungi significantly correlated (positively or negatively) with the abundance of H. fraxineus, among which Cladosporium sp. showed a negative correlation, suggesting that it could be a potential inhibitor of H. fraxineus. Investigated stands in natural F. excelsior habitats exhibited a higher diversity of fungal taxa (especially ash-specific) than progeny trials planted on former grasslands, indicating the potential impact and importance of natural habitats.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/f13101609/s1. Table S1. Occurrence and relative abundance of fungal taxa (shown as a proportion of all high-quality fungal sequences) in Fraxinus excelsior asymptomatic (A) and symptomatic (S) leaves from ten different sites (U1-U10) in Lithuania; Table S2. Occurrence and relative abundance of fungal taxa (shown as a proportion of all high-quality fungal sequences) in Fraxinus excelsior asymptomatic (A) and symptomatic (S) shoots from ten different sites (U1-U10) in Lithuania; Table S3. Occurrence and relative abundance of fungal taxa (shown as a proportion of all high-quality fungal sequences) in Fraxinus excelsior roots from ten different sites (U1-U10) in Lithuania; Table S4. Occurrence and relative abundance of fungal taxa (shown as a proportion of all high-quality fungal sequences) in Fraxinus excelsior organic (O) and mineral (M) soil from ten different sites (U1-U10) in Lithuania.
Author Contributions
Conceptualization, D.M., R.B. and A.M. (Audrius Menkis); methodology, D.M., R.B., A.M. (Adas Marčiulynas) and A.M. (Audrius Menkis); validation, R.B., A.P., G.B., J.L. and A.M. (Adas Marčiulynas); formal analysis, R.B., A.P. and A.M. (Audrius Menkis); investigation, R.B., A.P., A.M. (Audrius Menkis), J.L. and D.M.; resources, D.M.; data curation, A.P. and A.M. (Adas Marčiulynas); writing—original draft preparation, R.B.; writing—review and editing, A.M. (Audrius Menkis), A.P. and J.L.; visualization, A.P., A.M. (Audrius Menkis) and R.B.; supervision, A.M. (Audrius Menkis); project administration, D.M. All authors have read and agreed to the published version of the manuscript.
Funding
This project has received funding from European Social Fund (project No. 09.3.3-LMT-K-712-01-0039) under grant agreement with the Research Council of Lithuania (LMTLT).
Data Availability Statement
The data presented in this study are available in Supplementary Material.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Baral, H.O.; Queloz, V.; Hosoya, T. Hymenoscyphus fraxineus, the correct scientific name for the fungus causing ash dieback in Europe. IMA Fungus. 2014, 5, 79. [Google Scholar] [CrossRef] [PubMed]
- Vasiliauskas, A.; Juodvalkis, A.; Treigiene, A. Possible causes of a massive dieback of ash in forests of Lithuania. In Proceedings of the International Conference on Forest Pathology and Mycology; Storozhenko, V.G., Ed.; Russian Academy of Sciences: Moscow, Russian, 2002; pp. 35–37. (In Russian) [Google Scholar]
- Kowalski, T. Chalara fraxinea sp. nov. associated with dieback of ash (Fraxinus excelsior) in Poland. For. Pathol. 2006, 36, 264–270. [Google Scholar] [CrossRef]
- Timmermann, V.; Børja, I.; Hietala, A.M.; Kirisits, T.; Solheim, H. Ash dieback: Pathogen spread and diurnal patterns of ascospore dispersal, with special emphasis on Norway. OEPP/EOPP Bull. 2011, 41, 14–20. [Google Scholar] [CrossRef]
- Vasaitis, R.; Enderle, R. Dieback of European Ash (Fraxinus spp.)—Consequences and Guidelines for Sustainable Management; SLU Service/Repro: Uppsala, Sweden, 2017; 320p, ISBN 978-91-576-8697-8. [Google Scholar]
- CABI. Invasive Species Compendium—Hymenoscyphus fraxineus (Ash Dieback). Available online: https://www.cabi.org/isc/datasheet/108083#tosummaryOfInvasiveness (accessed on 10 June 2022).
