Chloroplast Genome Draft of Dryobalanops aromatica Generated Using Oxford Nanopore Technology and Its Potential Application for Phylogenetic Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Genomic DNA Extraction
2.3. DNA Library Preparation and Sequencing
2.4. Data Analysis
2.4.1. Sequence Raw Data Analysis
2.4.2. Chloroplast Marker Analysis
2.4.3. Phylogenetic Tree Construction
3. Results
3.1. Genome Sequencing and Assembly
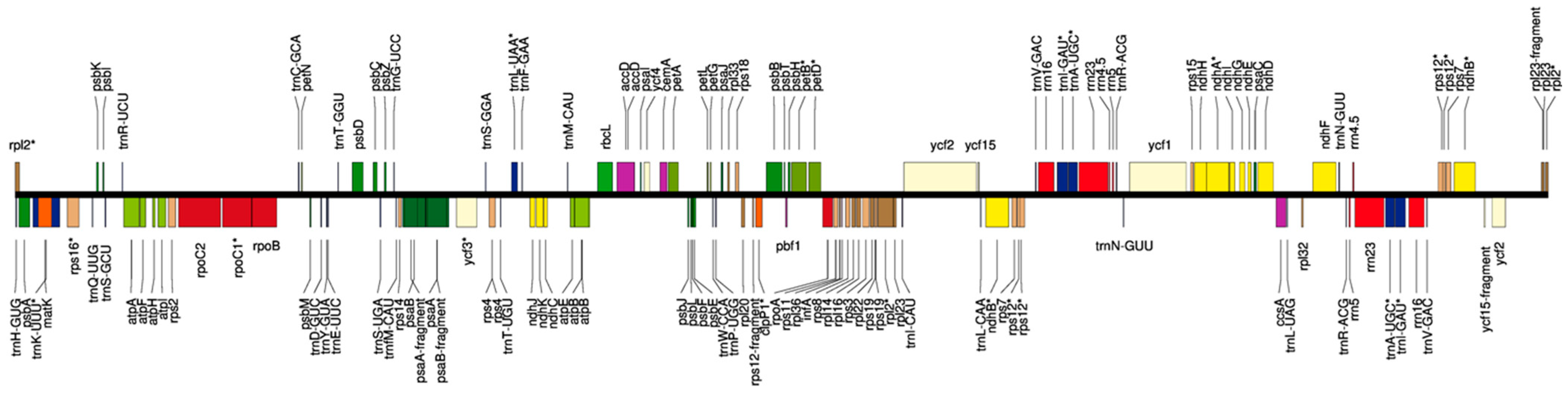
3.2. Chloroplast Genome Annotation
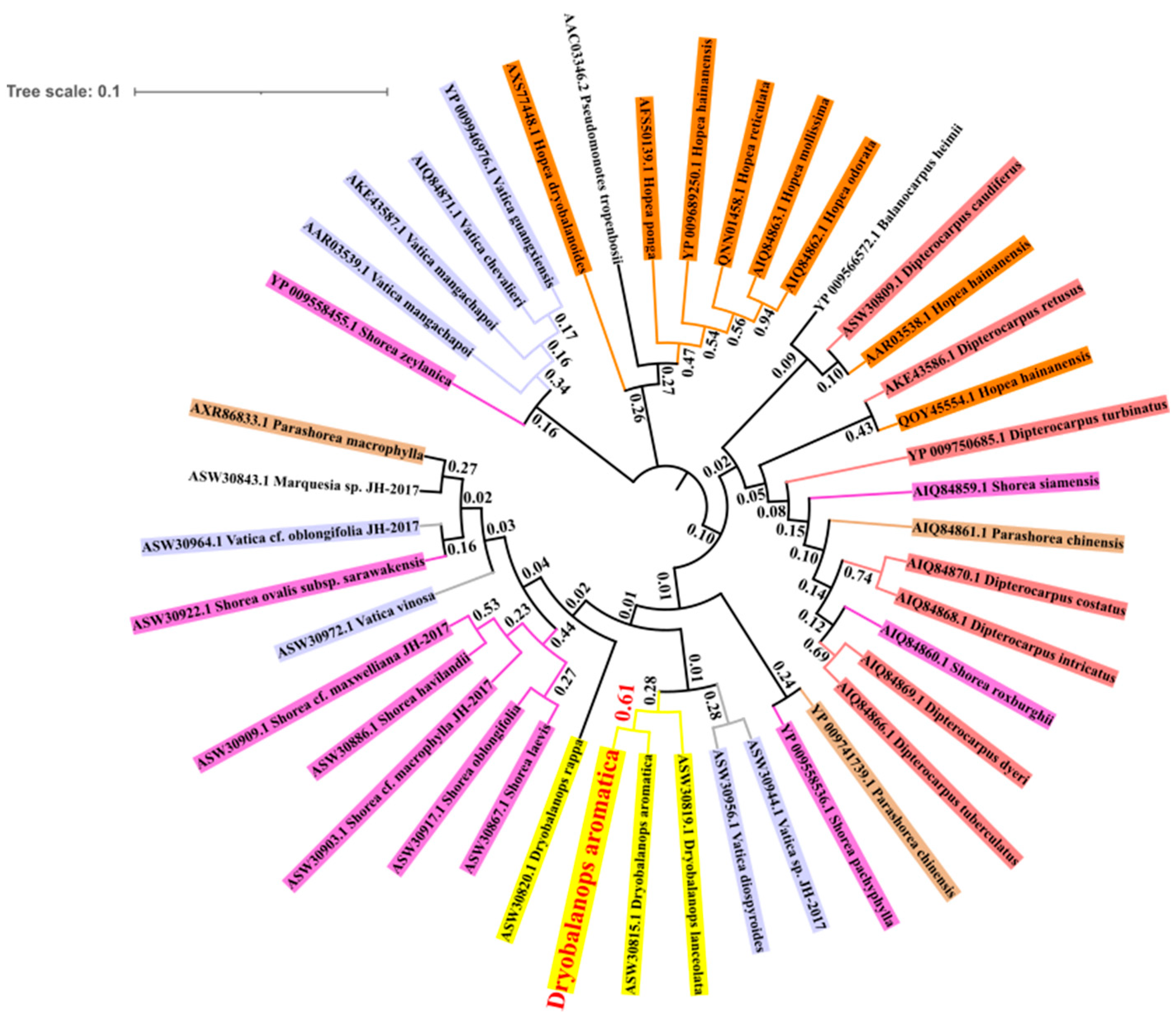
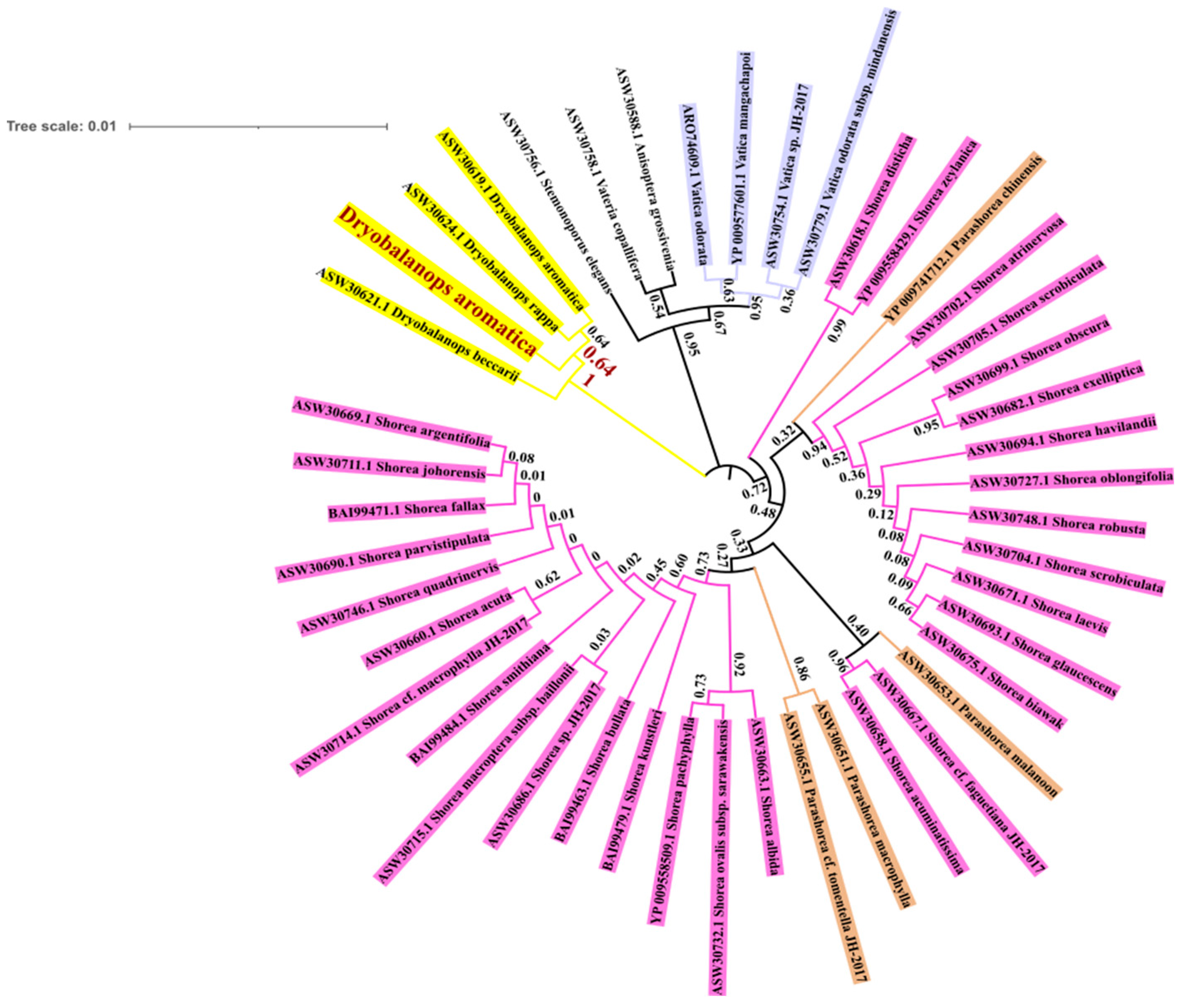
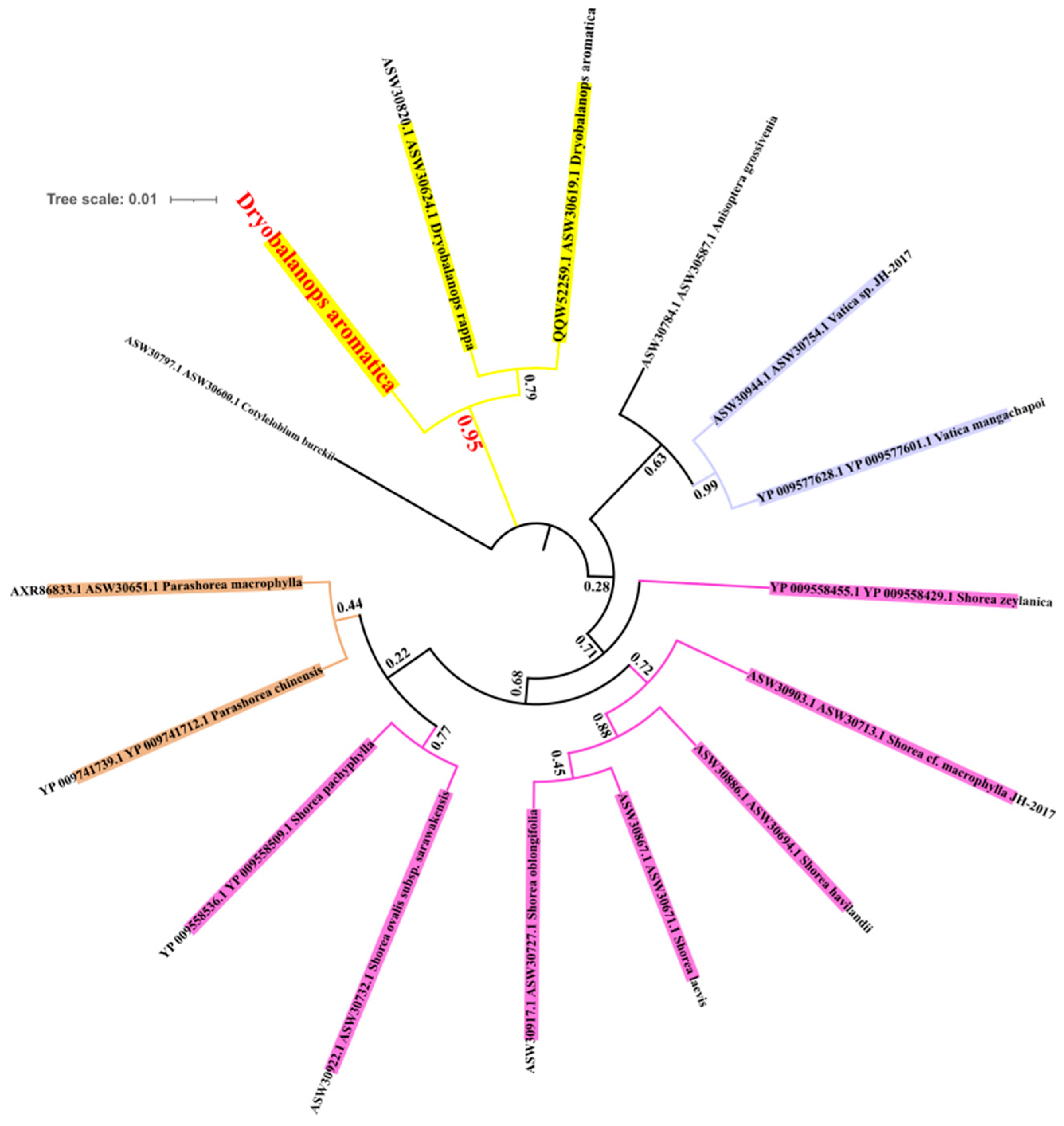
3.3. Phylogenetic Tree Construction
4. Discussion
4.1. Genome Sequencing and Assembly
4.2. Organization of a Partial Chloroplast Genome
4.3. Phylogenetic Inference
