Origins, Diversity and Naturalization of Eucalyptus globulus (Myrtaceae) in California
Abstract
1. Introduction
2. Materials and Methods
2.1. Collections and Naturalization
2.2. Naturalization Analysis
2.3. Chloroplast DNA
2.4. Nuclear DNA
2.5. Naturalization Analyses
3. Results
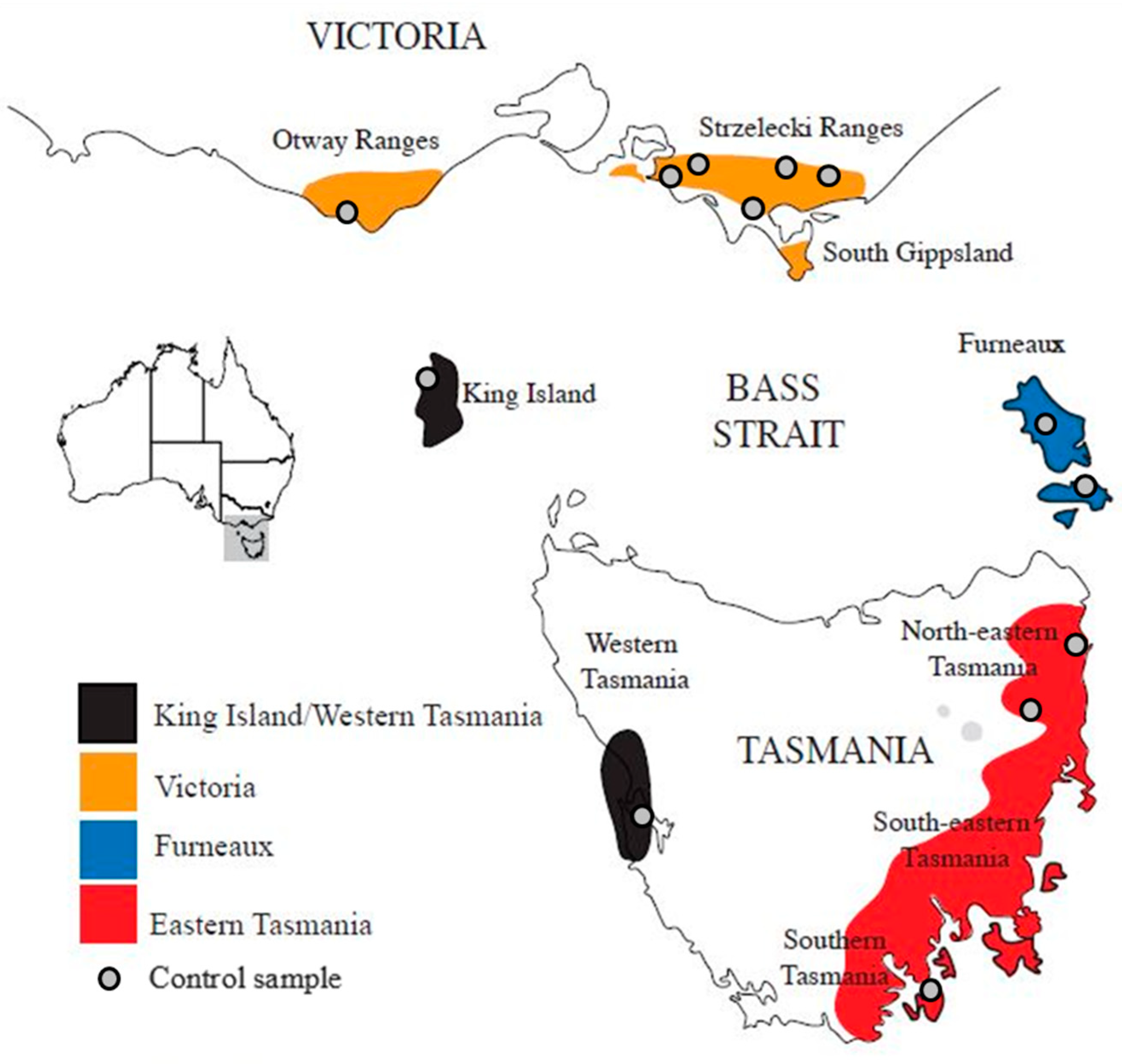
3.1. Native Origin of Californian E. Globulus
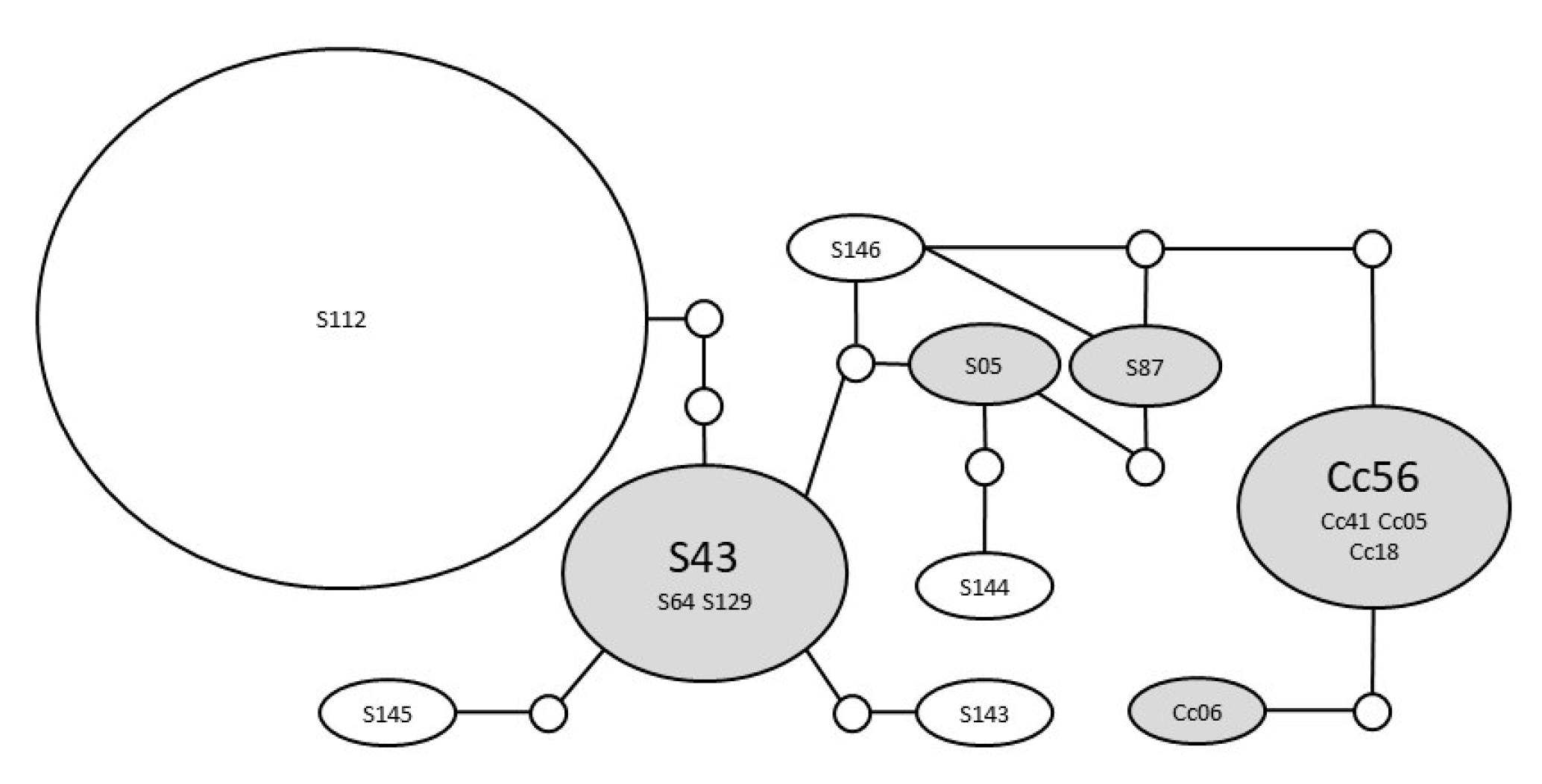
3.2. Genetic Diversity in California
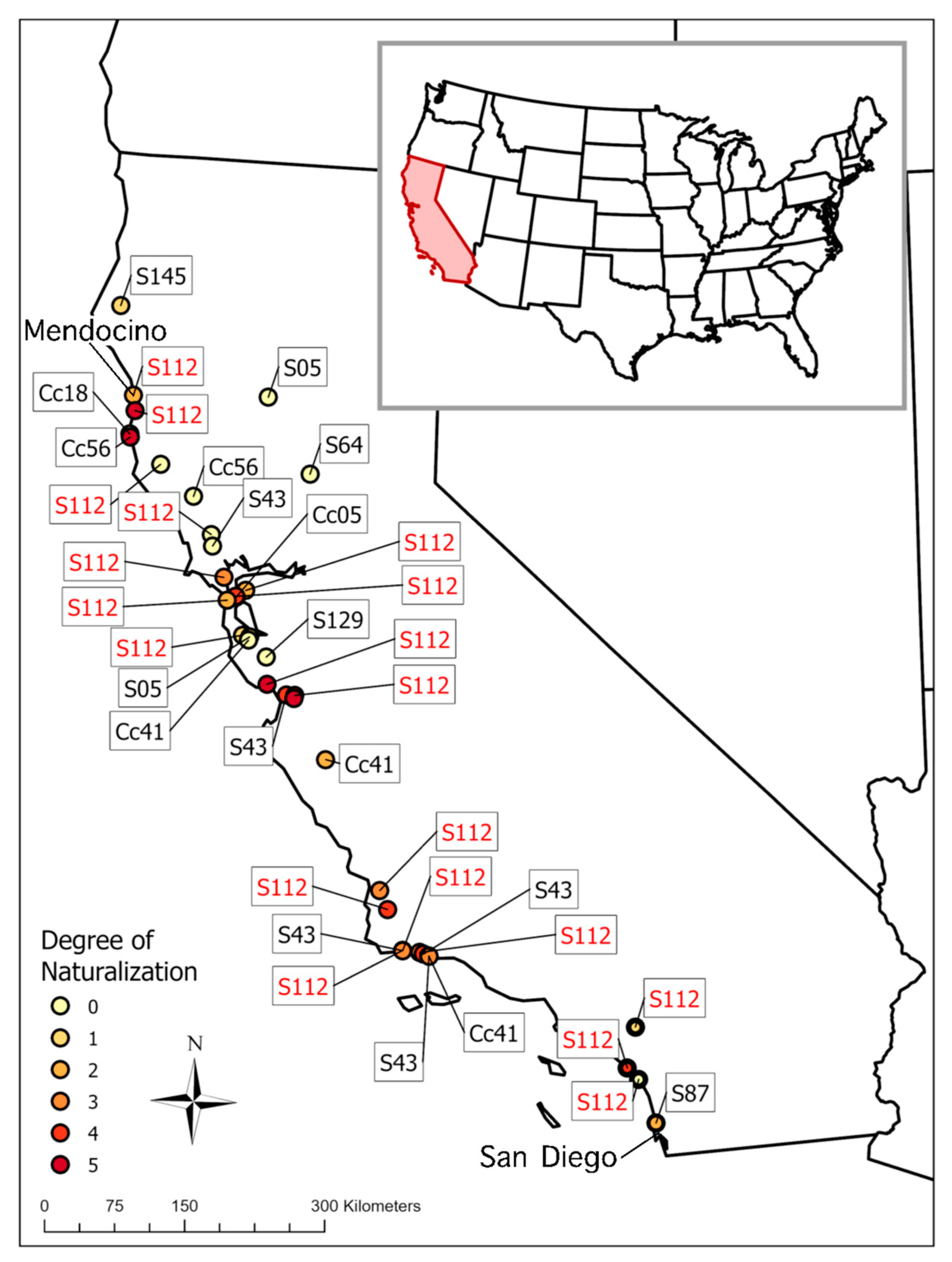
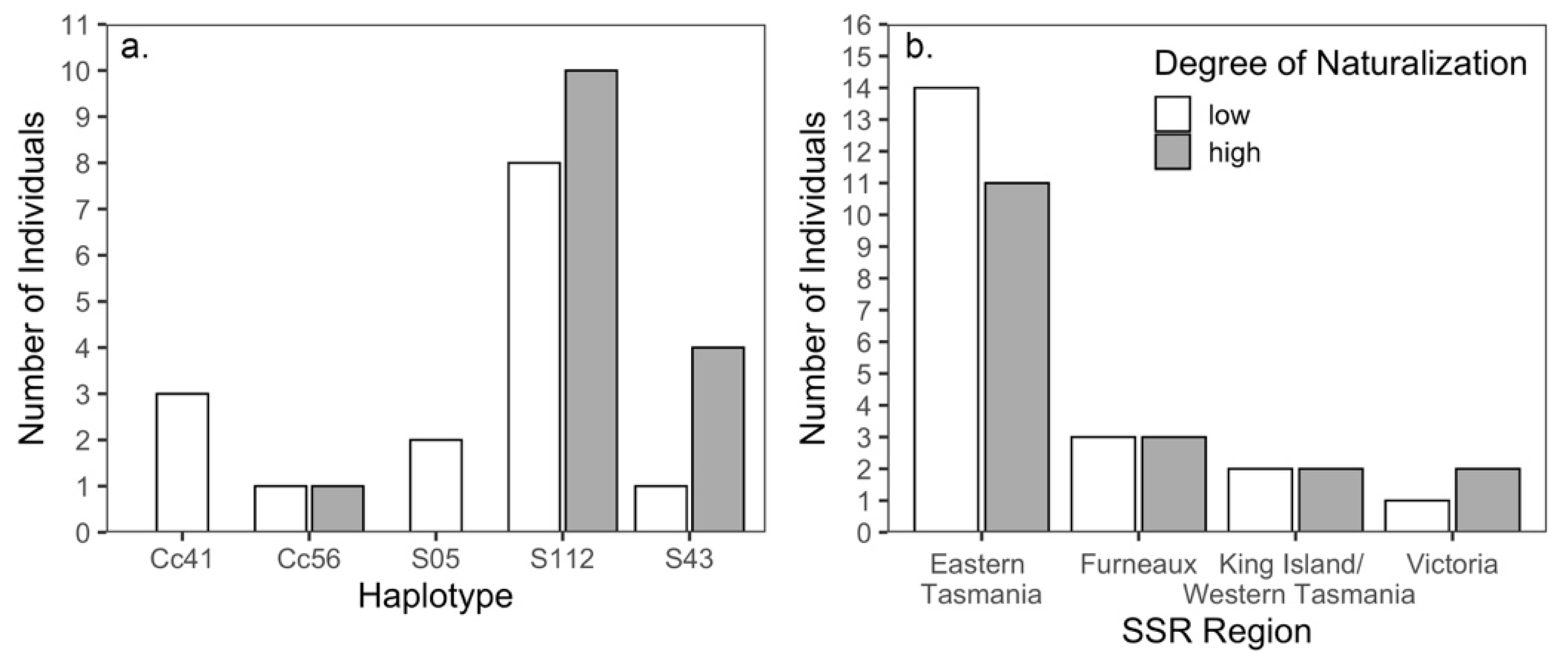
3.3. Naturalization in California
4. Discussion
4.1. Native Origin of Californian E. globulus
4.2. Genetic Diversity in California
4.3. Naturalization in California
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Character No. | 40 | 44 | 45 | 54 | 55 | 73 A | 84 | 91 | 112 B |
|---|---|---|---|---|---|---|---|---|---|
| Haplotype | Poly T | Point Mutation | Poly A | Poly A | Point Mutation | 1st Multistate Repeat | Poly A | Multistate Repeat C | 2nd Repeat |
| Location (bp) | 297–314 | 324 | 326–352 | 386–401 | 408 | 519–544 | 663–673 | 755–928 | 1066–1090 |
| Cc05 | 9T | - | 12A | 11A | C | Present | 10A | a | Absent |
| Cc06 | 9T | - | 11A | 11A | C | Absent | 9A | a | Absent |
| Cc18 | 9T | - | 12A | 12A | C | Absent | 9A | a | Present |
| Cc41 | 9T | - | 11A | 12A | C | Present | 9A | a | Present |
| Cc56 | 9T | - | 11A | 11A | C | Present | 10A | a | Absent |
| S05 | 15T (A at bp 10) | T | 21A | 7A | C | Absent | 10A | a and b | Absent |
| S43 | 14T (A at bp 9) | T | 19A (T at bp 6) | 7A | C | Absent | 10A | a and b | Absent |
| S64 | 14T (A at bp 9) | T | 18A (T at bp 6) | 7A | C | Absent | 11A | 3 a and 1 b | Absent |
