Heterozygous Trees Rebound the Fastest after Felling by Beavers to Positively Affect Arthropod Community Diversity
Abstract
1. Introduction
2. Materials and Methods
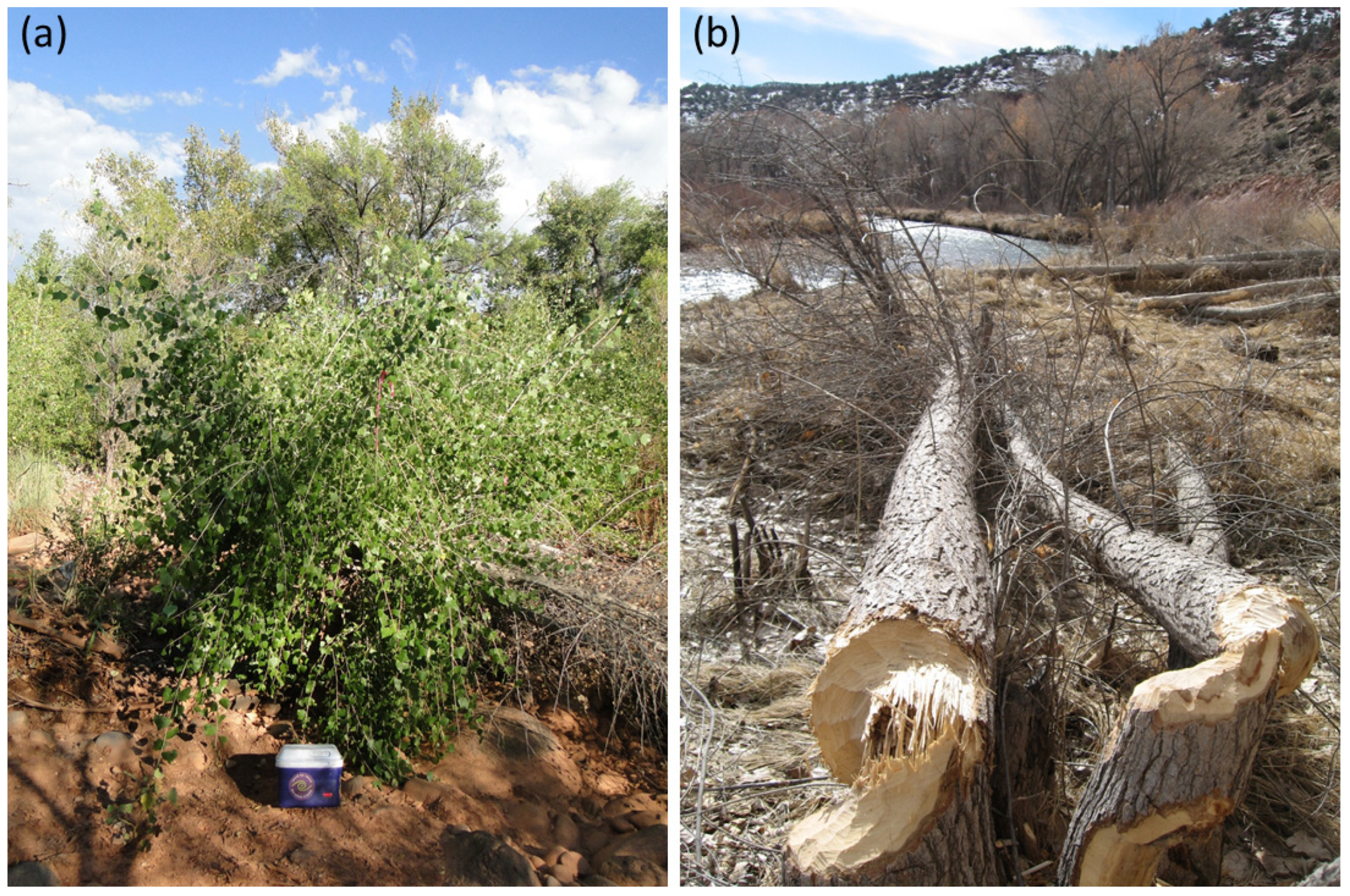
2.1. Study Site
2.2. Arthropod Surveys and Twig Chemistry
2.3. Microsatellite Genotyping
2.4. Productivity Measurement
2.5. Statistical Approach
2.6. Examination of Potentially Confounding Factors
3. Results
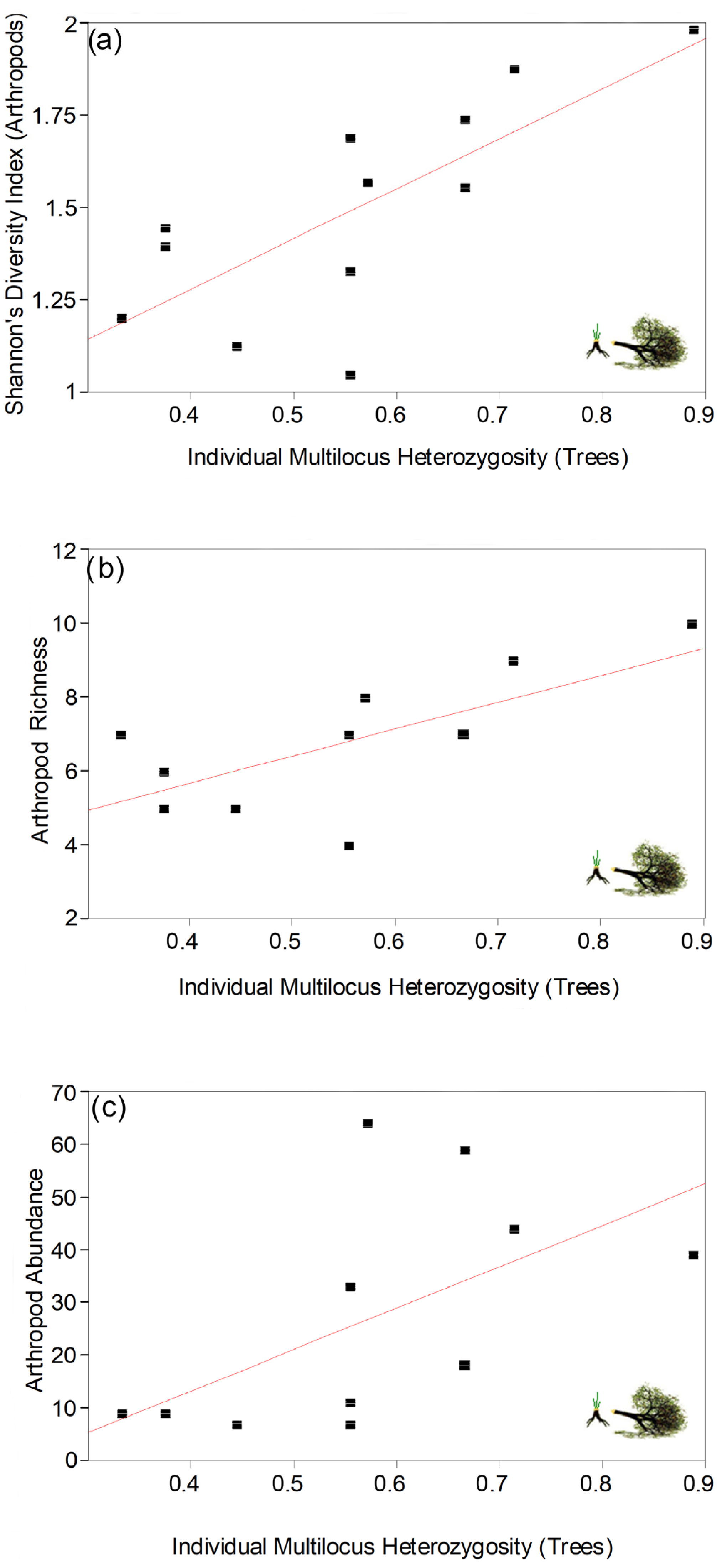
3.1. IMH Correlations for Felled Trees
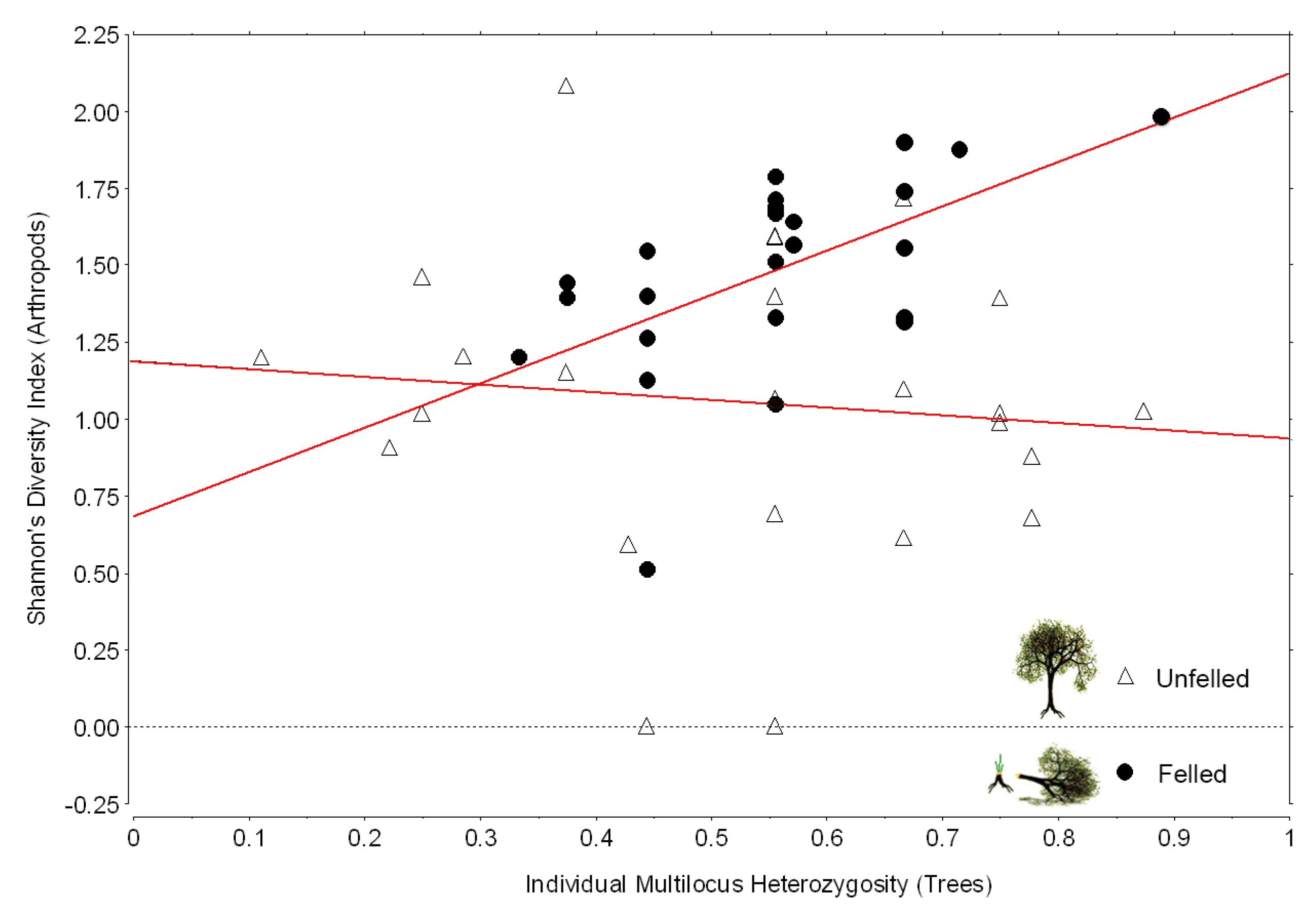
3.2. Contrasts of Felled and Unfelled Trees
3.3. Alternative Hypotheses
3.4. Locus Behavior
4. Discussion
4.1. The IMH–Arthropod Correlation as It Relates to Productivity and Plant Defense
4.2. Heterozygosity, Stress, and a Semi-Arid System
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Whitham, T.G.; Bailey, J.K.; Schweitzer, J.A.; Shuster, S.M.; Bangert, R.K.; Leroy, C.J.; Lonsdorf, E.V.; Allan, G.J.; DiFazio, S.P.; Potts, B.M.; et al. A framework for community and ecosystem genetics: From genes to ecosystems. Nat. Rev. Genet. 2006, 7, 510–523. [Google Scholar] [CrossRef] [PubMed]
- Hughes, A.R.; Inouye, B.D.; Johnson, M.T.J.; Underwood, N.; Vellend, M. Ecological consequences of genetic diversity. Ecol. Lett. 2008, 11, 609–623. [Google Scholar] [CrossRef] [PubMed]
- Johnson, M.T.; Stinchcombe, J.R. An emerging synthesis between community ecology and evolutionary biology. Trends Ecol. Evol. 2007, 22, 250–257. [Google Scholar] [CrossRef]
- Whitham, T.G.; Gehring, C.A.; Lamit, L.J.; Wojtowicz, T.; Evans, L.M.; Keith, A.R.; Smith, D.S. Community specificity: Life and afterlife effects of genes. Trends Plant Sci. 2012, 17, 271–281. [Google Scholar] [CrossRef] [PubMed]
- Whitham, T.G.; Allan, G.J.; Cooper, H.F.; Shuster, S.M. Intraspecific Genetic Variation and Species Interactions Contribute to Community Evolution. Annu. Rev. Ecol. Evol. Syst. 2020, 51, 587–612. [Google Scholar] [CrossRef]
- Crutsinger, G.M. A community genetics perspective: Opportunities for the coming decade. New Phytol. 2016, 210, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Roches, S.D.; Post, D.M.; Turley, N.E.; Bailey, J.K.; Hendry, A.P.; Kinnison, M.T.; Schweitzer, J.A.; Palkovacs, E.P. The ecological importance of intraspecific variation. Nat. Ecol. Evol. 2018, 2, 57–64. [Google Scholar] [CrossRef]
- Wade, M.J. The co-evolutionary genetics of ecological communities. Nat. Rev. Genet. 2007, 8, 185–195. [Google Scholar] [CrossRef]
- Reynolds, L.K.; McGlathery, K.J.; Waycott, M. Genetic Diversity Enhances Restoration Success by Augmenting Ecosystem Services. PLoS ONE 2012, 7. [Google Scholar] [CrossRef]