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Purba, P.R. Aspek ekologi dan potensi kapur (Dryobalanops aromatica) di Sumatera Utara. J. Akar 2020, 9, 115–126. [Google Scholar] [CrossRef]
- Dwiyanti, F.G.; Kamiya, K.; Harada, K. Phylogeographic structure of the commercially important tropical tree species, Dryobalanops aromatica Gaertn. F. Reinwardtia 2014, 14, 43–51. [Google Scholar] [CrossRef]
- Yahya, A.Z.; van Gardingen, P. Stand growth structure and yield of plantation grown Dryobalanops aromatica in Peninsular Malaysia: >50 years after planting. J. Trop. For. Sci. 1999, 11, 205–217. [Google Scholar]
- Kamariyah, A.S.; Ozek, T.; Demirci, B.; Baser, K.H.C. Chemical composition of leaf and seed oils of Dryobalanops aromatica Gaertn. (Dipterocarpaceae). ASEAN J. Sci. Technol. Dev. 2012, 29, 105–114. [Google Scholar] [CrossRef]
- Pasaribu, G.; Gusmailina, G.; Komarayati, S.; Zulnely, Z.; Dahlian, E. Analisis senyawa kimia Dryobalanops aromatica. J. Penelit. Has. Hutan 2014, 32, 21–26. (In Indonesian) [Google Scholar] [CrossRef]
- Le, T.; Ho, A.S.; Mah, S.; Wong, T.; Ong, H.C.; Loh, P.H.; Lim, Y. Chemical composition of essential oil of exudates of Dryobalanops aromatica. Trop. J. Pharm. Res. 2017, 16, 621–625. [Google Scholar] [CrossRef]
- Ritonga, F.N.; Dwiyanti, F.G.; Kusmana, C.; Siregar, U.J.; Siregar, I.Z. Population genetics and ecology of Sumatra camphor (Dryobalanops aromatica) in natural and community-owned forests in Indonesia. Biodiversitas J. Biol. Divers. 2018, 19, 2175–2182. [Google Scholar] [CrossRef]
- Rangkuti, A.B.; Susilowati, A.; Rachmat, H.H.; Lubis, T.S. DNA isolation and amplification of Dryobalanops oblongifolia Drey and Dryobalanops lanceolata Burck. In IOP Conference Series: Earth and Environmental Science; IOP Publishing: Bristol, UK, 2021; Volume 782, p. 042043. [Google Scholar] [CrossRef]
- Kong, Q.X.; Wu, Z.Y.; Chu, X.; Liang, R.Q.; Xia, M.; Li, L. Study on the anti-cerebral ischemia effect of Borneoland its mechanism. Afr. J. Tradit. Complementary Altern. Med. 2014, 11, 161–164. [Google Scholar] [CrossRef]
- Li, Y.; Ren, M.; Wang, J.; Ma, R.; Chen, H.; Xie, Q.; Li, H.; Li, J.; Wang, J. Progress in Borneol Intervention for Ischemic Stroke: A Systematic Review. Front. Pharmacol. 2021, 12, 606682. [Google Scholar] [CrossRef]
- Gusmailina, G. Borneol-future potential of essential oils. Pros. Semin. Nas. Masy. Biodiversitas Indones. 2015, 1, 259–264. (In Indonesian) [Google Scholar] [CrossRef]
- Barstow, M.R. The IUCN Red List of Threatened Species: Dryobalanops aromatica. Available online: https://doi.org/10.2305/IUCN.UK.20181.RLTS.T61998024A173026192.en (accessed on 10 January 2021).