| S87 | 14T (A at bp 9) | - | 20A | 7A | C | Absent | 10A | a and b | Absent |
| S112 | 14T (A at bp 9) | T | 26A (T at bp 6, 7 and 13) | 7A | T | Absent | 10A | a and b | Absent |
| S129 | 14T | T | 19A (T at bp 6) | 7A | C | Absent | 10A | a and b | Absent |
| S143 | 14T (A at bp 9) | T | 19A (T at bp 6) | 7A | C | Absent | 10A | a and c | Absent |
| S144 | 14T (A at bp 9 and 11) | T | 19A | 7A | C | Absent | 11A | a and b | Absent |
| S145 | 14T (A at bp 9) | C | 18A (T at bp 5) | 7A | C | Absent | 10A | a and b | Absent |
| S146 | 14T (A at bp 9) | - | 16A | 7A | C | Absent | 10A | a and b | Absent |
References
- Brooker, M.I.H. A new classification of the genus Eucalyptus L’Her. (Myrtaceae). Aust. Syst. Bot. 2000, 13, 79–148. [Google Scholar] [CrossRef]
- Slee, A.; Brooker, M.; Duffy, S.; West, J. EUCLID: Eucalypts of Australia; CSIRO Publishing: Canberra, Australia, 2006. [Google Scholar]
- Nicolle, D. Classification of the Eucalypts (Angophora, Corymbia and Eucalyptus) Version 4. Available online: http://www.dn.com.au/Classification-Of-The-Eucalypts.pdf (accessed on 2 April 2021).
- Eldridge, K.; Davidson, J.; Harwood, C.; van Wyk, G. Eucalypt Domestication and Breeding; Clarendon Press: Oxford, UK, 1993. [Google Scholar]
- Doughty, R.W. The Eucalyptus. A Natural and Commercial History of the Gum Tree; The Johns Hopkins University Press: Baltimore, MD, USA; London, UK, 2000; p. 237. [Google Scholar]
- Simberloff, D.; Rejmánek, M. Eucalypts. Encyclopedia of Biological Invasions; Simberloff, D., Rejmanek, M., Eds.; University of California Press: Berkeley, CA, USA, 2011; pp. 203–209. [Google Scholar]