- Bangert, R.K.; Turek, R.J.; Rehill, B.; Wimp, G.M.; Schweitzer, J.A.; Allan, G.J.; Bailey, J.K.; Martinsen, G.D.; Keim, P.; Lindroth, R.L.; et al. A genetic similarity rule determines arthropod community structure. Mol. Ecol. 2005, 15, 1379–1391. [Google Scholar] [CrossRef] [PubMed]
- Wimp, G.M.; Young, W.P.; Woolbright, S.A.; Martinsen, G.D.; Keim, P.; Whitham, T.G. Conserving plant genetic diver-sity for dependent animal communities. Ecol. Lett. 2004, 7, 776–780. [Google Scholar] [CrossRef]
- Crutsinger, G.M.; Reynolds, W.N.; Classen, A.T.; Sanders, N.J. Disparate effects of plant genotypic diversity on foliage and litter arthropod communities. Oecologia 2008, 158, 65–75. [Google Scholar] [CrossRef]
- Ferrier, S.M.; Bangert, R.K.; Hersch-Green, E.I.; Bailey, J.K.; Allan, G.J.; Whitham, T.G. Unique arthropod communities on different host-plant genotypes results in greater arthropod diversity. Arthropod-Plant Interact. 2012, 6, 187–195. [Google Scholar] [CrossRef]
- Castagneyrol, B.; Lagache, L.; Giffard, B.; Kremer, E.; Jactel, H. Genetic Diversity Increases Insect Herbivory on Oak Sap-lings. PLoS ONE 2012, 7. [Google Scholar] [CrossRef]
- Evans, L.M.; Kaluthota, S.; Pearce, D.W.; Allan, G.J.; Floate, K.; Rood, S.; Whitham, T.G. Bud phenology and growth are subject to divergent selection across a latitudinal gradient in Populus angustifolia and impact adaptation across the distributional range and associated arthropods. Ecol. Evol. 2016, 6, 4565–4581. [Google Scholar] [CrossRef] [PubMed]
- Keith, A.R.; Bailey, J.K.; Lau, M.K.; Whitham, T.G. Genetics-based interactions of foundation species affect community diversity, stability and network structure. Proc. R. Soc. B Boil. Sci. 2017, 284. [Google Scholar] [CrossRef] [PubMed]
- Holeski, L.M.; Hillstrom, M.L.; Whitham, T.G.; Lindroth, R.L. Relative importance of genetic, ontogenetic, induction, and seasonal variation in producing a multivariate defense phenotype in a foundation tree species. Oecologia 2012, 170, 695–707. [Google Scholar] [CrossRef]
- Lamit, L.J.; Busby, P.E.; Lau, M.K.; Compson, Z.G.; Wojtowicz, T.; Keith, A.R.; Zinkgraf, M.S.; Schweitzer, J.A.; Shuster, S.M.; Gehring, C.A.; et al. Tree genotype mediates covariance among communities from microbes to lichens and arthropods. J. Ecol. 2015, 103, 840–850. [Google Scholar] [CrossRef]
- Barbour, R.C.; Forster, L.G.; Baker, S.C.; Steane, D.A.; Potts, B.M. Biodiversity Consequences of Genetic Variation in Bark Characteristics within a Foundation Tree Species. Conserv. Biol. 2009, 23, 1146–1155. [Google Scholar] [CrossRef] [PubMed]
- Barbour, R.C.; O’Reilly-Wapstra, J.M.; De Little, D.W.; Jordan, G.J.; Steane, R.A.; Humphreys, J.R.; Bailey, J.K.; Whitham, T.G.; Potts, B.M. A geographic mosaic of genetic variation within a foundation tree species and its community-level consequences. Ecology 2009, 90, 1762–1772. [Google Scholar] [CrossRef]