- United Nations (UN). The United Nations Decade on Ecosystem Restoration. Available online: https://www.decadeonrestoration.org (accessed on 13 July 2021).
- Soenarno, S.; Astana, S. Lacak balak untuk verifikasi uji legalitas kayu pada pemanenan kayu hutan alam. J. Penelit. Has. Hutan 2018, 36, 47–58. (In Indonesian) [Google Scholar] [CrossRef]
- Vernesi, C.; Bruford, M. Recent developments in molecular tools for conservations. In Population Genetics for Animal Conservation (Conservation Biology, pp. 321–344); Bertorelle, G., Bruford, M., Hauffe, H., Rizzoli, A., Vernesi, C., Eds.; Cambridge University Press: Cambridge, UK, 2009. [Google Scholar] [CrossRef]
- Goodwin, S.; Gurtowski, J.; Ethe-Sayers, S.; Deshpande, P.; Schatz, M.C.; McCombie, W.R. Oxford Nanopore sequencing, hybrid error correction, and de novo assembly of a eukaryotic genome. Genome Res. 2015, 25, 1750–1756. [Google Scholar] [CrossRef]
- Wang, W.; Schalamun, M.; Morales-Suarez, A.; Kainer, D.; Schwessinger, B.; Lanfear, R. Assembly of chloroplast genomes with long-read and short-read data: A comparison of approaches using Eucalyptus pauciflora as a test case. BMC Genom. 2018, 19, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Oxford Nanopore Technologies (ONT). Nanopore Sequencing the Advantages of Long Reads for Genome Assembly. Available online: https://nanoporetech.com/sites/default/files/s3/white-papers/WGS_Assembly_white_paper.pdf?submissionGuid=40a7546b-9e51-42e7-bde9-b5ddef3c3512 (accessed on 12 January 2021).
- Breed, M.F.; Harrison, P.A.; Blyth, C.; Byrne, M.; Gaget, V.; Gellie, N.J.C.; Groom, S.V.C.; Hodgson, R.; Mills, J.G.; Prowse, T.A.A.; et al. The potential of genomics for restoring ecosystems and biodiversity. Nat. Rev. Genet. 2019, 20, 615–628. [Google Scholar] [CrossRef]
- Watsa, M.; Erkenswick, G.A.; Pomerantz, A.; Prost, S. Portable sequencing as a teaching tool in conservation and biodiversity research. PLOS Biol. 2020, 18, e3000667. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.S.; Chung, M.G.; Park, S. The complete chloroplast genome sequences of three Veroniceae species (Plantaginaceae): Comparative analysis and highly divergent regions. Front. Plant Sci. 2016, 7, 355. [Google Scholar] [CrossRef]
- Hollingsworth, M.L.; Clark, A.A.; Forrest, L.L.; Richardson, J.; Pennington, R.T.; Long, D.G.; Cowan, R.; Chase, M.W.; Gaudeul, M.; Hollingsworth, P.M. Selecting barcoding loci for plants: Evaluation of seven candidate loci with species-level sampling in three divergent groups of land plants. Mol. Ecol. Resour. 2009, 9, 439–457. [Google Scholar] [CrossRef] [PubMed]
- Webb, C.O.; Slik, J.W.F.; Triono, T. Biodiversity inventory and informatics in Southeast Asia. Biodivers. Conserv. 2010, 19, 955–972. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.L. Isolation of plant DNA from fresh tissue. Focus 1990, 12, 13–15. [Google Scholar]