- Husson, C.; Scala, B.; Cael, O.; Frey, P.; Feau, N.; Ioos, R.; Marçais, B. Chalara fraxinea is an invasive pathogen in France. Eur. J. Plant. Pathol. 2011, 130, 311–324. [Google Scholar] [CrossRef]
- Zhao, Y.J.; Hosoya, T.; Baral, H.O.; Hosaka, K.; Kakishima, M. Hymenoscyphus pseudoalbidus, the correct name for Lambertella albida reported from Japan. Mycotaxon 2012, 122, 25–41. [Google Scholar] [CrossRef]
- Timmermann, V.; Nagy, N.E.; Hietala, A.M.; Børja, I.; Solheim, H. Progression of ash dieback in Norway related to tree age, disease history and regional aspects. Balt. For. 2017, 23, 150–158. [Google Scholar]
- Skovsgaard, J.P.; Thomsen, I.M.; Skovgaard, I.M.; Martinussen, T. Associations among symptoms of dieback in even-aged stands of ash (Fraxinus excelsior L.). For. Pathol. 2010, 40, 7–18. [Google Scholar] [CrossRef]
- Bengtsson, S.B.; Vasaitis, R.; Kirisits, T.; Solheim, H.; Stenlid. J. Population structure of Hymenoscyphus pseudoalbidus and its genetic relationship to Hymenoscyphus albidus. Fungal Ecol. 2012, 5, 147–153. [Google Scholar] [CrossRef]
- Gross, A.; Grünig, C.R.; Queloz, V.; Holdenrieder, O. A molecular toolkit for population genetic investigations of the ash dieback pathogen Hymenoscyphus pseudoalbidus. For. Pathol. 2012, 42, 252–264. [Google Scholar] [CrossRef]
- Gross, A.; Zaffarano, P.L.; Duo, A.; Grünig, C.R. Reproductive mode and life cycle of the ash dieback pathogen Hymenoscyphus pseudoalbidus. Fungal Genet. Biol. 2012, 49, 977–986. [Google Scholar] [CrossRef]
- Kirisits, T.; Cech, T.L. Beobachtungen zum sexuellen Stadium des Eschentriebsterben-Erregers Chalara fraxinea in Österreich. (Observations on the sexual stage of the causative agent of ash dieback Chalara fraxinea in Austria). Forstschutz. Aktuell. 2009, 48, 21–25. [Google Scholar]
- Kowalski, T.; Holdenrieder, O. Pathogenicity of Chalara fraxinea. For. Pathol. 2009, 39, 1–7. [Google Scholar] [CrossRef]
- Schumacher, J.; Kehr, R.; Leonhard, S. Mycological and histological investigations of Fraxinus excelsior nursery saplings naturally infected by Chalara fraxinea. For. Pathol. 2010, 40, 419–429. [Google Scholar] [CrossRef]
- Gross, A.; Holdenrieder, O.; Pautasso, M.; Queloz, V.; Sieber, T.N. Hymenoscyphus pseudoalbidus, the causal agent of European ash dieback. Mol. Plant Pathol. 2014, 15, 5–21. [Google Scholar] [CrossRef]
- Enderle, R.; Peters, F.; Nakou, A.; Metzler, B. Temporal development of ash dieback symptoms and spatial distribution of collar rots in a provenance trial of Fraxinus excelsior. Eur. J. For. Res. 2013, 132, 865–876. [Google Scholar] [CrossRef]
- Enderle, R.; Sander, F.; Metzler, B. Temporal development of collar necroses and butt rot in association with ash dieback. iForest 2017, 10, 529–536. [Google Scholar] [CrossRef]
- Cleary, M.R.; Daniel, G.; Stenlid, J. Light and scanning electron microscopy studies of the early infection stages of Hymenoscyphus pseudoalbidus on Fraxinus excelsior. Plant Pathol. 2013, 62, 1294–1301. [Google Scholar] [CrossRef]