- Barker, H.L.; Holeski, L.M.; Lindroth, R.L. Genotypic variation in plant traits shapes herbivorous insect and ant communities on a foundation tree species. PLoS ONE 2018, 13. [Google Scholar] [CrossRef]
- Szulkin, M.; Bierne, N.; David, P. Heterozygosity-Fitness Correlations: A Time for Reappraisal. Evology 2010, 64, 1202–1217. [Google Scholar] [CrossRef] [PubMed]
- Hapman, J.R.C.; Nakagawa, S.; Oltman, D.W.C.; Slate, J.; Heldon, B.C.S. A quantitative review of heterozygosity-fitness correlations in animal populations. Mol. Ecol. 2009, 18, 2746–2765. [Google Scholar] [CrossRef] [PubMed]
- Hansson, B.; Westerberg, L. On the correlation between heterozygosity and fitness in natural populations. Mol. Ecol. 2002, 11, 2467–2474. [Google Scholar] [CrossRef]
- Coltman, D.W.; Slate, J. Microsatellite measures of inbreeding: A meta-analysis. Evolution 2003, 57, 971–983. [Google Scholar] [CrossRef]
- Ohta, T.; Cockerham, C.C. Detrimental genes with partial selfing and effects on a neutral locus. Genet. Res. 1974, 23, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Ohta, T. Associative overdominance caused by linked detrimental mutations. Genet. Res. 1971, 18, 277–286. [Google Scholar] [CrossRef]
- Slate, J.; Pemberton, J.M. Admixture and patterns of linkage disequilibrium in a free-living vertebrate population. J. Evol. Biol. 2007, 20, 1415–1427. [Google Scholar] [CrossRef]
- Charlesworth, D.; Charlesworth, B. Inbreeding depression and its evolutionary consequences. Ann. Rev. Ecol. Syst. 1987, 18, 237–268. [Google Scholar] [CrossRef]
- Pérez-González, J.; Carranza, J.; Torres-Porras, J.; Fernández-García, J.L. Low Heterozygosity at Microsatellite Markers in Iberian Red Deer with Small Antlers. J. Hered. 2010, 101, 553–561. [Google Scholar] [CrossRef]
- Acevedo-Whitehouse, K.; Petetti, L.; Duignan, P.; Castinel, A. Hookworm infection, anaemia and genetic variability of the New Zealand sea lion. Proc. R. Soc. B Boil. Sci. 2009, 276, 3523–3529. [Google Scholar] [CrossRef] [PubMed]
- Coltman, D.W.; Pilkington, J.G.; Smith, J.A.; Pemberton, J.M. Parasite-Mediated Selection against Inbred Soay Sheep in a Free-Living, Island Population. Evolution 1999, 53, 1259. [Google Scholar] [CrossRef] [PubMed]
- Gompper, M.E.; Monello, R.J.; Eggert, L.S. Genetic variability and viral seroconversion in an outcrossing vertebrate population. Proc. R. Soc. B Boil. Sci. 2010, 278, 204–210. [Google Scholar] [CrossRef] [PubMed]
- Cohas, A.; Bonenfant, C.; Kempenaers, B.; Allainé, D. Age-specific effect of heterozygosity on survival in alpine mar-mots, Marmota marmota. Mol. Ecol. 2009, 18, 1491–1503. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, A.; Gaillard, J.-M.; Yoccoz, N.G.; Hewison, A.J.M.; Galan, M.; Coulson, T.; Allainé, D.; Vial, L.; Delorme, D.; Van Laere, G.; et al. Heterozygisity-fitness correlations revealed by neutral and candidate gene markers in roe deer from a long-term study. Evolution 2009, 63, 403–417. [Google Scholar] [CrossRef]