- Tomé, M.; Almeida, M.H.; Barreiro, S.; Branco, M.R.; Deus, E.; Pinto, G.; Silva, J.S.; Soares, P.; Rodríguez-Soalleiro, R. Opportunities and challenges of Eucalyptus plantations in Europe: The Iberian Peninsula experience. Eur. J. For. Res. 2021, 140, 489–510. [Google Scholar] [CrossRef]
- Potts, B.M.; Vaillancourt, R.E.; Jordan, G.J.; Dutkowski, G.W.; Costa e Silva, J.; McKinnon, G.E.; Steane, D.A.; Volker, P.W.; Lopez, G.A.; Apiolaza, L.A.; et al. Exploration of the Eucalyptus globulus gene pool. In Proceedings of the Eucalyptus in a Changing World, Aveiro, Portugal, 11–15 October 2004; pp. 46–61. [Google Scholar]
- Ritter, M.; Yost, J. Diversity, reproduction, and potential for invasiveness of Eucalyptus in California. Madrono 2009, 56, 155–167. [Google Scholar] [CrossRef]
- Richardson, D.M.; Rejmánek, M. Trees and shrubs as invasive alien species—A global review. Divers. Distrib. 2011, 17, 788–809. [Google Scholar] [CrossRef]
- Larcombe, M.J.; Silva, J.S.; Vaillancourt, R.E.; Potts, B.M. Assessing the invasive potential of Eucalyptus globulus in Australia: Quantification of wildling establishment from plantations. Biol. Invasions 2013, 15, 2763–2781. [Google Scholar] [CrossRef]
- Catry, F.X.; Moreira, F.; Deus, E.; Silva, J.S.; Aguas, A. Assessing the extent and the environmental drivers of Eucalyptus globulus wildling establishment in Portugal: Results from a countrywide survey. Biol. Invasions 2015, 17, 3163–3181. [Google Scholar] [CrossRef]
- dos Santos, P.; Matias, H.; Deus, E.; Aguas, A.; Silva, J.S. Fire effects on capsules and encapsulated seeds from Eucalyptus globulus in Portugal. Plant Ecol. 2015, 216, 1611–1621. [Google Scholar] [CrossRef]