- Haňáčková, Z.; Koukol, O.; Čmoková, A.; Zahradník, D.; Havrdová, L. Direct evidence of Hymenoscyphus fraxineus infection pathway through the petiole-shoot junction. For. Pathol. 2017, 47, 12370. [Google Scholar] [CrossRef]
- Lygis, V.; Bakys, R.; Gustienė, A.; Burokienė, D.; Matelis, A.; Vasaitis, R. Forest self-regeneration following clear-felling of dieback-affected Fraxinus excelsior: Focus on ash. Eur. J. Forest. Res. 2014, 133, 501–510. [Google Scholar] [CrossRef]
- Juodvalkis, A.; Vasiliauskas, A. Lietuvos uosynų džiûvimo apimtys ir jas lemiantys veiksniai [The extent and possible causes of dieback of ash stands in Lithuania]. LŽŪU Mokslo Darbai. Biomed. Mokslai. 2002, 56, 17–22, (In Lithuanian with English Summary). [Google Scholar]
- Anonymous. Lietuvos Miškų Ūkio Statistika. In Lithuanian Statistical Yearbook of Forestry; L.R. Aplinkos Ministerija, Valstybinė Miškų Tarnyba: Lututė, Kaunas, 2001. [Google Scholar]
- Anonymous. Lietuvos Miškų Ūkio Statistika. In Lithuanian Statistical Yearbook of Forestry; L.R. Aplinkos Ministerija, Valstybinė Miškų Tarnyba: Lututė, Kaunas, 2021. [Google Scholar]
- Marigo, G.; Peltier, J.P.; Girel, J.; Pautou, G. Success in the demographic expansion of Fraxinus excelsior L. Trees 2000, 15, 1–13. [Google Scholar] [CrossRef]
- Dobrowolska, D.; Hein, S.; Oosterbaan, A.; Wagner, S.; Clark, J.; Skovsgaard, J.P. A review of European ash (Fraxinus excelsior L.): Implications for silviculture. Forestry 2011, 84, 133–148. [Google Scholar] [CrossRef]
- Pautasso, M.; Aas, G.; Queloz, V.; Holdenrieder, O. European ash (Fraxinus excelsior) dieback—A conservation biology challenge. Biol. Conserv. 2013, 158, 37–49. [Google Scholar] [CrossRef]
- Farr, D.F.; Rossman, A.Y. Fungal Databases. Systematic Mycology and Microbiology Laboratory, ARS, USDA. Available online: https://nt.ars-grin.gov/fungaldatabases/ (accessed on 10 June 2022).
- Mitchell, R.J.; Bailey, S.; Beaton, J.K.; Bellamy, P.E.; Brooker, R.W.; Broome, A.; Chetcuti, J.; Eaton, S.; Ellis, C.J.; Faren, J.; et al. The Potential Ecological Impact of Ash Dieback in the UK, JNCC Report No. 483; JNCC: Peterborough, UK, 2014; ISSN 0963-8091. [Google Scholar]
- Barklund, P. Unknown fungus causes dieback of ash shoots. SkogsEko 2006, 3, 10–11. (In Swedish) [Google Scholar]
- Bakys, R.; Vasaitis, R.; Barklund, P.; Ihrmark, K.; Stenlid, J. Investigations concerning the role of Chalara fraxinea in declining Fraxinus excelsior. Plant Pathol. 2009, 58, 284–292. [Google Scholar] [CrossRef]
- Hietala, A.M.; Børja, I.; Cross, H.; Nagy, N.E.; Solheim, H.; Timmermann, V.; Vivian-Smith, A. Dieback of European ash: What can we learn from the microbial community and species-specific traits of endophytic fungi associated with ash? In Endophytes of Forest Trees; Pirttilä, A., Frank, A., Eds.; Springer: Cham, Switzerland, 2018; pp. 229–258. [Google Scholar]
- Cross, H.; Sønstebø, J.H.; Nagy, N.E.; Timmermann, V.; Solheim, H.; Børja, I.; Kauserud, H.; Carlsen, T.; Rzepka, B.; Wasak, K.; et al. Fungal diversity and seasonal succession in ash leaves infected by the invasive ascomycete Hymenoscyphus fraxineus. New Phytol. 2017, 213, 1405–1417. [Google Scholar] [CrossRef]