- Küpper, C.; Kosztolányi, A.; Augustin, J.; Dawson, D.; Burke, T.; Székely, T. Heterozygosity-fitness correlations of con-served microsatellite markers in Kentish plovers Charadrius alexandrinus. Mol. Ecol. 2010, 19, 5172–5185. [Google Scholar] [CrossRef]
- Selonen, V.; Hanski, I.K. Condition-dependent, phenotype-dependent and genetic dependent factors in the natal disper-sal of a solitary rodent. J. Anim. Ecol. 2010, 79, 1093–1100. [Google Scholar] [CrossRef]
- Williams, S.L. Reduced genetic diversity in eelgrass transplantations affects both population growth and individual fit-ness. Ecol. Appl. 2001, 11, 1472–1488. [Google Scholar] [CrossRef]
- Cole, C.T.; Morrow, C.J.; Barker, H.L.; Rubert-Nason, K.F.; Riehl, J.F.L.; Köllner, T.G.; Lackus, N.D.; Lindroth, R.L. Growing up aspen: Ontogeny and trade-offs shape growth, defence and reproduction in a foundation species. Ann. Bot. 2021, 127, 505–517. [Google Scholar] [CrossRef]
- Hammerli, A.; Reusch, T.B.H. Inbreeding depression influences genet size distribution in a marine angiosperm. Mol. Ecol. 2003, 12, 619–629. [Google Scholar] [CrossRef]
- Cole, C.T.; Stevens, M.T.; Anderson, J.E.; Lindroth, R.L. Heterozygosity, gender, and the growth-defense trade-off in quaking aspen. Oecologia 2016, 181, 381–390. [Google Scholar] [CrossRef]
- Cole, C.T.; Anderson, J.E.; Lindroth, R.L.; Waller, D.M. Rising concentrations of atmospheric CO2 have increased growth in natural stands of quaking aspen (Populus tremuloides). Glob. Chang. Biol. 2009, 16, 2186–2197. [Google Scholar] [CrossRef]
- Stilwell, K.L.; Wilbur, H.M.; Werth, C.R.; Taylor, D.R. Heterozygote advantage in the American chestnut, Castanea dentata (Fagaceae). Am. J. Bot. 2003, 90, 207–213. [Google Scholar] [CrossRef]
- Pérez-Harguindeguy, N.; Díaz, S.; Vendramini, F.; Cornelissen, J.H.C.; Gurvich, D.E.; Cabido, M. Leaf Traits and herbi-vore selection in the field and cafeteria experiments. Austral Ecol. 2003, 28, 642–650. [Google Scholar] [CrossRef]
- Wright, I.J.; Cooke, J.; Cernusak, L.A.; Hutley, L.B.; Scalon, M.; Tozer, W.C.; Lehmann, C. Stem diameter growth rates in a fire-prone savanna correlate with photosynthetic rate and branch-scale biomass allocation, but not specific leaf area. Austral Ecol. 2019, 44, 339–350. [Google Scholar] [CrossRef]
- Cornelissen, J.H.C.; Lavorel, S.; Garnier, E.; Díaz, S.; Buchmann, N.; Gurvich, D.E.; Reich, P.B.; ter Steege, H.; Morgan, H.D.; van der Heijden, M.G.A.; et al. A handbook of protocols for standardized and easy measurement of plant func-tional traits worldwide. Am. J. Bot. 2003, 51, 335–380. [Google Scholar] [CrossRef]