- Águas, A.; Larcombe, M.J.; Matias, H.; Deus, E.; Potts, B.M.; Rego, F.C.; Silva, J.S. Understanding the naturalization of Eucalyptus globulus in Portugal: A comparison with Australian plantations. Eur. J. For. Res. 2017, 136, 433–446. [Google Scholar] [CrossRef]
- Anjos, A.; Fernandes, P.; Marques, C.; Borralho, N.; Valente, C.; Correia, O.; Máguas, C.; Chozas, S. Management and fire, a critical combination for Eucalyptus globulus dispersal. For. Ecol. Manag. 2021, 490, 119086. [Google Scholar] [CrossRef]
- Forsyth, G.G.; Richardson, D.M.; Brown, P.J.; Wilgen, B.W.V. A rapid assessment of the invasive status of Eucalyptus species in two South African provinces: Working for water. S. Afr. J. Sci. 2004, 100, 75–77. [Google Scholar] [CrossRef]
- Águas, A.; Ferreira, A.; Maia, P.; Fernandes, P.M.; Roxo, L.; Keizer, J.; Silva, J.S.; Rego, F.C.; Moreira, F. Natural establishment of Eucalyptus globulus Labill. in burnt stands in Portugal. For. Ecol. Manag. 2014, 323, 47–56. [Google Scholar] [CrossRef]
- Fernandes, P.; Antunes, C.; Pinho, P.; Máguas, C.; Correia, O. Natural regeneration of Pinus pinaster and Eucalyptus globulus from plantation into adjacent natural habitats. For. Ecol. Manag. 2016, 378, 91–102. [Google Scholar] [CrossRef]
- Deus, E.; Silva, J.S.; Larcombe, M.J.; Catry, F.X.; Queirós, L.; dos Santos, P.; Matias, H.; Águas, A.; Rego, F.C. Investigating the invasiveness of Eucalyptus globulus in Portugal: Site-scale drivers, reproductive capacity and dispersal potential. Biol. Invasions 2019, 21, 2027–2044. [Google Scholar] [CrossRef]
- Richardson, D.M.; Pyšek, P.; Rejmánek, M.; Barbour, M.G.; Panetta, F.D.; West, C.J. Naturalization and invasion of alien plants: Concepts and definitions. Divers. Distrib. 2000, 6, 93–107. [Google Scholar] [CrossRef]