- Reiher, D.B.A. Leaf-inhabiting endophytic fungi in the canopy of the Leipzig floodplain forest. Ph.D. Thesis, University of Leipzig, Leipzig, Germany, 10 June 2011; pp. 1–133. [Google Scholar]
- Scholtysik, A.; Unterseher, M.; Otto, P.; Wirth, C. Spatio-temporal dynamics of endophyte diversity in the canopy of European ash (Fraxinus excelsior). Mycol. Prog. 2013, 12, 291–334. [Google Scholar] [CrossRef]
- Trapiello, E.; Schoebel, C.N.; Rigling, D. Fungal community in symptomatic ash leaves in Spain. Balt. For. 2017, 23, 68–73. [Google Scholar]
- Lygis, V.; Vasiliauskas, R.; Larsson, K.; Stenlid, J. Wood-inhabiting fungi in stems of Fraxinus excelsior in declining ash stands of northern Lithuania, with particular reference to Armillaria cepistipes. Scand. J. For. Res. 2005, 20, 337–346. [Google Scholar] [CrossRef]
- Agostinelli, M.; Nguyen, D.; Witzell, J.; Cleary, M. Mycobiome of Fraxinus excelsior with different phenotypic susceptibility to ash dieback. Front. For. Glob. Chang. 2021, 4, 580514. [Google Scholar] [CrossRef]
- Witzell, J.; Martín, J.A. Endophytes and forest health. In Endophytes of Forest Trees; Pirttilä, A., Frank, A., Eds.; Springer: Cham, Switzerland, 2018; pp. 261–282. [Google Scholar]
- Schulz, B.; Boyle, C. The endophytic continuum. Mycol. Res. 2005, 109, 661–686. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, R.J.; White, J.F., Jr.; Arnold, A.E.; Redman, R.S. Fungal endophytes: Diversity and functional roles. New Phytol. 2009, 182, 314–330. [Google Scholar] [CrossRef] [PubMed]
- Bhardwaj, A.; Agrawal, P.K. A review fungal endophytes: As a store house of bioactive compound. World. J. Pharm. Pharm. Sci. 2014, 3, 228–237. [Google Scholar]
- Thongsandee, W.; Matsuda, Y.; Shimizu, M.; Ehara, H.; Ito, S. Isolation of endophytic Streptomycetes from above- and belowground organs of Quercus serrata. J. For. Res. 2013, 18, 179–189. [Google Scholar] [CrossRef]
- Strobel, G. The emergence of endophytic microbes and their biological promise. J. Fungi 2018, 4, 57. [Google Scholar] [CrossRef] [PubMed]
- Terhonen, E.; Blumenstein, K.; Kovalchuk, A.; Asiegbu, F.O. 2019. Forest tree microbiomes and associated fungal endophytes: Functional roles and impact on forest health. Forests 2019, 10, 42. [Google Scholar] [CrossRef]
- Kennedy, T.A.; Naeem, S.; Howe, K.M.; Knops, J.M.H.; Tilman, D.; Reich, P. Biodiversity as a barrier to ecological invasion. Nature 2002, 417, 636–638. [Google Scholar] [CrossRef]
- Bairey, E.; Kelsic, E.D.; Kishony, R. High-order species interactions shape ecosystem diversity. Nat. Commun. 2016, 7, 12285. [Google Scholar] [CrossRef]
- Stener, L.G. Clonal differences in susceptibility to the dieback of Fraxinus excelsior in southern Sweden. Scand. J. For. Res. 2013, 28, 205–216. [Google Scholar] [CrossRef]
- Pliūra, A.; Lygis, V.; Suchockas, V.; Bartkevičius, E. Performance of twenty-four European Fraxinus excelsior populations in three Lithuanian progeny trials with a special emphasis on resistance to Chalara fraxinea. Balt. For. 2011, 17, 17–34. [Google Scholar]