- Patyk, K.; Turmelle, A.; Blanton, J.D.; Rupprecht, C.E. Trends in National Surveillance Data for Bat Rabies in the United States: 2001–2009. Vector-Borne Zoonotic Dis. 2012, 12, 666–673. [Google Scholar] [CrossRef]
- Diner, B.; Berteaux, M.; Fyles, J.; Lindroth, R.L. Behavioral archives link the chemistry and clonal structure of trembling aspen to the food choice of North American porcupine. Oecologia 2009, 160, 687–695. [Google Scholar] [CrossRef]
- Stuart, C.W.; Scott, W.; Jason, V.; Richard, L.L. Aspen Decline, Aspen Chemistry, and Elk Herbivory: Are They Linked? Rangelands 2008, 30, 17–21. [Google Scholar] [CrossRef]
- Axelsson, E.P.; Senior, J.K. The extended consequences of genetic conductivity: Mating distance affects community phe-notypes in Norway spruce. Ecol. Evol. 2018, 8, 11645–11655. [Google Scholar] [CrossRef]
- Campbell, S.A.; Thaler, J.S.; Kessler, A. Plant chemistry underlies herbivore-mediated inbreeding depression in nature. Ecol. Lett. 2013, 16, 252–260. [Google Scholar] [CrossRef]
- Schrieber, K.; Schweiger, R.; Kroner, L.; Muller, C. Inbreeding diminishes herbivore-induced metabolic responses in na-tive and invasive plant populations. J. Ecol. 2019, 107, 923–936. [Google Scholar] [CrossRef]
- Williams, A.P.; Cook, E.R.; Smerdon, J.E.; Cook, B.I.; Abatzoglou, J.T.; Bolles, K.; Baek, S.H.; Badger, A.M.; Livneh, B. Large contribution from anthropogenic warming to an emerging North American megadrought. Science 2020, 368, 314–318. [Google Scholar] [CrossRef] [PubMed]
- Durben, R.M.; Walker, F.M.; Holeski, L.; Keith, A.; Kovacs, Z.; Hurteau, S.R.; Lindroth, R.L.S.; Shuster, S.M.; Whitham, T.G. Beavers, bugs and chemistry: A mammalian herbivore changes chemistry composition and arthropod communities in foundation tree species. Forests 2021. [Google Scholar]
- Bailey, J.K.; Schweitzer, J.A.; Rehill, B.J.; Lindroth, R.L.; Martinsen, G.D.; Whitham, T.G. Beavers as Molecular Geneticists: A Genetic Basis to the Foraging of An Ecosystem Engineer. Ecology 2004, 85, 603–608. [Google Scholar] [CrossRef]
- Waltz, A.M.; Whitham, T.G. Plant development affects arthropod communities: Opposing impacts of species removal. Ecology 1997, 78, 2133–2144. [Google Scholar] [CrossRef]
- Wimp, G.M.; Martinsen, G.D.; Floate, K.D.; Bangert, R.K.; Whitham, T.G. Plant genetic determinants of arthropod com-munity structure and diversity. Evolution 2005, 59, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Wimp, G.M.; Wooley, S.; Bangert, R.K.; Young, W.P.; Martinsen, G.D.; Keim, P.; Rehill, B.; Lindroth, R.L.; Whitham, T.G. Plant genetics predicts intra-annual variation in phytochemistry and arthropod community structure. Mol. Ecol. 2007, 16, 5057–5069. [Google Scholar] [CrossRef] [PubMed]
- Martinsen, G.D.; Driebe, E.M.; Whitham, T.G. Indirect interactions mediated by changing plant chemistry: Beaver browsing benefits beetles. Ecology 1998, 79, 192–200. [Google Scholar] [CrossRef]