- Santos, R.L. The Eucalyptus of California: Seeds of Good or Seeds of Evil; California State University: Stanislaus, CA, USA, 1997. [Google Scholar]
- Groenendaal, G.M. Eucalyptus helped solve a timber problem: 1853–1880. In Proceedings of the Workshop on Eucalyptus in California, Sacramento, CA, USA, 14–16 June 1983; pp. 1–8. [Google Scholar]
- Thompson, K. The Australian fever tree in California: Eucalypts and malaria prophylaxis. Ann. Assoc. Am. Geogr. 1970, 60, 230–244. [Google Scholar] [CrossRef] [PubMed]
- Butterfield, H.M. The introduction of Eucalyptus into California. Madrono 1935, 3, 149–154. [Google Scholar]
- Bracewell, R.N. Trees of Stanford and Environs. Available online: https://trees.stanford.edu/ENCYC/EUCglo.htm (accessed on 16 July 2018).
- Farmer, J. Trees in Paradise: A California History; WW Norton & Company: New York, NY, USA, 2013. [Google Scholar]
- Wolf, K.M.; DiTomaso, J. Management of blue gum eucalyptus in California requires region-specific consideration. Calif. Agric. 2016, 70, 39–47. [Google Scholar] [CrossRef]
- Kirkpatrick, J.B. Eucalypt invasion in Southern California. Aust. Geogr. 1977, 13, 387–393. [Google Scholar] [CrossRef]
- Ritter, M. Myrtaceae: Eucalyptus. In The Jepson Manual of Higher Plants in California; Baldwin, B.G., Goldman, D.H., Keil, D.J., Patterson, R., Rosatti, T.J., Wilken, D.H., Eds.; University of California Press: Berkeley, CA, USA, 2012; pp. 913–914. [Google Scholar]
- California Invasive Plant Council. Cal-IPC Publication 2015. Available online: http://cal-ipc.org/paf/site/paf/538 (accessed on 9 July 2018).
- California Invasive Plant Council. CalWeedMapper. Available online: https://www.cal-ipc.org/resources/calweedmapper/ (accessed on 11 June 2021).