- McKinney, L.V.; Nielsen, L.R.; Collinge, D.B.; Thomsen, I.M.; Hansen, J.K.; Kjaer, E.D. The ash dieback crisis: Genetic variation in resistance can prove a long-term solution. Plant. Pathol. 2014, 63, 485–499. [Google Scholar] [CrossRef]
- Rosling, A.; Landeweert, R.; Lindahl, B.D.; Larsson, K.-H.; Kuyper, T.W.; Taylor, A.F.S.; Finlay, R.D. Vertical distribution of ectomycorrhizal fungal taxa in a podzol soil profile. New Phytol. 2003, 159, 775–783. [Google Scholar] [CrossRef]
- Ihrmark, K.; Bodeker, T.M.; Cruz-Martinez, K.; Friberg, H.; Kubartova, A.; Schenck, J.; Strid, Y.; Stenlid, J.; Brandstrom-Durling, M.; Clemmensen, K.E.; et al. New primers to amplify the fungal ITS2 region—Evaluation by 454-sequencing of artificial and natural communities. FEMS Microbiol. Ecol. 2012, 82, 666–677. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. In PCR Protocols, A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Snisky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 2018; pp. 315–322. [Google Scholar]
- Ter Braak, C.J.F.; Šmilauer, P. Canoco Reference Manual and User’s Guide: Software for Ordination, Version 5.10. Microcomputer Power: Ithaca, NY, USA, 2018; 536p. [Google Scholar]
- Bakys, R.; Vasiliauskas, A.; Ihrmark, K.; Stenlid, J.; Menkis, A.; Vasaitis, R. Root rot, associated fungi and their impact on health condition of declining Fraxinus excelsior stands in Lithuania. Scand. J. For. Res. 2011, 26, 128–135. [Google Scholar] [CrossRef]
- Díaz-Yáñez, O.; Mola-Yudego, B.; Timmermann, V.; Tollefsrud, M.M.; Hietala, A.M.; Oliva, J. The invasive forest pathogen Hymenoscyphus Fraxineus boosts mortality and triggers niche replacement of European ash (Fraxinus excelsior). Sci. Rep. 2020, 10, 5310. [Google Scholar] [CrossRef] [PubMed]
- Hietala, A.M.; Børja, I.; Solheim, H.; Nagy, N.E.; Timmermann, V. Propagule pressure build-up by the invasive Hymenoscyphus fraxineus following its introduction to an ash forest inhabited by the native Hymenoscyphus albidus. Front. Plant Sci. 2018, 9, 1087. [Google Scholar] [CrossRef] [PubMed]
- Lahiri, A.; Murphy, B.R.; Hodkinson, T.R. Assessing genotypic and environmental effects on endophyte communities of Fraxinus (Ash) using culture dependent and independent DNA sequencing. J. Fungi 2021, 7, 565. [Google Scholar] [CrossRef]
- Kowalski, T.; Bilański, P. Fungi detected in the previous year’s leaf petioles of Fraxinus excelsior and their antagonistic potential against Hymenoscyphus fraxineus. Forests 2021, 12, 1412. [Google Scholar] [CrossRef]
- Agan, A.; Drenkhan, R.; Adamson, K.; Tedersoo, L.; Solheim, H.; Børja, I.; Matsiakh, I.; Timmermann, V.; Nagy, N.E.; Hietala, A.M. The relationship between fungal diversity and invasibility of a foliar niche—the case of ash dieback. J. Fungi 2020, 6, 150. [Google Scholar] [CrossRef]
- Schlegel, M.; Dubach, V.; von Buol, L.; Sieber, T.N. Effects of endophytic fungi on the ash dieback pathogen. FEMS Microbiol. Ecol. 2016, 92, 142. [Google Scholar] [CrossRef]