- Lindroth, R.; Kinney, K.K.; Platz, C.L. Responses of Diciduous Trees to Elevated Atmospheric CO2: Productivity, Phytochemistry, and Insect Performance. Ecology 1993, 74, 763–777. [Google Scholar] [CrossRef]
- Parsons, W.F.J.; Bockheim, J.G.; Lindroth, R.L. Independent, Interactive, and Species-Specific Responses of Leaf Litter Decomposition to Elevated CO2 and O3 in a Northern Hardwood Forest. Ecosystems 2008, 11, 505–519. [Google Scholar] [CrossRef]
- Liston, A.; Rieseberg, L.H.; Adams, R.P.; Do, N.; Ge-Lin, Z. A Method for Collecting Dried Plant Specimens for DNA and Isozyme Analyses, and the Results of a Field Test in Xinjiang, China. Ann. Mo. Bot. Gard. 1990, 77, 859–863. [Google Scholar] [CrossRef]
- Woolbright, S.A.; DiFazio, S.P.; Yin, T.; Martinsen, G.D.; Zhang, X.; Allan, G.J.; Whitham, T.G.; Keim, P. A dense linkage map of hybrid cottonwood (Populus fremontii_P. angustifolia) contributes to long-term ecological research and compari-son mapping in a model forest tree. Heredity 2008, 100, 59–70. [Google Scholar] [CrossRef]
- Tuskan, G.A.; DiFazio, S.; Jansson, S.; Bohlmann, J.; Grigoriev, I.; Hellsten, U.; Putnam, N.; Ralph, S.; Rombauts, S.; Salamov, A.; et al. The Genome of Black Cottonwood, Populus trichocarpa (Torr. & Gray). Science 2006, 313, 1596–1604. [Google Scholar] [CrossRef] [PubMed]
- Dunlap, J.M.; Stettler, R.F. Genetic variation and productivity of Populus trichocarpa and its hybrids. X. Trait correlations in young black cottonwood from four river valleys in Washington. Trees 1998, 13, 28–39. [Google Scholar] [CrossRef]
- Imada, S.; Yamanaka, N.; Tamai, S. Contribution of root growth responses to leaf traits and relative growth rate of Pop-ulus alba under different water-table conditions. Trees 2010, 24, 1163–1172. [Google Scholar] [CrossRef]
- Poorter, H.; Niinemets, Ü.; Poorter, L.; Wright, I.J.; Villar, R. Causes and consequences of variation in leaf mass per area (LMA): A meta-analysis. New Phytol. 2009, 182, 565–588. [Google Scholar] [CrossRef]
- Coulon, A. GENHET: An easy-to-use R function to estimate individual heterozygosity. Mol. Ecol. Resour. 2010, 10, 167–169. [Google Scholar] [CrossRef]
- Magurran, A.E. Ecological Diversity and Its Measurement; Princeton University Press: Princeton, NJ, USA, 1988. [Google Scholar]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software structure: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of Population Structure Using Multilocus Genotype Data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Wen, W. Documentation for Structure Software, Version 2; Department of Human Genetics, University of Chicago: Chicago, IL, USA, 2003. [Google Scholar]