- Queirós, L.; Deus, E.; Silva, J.S.; Vicente, J.; Ortiz, L.; Fernandes, P.M.; Castro-Díez, P. Assessing the drivers and the recruitment potential of Eucalyptus globulus in the Iberian Peninsula. For. Ecol. Manag. 2020, 466, 118147. [Google Scholar] [CrossRef]
- Jordan, G.; Potts, B.M.; Wiltshire, R. Strong, independent quantitative genetic control of vegetative phase change and first flowering in Eucalyptus globulus ssp. globulus. Heredity 1999, 83, 179–187. [Google Scholar] [CrossRef] [PubMed]
- Dutkowski, G.W.; Potts, B.M. Genetic variation in the susceptibility of Eucalyptus globulus to drought damage. Tree Genet. Genomes 2012, 8, 757–773. [Google Scholar] [CrossRef]
- Freeman, J.S.; Hamilton, M.G.; Lee, D.J.; Pegg, G.S.; Brawner, J.T.; Tilyard, P.A.; Potts, B.M. Comparison of host susceptibilities to native and exotic pathogens provides evidence for pathogen-imposed selection in forest trees. New Phytol. 2018, 221, 2261–2272. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.T.; Hui, C.; Thornhill, A.H.; Gallien, L.; Le Roux, J.J.; Richardson, D.M. Is invasion success of Australian trees mediated by their native biogeography, phylogenetic history, or both? AoB Plants 2016, 9. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zenni, R.D.; Dickie, I.A.; Wingfield, M.J.; Hirsch, H.; Crous, C.J.; Meyerson, L.A.; Burgess, T.I.; Zimmermann, T.G.; Klock, M.M.; Siemann, E.; et al. Evolutionary dynamics of tree invasions: Complementing the unified framework for biological invasions. AoB Plants 2016, 9, plw085. [Google Scholar] [CrossRef] [PubMed]
- Zenni, R.D.; Bailey, J.K.; Simberloff, D. Rapid evolution and range expansion of an invasive plant are driven by provenance–environment interactions. Ecol. Lett. 2014, 17, 727–735. [Google Scholar] [CrossRef]
- Dutkowski, G.W.; Potts, B.M. Geographic patterns of genetic variation in Eucalyptus globulus ssp. globulus and a revised racial classification. Aust. J. Bot. 1999, 47, 237–263. [Google Scholar]
- Jones, T.H.; Steane, D.A.; Jones, R.C.; Pilbeam, D.; Vaillancourt, R.E.; Potts, B.M. Effects of domestication on genetic diversity in Eucalyptus globulus. For. Ecol. Manag. 2006, 234, 78–84. [Google Scholar] [CrossRef]
- Yeoh, S.H.; Bell, J.C.; Foley, W.J.; Wallis, I.R.; Moran, G.F. Estimating population boundaries using regional and local-scale spatial genetic structure: An example in Eucalyptus globulus. Tree Genet. Genomes 2012, 8, 695–708. [Google Scholar] [CrossRef]
- Jones, R.C.; Steane, D.A.; Lavery, M.; Vaillancourt, R.E.; Potts, B.M. Multiple evolutionary processes drive the patterns of genetic differentiation in a forest tree species complex. Ecol. Evol. 2013, 3, 1–17. [Google Scholar] [CrossRef]
- Costa, J.; Vaillancourt, R.E.; Steane, D.A.; Jones, R.C.; Marques, C. Microsatellite analysis of population structure in Eucalyptus globulus. Genome 2017, 60, 770–777. [Google Scholar] [CrossRef]
- Freeman, J.S.; Jackson, H.D.; Steane, D.A.; McKinnon, G.E.; Dutkowski, G.W.; Potts, B.M.; Vaillancourt, R.E. Chloroplast DNA phylogeography of Eucalyptus globulus. Aust. J. Bot. 2001, 49, 585–596. [Google Scholar] [CrossRef]
- Freeman, J.S.; Marques, C.M.P.; Carocha, V.; Borralho, N.; Potts, B.M.; Vaillancourt, R.E. Origins and diversity of the Portuguese Landrace of Eucalyptus globulus. Ann. For. Sci. 2007, 64, 639–647. [Google Scholar] [CrossRef]
- McKinnon, G.E.; Vaillancourt, R.E.; Steane, D.A.; Potts, B.M. The rare silver gum, Eucalyptus cordata, is leaving its trace in the organellar gene pool of Eucalyptus globulus. Mol. Ecol. 2004, 13, 3751–3762. [Google Scholar] [CrossRef] [PubMed]
- McKinnon, G.E.; Vaillancourt, R.E.; Tilyard, P.A.; Potts, B.M. Maternal inheritance of the chloroplast genome in Eucalyptus globulus and interspecific hybrids. Genome 2001, 44, 831–835. [Google Scholar] [CrossRef] [PubMed]
- Jones, T.H.; Vaillancourt, R.E.; Potts, B.M. Detection and visualization of spatial genetic structure in continuous Eucalyptus globulus forest. Mol. Ecol. 2007, 16, 697–707. [Google Scholar] [CrossRef]
- Calviño-Cancela, M.; Rubido-Bara, M. Invasive potential of Eucalyptus globulus: Seed dispersal, seedling recruitment and survival in habitats surrounding plantations. For. Ecol. Manag. 2013, 305, 129–137. [Google Scholar] [CrossRef]
- Byrne, M. Phylogeny, diversity and evolution of eucalypts. In Plant Genome: Biodiversity and Evolution; Sharma, A.K., Sharma, A., Eds.; Science Publishers: Enfield, NH, USA, 2008; Volume 1, pp. 303–346. [Google Scholar]
- McKinnon, G.E.; Jordan, G.J.; Vaillancourt, R.E.; Steane, D.A.; Potts, B.M. Glacial refugia and reticulate evolution: The case of the Tasmanian eucalypts. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2004, 359, 275–284. [Google Scholar] [CrossRef]
- Swofford, D. PAUP: Phylogenetic Analysis Using Parsimony; Illinois Natural History Survey: Champaign, IL, USA, 1991. [Google Scholar]
- Clement, M.; Posada, D.; Crandall, K.A. TCS: A computer program to estimate gene genealogies. Mol. Ecol. 2000, 9, 1657–1659. [Google Scholar] [CrossRef]
- R-Core-Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013; Available online: https://www.R-project.org/ (accessed on 19 August 2021).
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; Stevens, M.R.H.; Wagner, H. Vegan: Community Ecology Package. R Package Version 2.0-2. 2012. Available online: http://cran.r-project.org/ (accessed on 19 August 2021).