- Dvořák, M.; Rotkova, G.; Botella, L. Detection of airborne inoculum of Hymenoscyphus fraxineus and H. albidus during seasonal fluctuations associated with absence of apothecia. Forests 2015, 7, 1. [Google Scholar] [CrossRef]
- Koukol, O.; Haňáčková, Z.; Dvořák, M.; Havrdová, L. Unseen, but still present in Czechia: Hymenoscyphus albidus detected by real-time PCR, but not by intensive sampling. Mycol. Prog. 2016, 15, 6. [Google Scholar] [CrossRef]
- Baral, H.-O.; Bemmann, M. Hymenoscyphus fraxineus vs. Hymenoscyphus albidus—A comparative light microscopic study on the causal agent of European ash dieback and related foliicolous, stroma-forming species. Mycology 2014, 5, 228–290. [Google Scholar] [CrossRef] [PubMed]
- Queloz, V.; Grünig, C.R.; Berndt, R.; Kowalski, T.; Sieber, T.N.; Holdenrieder, O. Cryptic speciation in Hymenoscyphus albidus. For. Pathol. 2011, 41, 133–142. [Google Scholar] [CrossRef]
- Cleary, M.; Nguyen, D.; Marčiulynienė, D.; Berlin, A.; Vasaitis, R.; Stenlid, J. Friend or foe? Biological and ecological traits of the European ash dieback pathogen Hymenoscyphus fraxineus in its native environment. Sci. Rep. 2016, 6, 21895. [Google Scholar] [CrossRef]
- Davydenko, K.; Vasaitis, R.; Stenlid, J.; Menkis, A. Fungi in foliage and shoots of Fraxinus excelsior in eastern Ukraine: A first report on Hymenoscyphus pseudoalbidus. For. Pathol. 2013, 43, 462–467. [Google Scholar] [CrossRef]
- Kowalski, T.; Kraj, W.; Bednarz, B. Fungi on stems and twigs in initial and advanced stages of dieback of European ash (Fraxinus excelsior) in Poland. Eur. J. For. Res. 2016, 135, 565–579. [Google Scholar] [CrossRef]
- Przybyl, K. Fungi associated with necrotic apical parts of Fraxinus excelsior shoots. For. Pathol. 2002, 32, 387–394. [Google Scholar] [CrossRef]
- Linaldeddu, B.T.; Bottecchia, F.; Bregant, C.; Maddau, L.; Montecchio, L. Diplodia fraxini and Diplodia subglobosa: The main species associated with cankers and dieback of Fraxinus excelsior in north-eastern Italy. Forests 2020, 11, 883. [Google Scholar] [CrossRef]
- Zimmerman, N.B.; Vitousek, P.M. Fungal endophyte communities reflect environmental structuring across a Hawaiian landscape. Proc. Natl. Acad. Sci. USA 2012, 109, 13022–13027. [Google Scholar] [CrossRef]
- Borrell, A.N.; Shi, Y.; Gan, Y.; Bainard, L.D.; Germida, J.J.; Hamel, C. Fungal diversity associated with pulses and its influence on the subsequent wheat crop in the Canadian prairies. Plant Soil 2017, 414, 13–31. [Google Scholar] [CrossRef]
- Weber, G.; Claus, M. The influence of chemical soil factors on the development of VA mycorrhizas of ash (Fraxinus excelsior) L. and sycamore (Acer pseudoplatanus L.) in pot experiments. J. Plant Nutr. Soil Sci. 2000, 163, 609–616. [Google Scholar] [CrossRef]
- Lang, C.; Seven, J.; Polle, A. Host preferences and differential contributions of deciduous tree species shape mycorrhizal species richness in a mixed central European forest. Mycorrhiza 2011, 21, 297–308. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).