- Excoffier, L.; Lischer, H.E.L. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Queller, D.C.; Goodnight, K.F. Estimating relatedness using genetic markers. Evolution 1989, 43, 258–275. [Google Scholar] [CrossRef]
- Keim, P.; Paige, K.N.; Whitham, T.G.; Lark, K.G. Genetic analysis of an interspecific hybrid swarm of Populus: Occurrence of unidirectional introgression. Genetics 1989, 123, 557–565. [Google Scholar] [CrossRef] [PubMed]
- Martinsen, G.D.; Whitham, T.G.; Turek, R.J.; Keim, P. Hybrid populations selectively filter gene introgression between species. Evolution 2001, 55, 1325–1335. [Google Scholar] [CrossRef] [PubMed]
- Bothwell, H.M.; Cushman, S.A.; Woolbright, S.A.; Hersch-Green, E.I.; Evans, L.M.; Whitham, T.G.; Allan, G.J. Conserving threatened riparian ecosystems in the American West: Precipitation gradients and river networks drive genetic connectivity and diversity in a foundation riparian tree (Populus angustifolia). Mol. Ecol. 2017, 26, 5114–5132. [Google Scholar] [CrossRef]
- Waits, L.; Luikart, G.; Taberlet, P. Estimating the probability of identity among genotypes in natural populations: Cau-tions and guidelines. Mol. Ecol. 2001, 10, 249–256. [Google Scholar] [CrossRef]
- Raymond, M.; Rousset, F. GENEPOP (Version 1.2): Population Genetics Software for Exact Tests and Ecumenicism. J. Hered. 1995, 86, 248–249. [Google Scholar] [CrossRef]
- Louault, F.; Pillar, V.D.; Aufrère, J.; Garnier, E.; Soussana, J.F. Plant traits and functional types in response to reduced disturbance in a semi-natural grassland. J. Veg. Sci. 2005, 16, 151–160. [Google Scholar] [CrossRef]
- Wright, I.J.; Reich, P.B.; Westoby, M.; Ackerly, D.; Baruch, Z.; Bongers, F.; Cavender-Bares, J.; Chapin, T.; Cornelissen, J.H.C.; Diemer, M.; et al. The worldwide leaf economics spectrum. Nature 2004, 428, 821–827. [Google Scholar] [CrossRef] [PubMed]
- Bush, R.M.; Smouse, P.E. Evidence for the adaptives ignificance of allozymes in forest trees. New For. 1992, 6, 179–196. [Google Scholar] [CrossRef]
- Mitton, J.B.; Grant, M.C. Observations on the ecology and evolution of quaking aspen, Populus tremuloides, in the Colo-rado front range. Am. J. Bot. 1980, 67, 202–209. [Google Scholar] [CrossRef]
- Jelinski, D.E. Associations between environmental heterogeneity, heterozygosity, and growth rates of Populus tremuloides in a cordilleran landscape. Arct. Alp. Res. 1993, 25, 183–188. [Google Scholar] [CrossRef]
- González-Díaz, P.; Gazol, A.; Valbuena-Carabaña, M.; Sangüesa-Barreda, G.; Moreno-Urbano, A.; Zavala, M.A.; Camarero, J.J. Remaking a stand: Links between genetic diversity and tree growth in expanding Mountain pine populations. For. Ecol. Manag. 2020, 472. [Google Scholar] [CrossRef]
- Lesbarréres, D.; Primmer, C.R.; Laurila, A.; Merilä, J. Environmental and population dependency of genetic variabil-ity-fitness correlations in Rana temporaria. Mol. Ecol. 2005, 14, 311–323. [Google Scholar] [CrossRef]