- Brondani, R.P.V.; Brondani, C.; Tarchini, R.; Grattapaglia, D. Development, characterization and mapping of microsatellite markers in Eucalyptus grandis and E. urophylla. Theor. Appl. Genet. 1998, 97, 816–827. [Google Scholar] [CrossRef]
- Brondani, R.P.V.; Brondani, C.; Grattapaglia, D. Towards a genus-wide reference linkage map for Eucalyptus based exclusively on highly informative microsatellite markers. Mol. Genet. Genom. 2002, 267, 338–347. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, M.M.; Sanchez, L.; Ribeiro, C.; Cunha, F.; Araujo, J.; Borralho, N.M.G.; Marques, C. A case study of Eucalyptus globulus fingerprinting for breeding. Ann. For. Sci. 2011, 68, 701–714. [Google Scholar] [CrossRef]
- Freeman, J.S.; Potts, B.M.; Shepherd, M.; Vaillancourt, R.E. Parental and consensus linkage maps of Eucalyptus globulus using AFLP and microsatellite markers. Silvae Genet. 2006, 55, 202–217. [Google Scholar] [CrossRef]
- Steane, D.A.; Vaillancourt, R.E.; Russell, J.; Powell, W.; Marshall, D.; Potts, B.M. Microsatellite variation in a forest tree, Eucalyptus globulus ssp. globulus (Myrtaceae). Silvae Genet. 2001, 50, 89–91. [Google Scholar]
- Schuelke, M. An economic method for the fluorescent labeling of PCR fragments. Nat. Biotechnol. 2000, 18, 233–234. [Google Scholar] [CrossRef]
- Steane, D.A.; Conod, N.; Jones, R.C.; Vaillancourt, R.E.; Potts, B.M. A comparative analysis of population structure of a forest tree, Eucalyptus globulus (Myrtaceae), using microsatellite markers and quantitative traits. Tree Genet. Genomes 2006, 2, 30–38. [Google Scholar] [CrossRef]
- Yeh, F.C.; Boyle, T.J.B. Population genetic analysis of co-dominant and dominant markers and quantitative traits. Belg. J. Bot. 1997, 129, 156–159. [Google Scholar]
- Goudet, J. FSTAT, a Program to Estimate and Test Gene Diversities and Fixation Indices Version 2.9.3.2. 2002. Available online: https://www.scienceopen.com/document?vid=79097bb4-ec3c-47c3-94a1-47085d721e6b (accessed on 19 August 2021).
- Peakall, R.; Smouse, P.E. GENALEX 6: Genetic Analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Rosenberg, N.A.; Donnelly, P. Association mapping in structured populations. Am. J. Hum. Genet. 2000, 67, 170–181. [Google Scholar] [CrossRef]
- Jordan, G.J.; Potts, B.M.; Kirkpatrick, J.B.; Gardiner, C. Variation in the Eucalyptus globulus complex revisited. Aust. J. Bot. 1993, 41, 763–785. [Google Scholar] [CrossRef]
- Astorga, R.; Soria, F.; Basurco, F.; Toval, G. Diversity analysis and genetic structure of Eucalyptus globulus Labill. In Proceedings of the Eucalyptus in a Changing World: An IUFRO Conference, Aveiro, Portugal, 11–15 October 2004; pp. 351–363. [Google Scholar]
- Zang, D.; Wang, H.; You, Y. Early results from progeny trial of Eucalyptus globulus in Yunnan Province, China. In Proceedings of the Australian Tree Species Research in China Proc. CRCTHF-IUFRO Workshop, Zhangzhou, China, 2–5 November 1992; pp. 64–69. [Google Scholar]
- López, G.A.; Potts, B.M.; Dutkowski, G.W.; Traverso, J.M.R. Quantitative genetics of Eucalyptus globulus: Affinities of land race and native stand localities. Silvae Genet. 2001, 50, 244–252. [Google Scholar]
- Kirkpatrick, J.B. The numerical intraspecific taxonomy of Eucalyptus globulus Labill. (Myrtaceae). Bot. J. Linn. Soc. 1974, 69, 89–104. [Google Scholar] [CrossRef]
- Anon. Landmark Trees of the City of Fremont; Landscape Architecture Division, Division of Community Services: Fremont, CA, USA, 2011. [Google Scholar]
- Spencer, R.D. Horticultural Flora of South-Eastern Australia, Flowering Plants: Dicotyledons; University of New South Wales Press: Sydney, Australia, 2002; Volume 3. [Google Scholar]
- Patterson, B.; Vaillancourt, R.E.; Potts, B.M. Eucalypt seed collectors: Beware of sampling seed lots from low in the canopy! Aust. For. 2001, 64, 139–142. [Google Scholar] [CrossRef]
- Hardner, C.M.; Vaillancourt, R.E.; Potts, B.M. Stand density influences outcrossing rate and growth of open-pollinated families of Eucalyptus globulus. Silvae Genet. 1996, 45, 226–228. [Google Scholar]