- Schmeller, D.S.; Schregel, J.; Veith, M. The importance of heterozygosity in a frog’s life. Naturwissenschaften 2007, 94, 360–366. [Google Scholar] [CrossRef]
- Mopper, S.; Mitton, J.B.; Whitham, T.G.; Cobb, N.S.; Christensen, K.M. Genetic Differentiation and Heterozygosity in Pinyon Pine Associated with Resistance to Herbivory and Environmental Stress. Evology 1991, 45, 989–999. [Google Scholar] [CrossRef] [PubMed]
- Stone, A.C.; Gehring, C.A.; Whitham, T.G. Drought negatively affects communities on a foundation tree: Growth rings predict diversity. Oecologia 2010, 164, 751–761. [Google Scholar] [CrossRef] [PubMed]
- Stone, A.C.; Gehring, C.A.; Cobb, N.S.; Whitham, T.G. Genetic-Based Susceptibility of a Foundation Tree to Herbivory Interacts With Climate to Influence Arthropod Community Composition, Diversity, and Resilience. Front. Plant Sci. 2018, 9, 1831. [Google Scholar] [CrossRef] [PubMed]
- Trotter, R.T.; Cobb, N.S.; Whitham, T.G. Arthropod community diversity and trophic structure: A comparison between extremes of drought-stress. Ecol. Entomol. 2008, 33, 1–11. [Google Scholar] [CrossRef]
- Price, P.W. The Plant Vigor Hypothesis and Herbivore Attack. Oikos 1991, 62, 244. [Google Scholar] [CrossRef]
- David, P. Heterozygosity-fitness correlations: New perspective on old problems. Heredity 1998, 80, 531–537. [Google Scholar] [CrossRef] [PubMed]
- Nickolas, H.; Harrison, P.A.; Tilyard, P.; Vaillancourt, R.E.; Potts, B.M. Inbreeding depression and differential maladap-tation shape the fitness trajectory of two co-occurring Eucalyptus species. Ann. For. Sci. 2019, 76, 10–23. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Walker, F.M.; Durben, R.; Shuster, S.M.; Lindroth, R.L.; Whitham, T.G. Heterozygous Trees Rebound the Fastest after Felling by Beavers to Positively Affect Arthropod Community Diversity. Forests 2021, 12, 694. https://doi.org/10.3390/f12060694
Walker FM, Durben R, Shuster SM, Lindroth RL, Whitham TG. Heterozygous Trees Rebound the Fastest after Felling by Beavers to Positively Affect Arthropod Community Diversity. Forests. 2021; 12(6):694. https://doi.org/10.3390/f12060694
Chicago/Turabian StyleWalker, Faith M., Rachel Durben, Stephen M. Shuster, Richard L. Lindroth, and Thomas G. Whitham. 2021. "Heterozygous Trees Rebound the Fastest after Felling by Beavers to Positively Affect Arthropod Community Diversity" Forests 12, no. 6: 694. https://doi.org/10.3390/f12060694
APA StyleWalker, F. M., Durben, R., Shuster, S. M., Lindroth, R. L., & Whitham, T. G. (2021). Heterozygous Trees Rebound the Fastest after Felling by Beavers to Positively Affect Arthropod Community Diversity. Forests, 12(6), 694. https://doi.org/10.3390/f12060694