- Hardner, C.M.; Potts, B.M.; Gore, P.L. The relationship between cross success and spatial proximity of Eucalyptus globulus ssp. globulus parents. Evolution 1998, 52, 614–618. [Google Scholar]
- Leberg, P.L. Estimating allelic richness: Effects of sample size and bottlenecks. Mol. Ecol. 2002, 11, 2445–2449. [Google Scholar] [CrossRef] [PubMed]
- McKinnon, G.E.; Vaillancourt, R.E.; Jackson, H.D.; Potts, B.M. Chloroplast sharing in the Tasmanian eucalypts. Evolution 2001, 55, 703–711. [Google Scholar] [CrossRef]
- Fork, S.; Woolfolk, A.; Akhavan, A.; Van Dyke, E.; Murphy, S.; Candiloro, B.; Newberry, T.; Schreibman, S.; Salisbury, J.; Wasson, K. Biodiversity effects and rates of spread of nonnative eucalypt woodlands in central California. Ecol. Appl. 2015, 25, 2306–2319. [Google Scholar] [CrossRef] [PubMed]
- Nereu, M.; Silva, J.S.; Deus, E.; Nunes, M.; Potts, B. The effect of management operations on the demography of Eucalyptus globulus seedlings. For. Ecol. Manag. 2019, 453, 117630. [Google Scholar] [CrossRef]
- Silva, J.S.; Nereu, M.; Pinho, S.; Queirós, L.; Jesús, C.; Deus, E. Post-fire demography, drowth, and dontrol of Eucalyptus globulus wildlings. Forests 2021, 12, 156. [Google Scholar] [CrossRef]
- Deus, E.; Silva, J.S.; Marchante, H.; Marchante, E.; Felix, C. Are post-dispersed seeds of Eucalyptus globulus predated in the introduced range? Evidence from an experiment in Portugal. Web Ecol. 2018, 18, 67–79. [Google Scholar] [CrossRef]


| Inferred Clusters | N | ||||
|---|---|---|---|---|---|
| Given Population | Victoria | Furneaux | King Is./ W Tas | Eastern Tasmania | |
| California | 0.187 | 0.240 | 0.197 | 0.377 | 70 |
| Victoria | 0.974 | 0.007 | 0.010 | 0.009 | 222 |
| Furneaux | 0.008 | 0.981 | 0.007 | 0.005 | 50 |
| King Island/W Tas | 0.004 | 0.006 | 0.974 | 0.016 | 107 |
| Eastern Tasmania | 0.007 | 0.009 | 0.010 | 0.974 | 211 |
| Population/Region | Victoria | Furneaux | King Island/Western Tasmania | Eastern Tasmania |
|---|---|---|---|---|
| California | 0.099 | 0.089 | 0.116 | 0.043 |
| Victoria | 0.054 | 0.064 | 0.045 | |
| Furneaux | 0.102 | 0.040 | ||
| King Island/ Western Tasmania | 0.070 |
| Nuclear SSRs | cpDNA Haplotypes | |||||||
|---|---|---|---|---|---|---|---|---|
| Region | Na | Ne | AR | Ho | He | FIS | n¹ | d |
| California | 10.33 | 4.54 | 9.38 ± 3.95 | 0.58 | 0.73 | 0.19 | 67 | 6.20 ± 1.38 |
| Eastern Tasmania | 17.67 | 7.09 | 12.92 ± 3.16 | 0.71 | 0.84 | 0.15 | 342 | 12.82 ± 1.32 |
| King Island/ Western Tasmania | 13.83 | 4.51 | 10.91 ± 3.63 | 0.62 | 0.74 | 0.16 | 16 | 6.88 ± 0.33 |
| Furneaux | 10.50 | 5.34 | 10.04 ± 2.94 | 0.69 | 0.82 | 0.15 | 20 | 9.38 ± 0.90 |
| Victoria | 17.17 | 6.54 | 12.32 ± 3.84 | 0.68 | 0.81 | 0.17 | 87 | 10.31 ± 1.46 |
| Mean per native region | 14.79 | 5.87 | 11.55 | 0.69 | 0.80 | 0.15 | 116 | 9.85 |
| SE | 1.66 | 0.58 | 0.66 | 0.01 | 0.02 | 0.00 | 77 | 1.23 |
| Across all native E. globulus | 22.83 | 6.76 | 13.72 | 0.68 | 0.83 | 0.15 | 465 | 13.28 |
| SE | 2.21 | 0.99 | 1.42 | 0.05 | 0.03 | 1.20 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yost, J.M.; Wise, S.L.; Love, N.L.R.; Steane, D.A.; Jones, R.C.; Ritter, M.K.; Potts, B.M. Origins, Diversity and Naturalization of Eucalyptus globulus (Myrtaceae) in California. Forests 2021, 12, 1129. https://doi.org/10.3390/f12081129
Yost JM, Wise SL, Love NLR, Steane DA, Jones RC, Ritter MK, Potts BM. Origins, Diversity and Naturalization of Eucalyptus globulus (Myrtaceae) in California. Forests. 2021; 12(8):1129. https://doi.org/10.3390/f12081129
Chicago/Turabian StyleYost, Jennifer M., Sascha L. Wise, Natalie L. R. Love, Dorothy A. Steane, Rebecca C. Jones, Matt K. Ritter, and Brad M. Potts. 2021. "Origins, Diversity and Naturalization of Eucalyptus globulus (Myrtaceae) in California" Forests 12, no. 8: 1129. https://doi.org/10.3390/f12081129
APA StyleYost, J. M., Wise, S. L., Love, N. L. R., Steane, D. A., Jones, R. C., Ritter, M. K., & Potts, B. M. (2021). Origins, Diversity and Naturalization of Eucalyptus globulus (Myrtaceae) in California. Forests, 12(8), 1129. https://doi.org/10.3390/f12081129