- Avila, C.; Puente, D.L.; Tobaruela, P.; Quesada, A.; Medina, R.; Rodriguez, C.; Vallejo, E. A new procedure for DNA isolation from saliva samples and comparative analysis of quality indicators. J. Bioprocess. Biotech. 2019, 9, 2–6. [Google Scholar]
- Amarasinghe, S.L.; Shian, S.; Xueyi, D.; Zappia, L.; Ritchiel, M.E.; Gouil, Q. Opportunities and challenges in long-read sequencing data analysis. Genome Biol. 2020, 21, 2–16. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Giordano, F.; Ning, Z. Oxford Nanopore MinION sequencing and genome assembly. Genom. Proteom. Bioinform. 2016, 14, 265–279. [Google Scholar] [CrossRef] [PubMed]
- Urban, L.; Holzer, A.; Baronas, J.J.; Hall, M.B.; Braeuninger-Weimer, P.; Scherm, M.J.; Kunz, D.J.; Perera, S.N.; Martin-Herranz, D.E.; Tipper, E.T.; et al. Freshwater monitoring by nanopore sequencing. eLife 2021, 10, 1–27. [Google Scholar] [CrossRef]
- Giraldo, P.A.; Shinozuka, H.; Spangenberg, G.C.; Smith, K.F.; Cogan, N.O.I. Rapid and detailed characterization of transgene insertion sites in genetically modified plants via nanopore sequencing. Front. Plant Sci. 2020, 11, 602313. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Nguyen, L.T.; Hayes, B.J.; Ross, E. Prowler: A novel trimming algorithm for Oxford Nanopore sequence data. Bioinformatics 2021. [Google Scholar] [CrossRef]
- Van der Nest, M.A.; Chávez, R.; De Vos, L.; Duong, T.A.; Gil-Durán, C.; Ferreira, M.A.; Lane, F.A.; Levicán, G.; Santana, Q.C.; Steenkamp, E.T.; et al. IMA genome—F14: Draft genome sequences of Penicillium roqueforti, Fusarium sororula, Chrysoporthe puriensis, and Chalaropsis populi. IMA Fungus 2021, 12, 1–12. [Google Scholar] [CrossRef]
- Ci, X.; Peng, J.; Shi, C.; Zhu, Z.; Cai, N.; Duan, A.; Wang, D. The complete chloroplast genome of Dipterocarpus turbinatus Gaertn. F. Mitochondrial DNA Part B 2019, 4, 3636–3637. [Google Scholar] [CrossRef]
- Oosterbroek, S.; Doorenspleet, K.; Nijland, R.; Jansen, L. Decona: From demultiplexing to consensus for nanopore amplicon data. ARPHA Conf. Abstr. 2021, 4, e65029. [Google Scholar] [CrossRef]
- Menghini, D.; Aubry, S. De novo transcriptome assembly data of the marine bioluminescent dinoflagellate Pyrocystis lunula. Data Brief 2021, 37, 107254. [Google Scholar] [CrossRef]
- Tillich, M.; Lehwark, P.; Pellizzer, T.; Ulbricht-Jones, E.S.; Fischer, A.; Bock, R.; Greiner, S. GeSeq—versatile and accurate an-notation of organelle genomes. Nucleic. Acids. Res. 2017, 45, W6–W11. [Google Scholar] [CrossRef]
- Wahyuni, F.D.; Saraswati, H.; Dewi, K.S. In-Silico Analysis for cryI gene amplification from Bacillus thuringiensis. BIOEDUKASI 2020, 18, 8–14. [Google Scholar] [CrossRef]
- Schoch, C.L.; Ciufo, S.; Domrachev, M.; Hotton, C.L.; Kannan, S.; Khovanskaya, R.; Leipe, D.; McVeigh, R.; O’Neill, K.; Robbertse, B.; et al. NCBI Taxonomy: A comprehensive update on curation, resources and tools. Database 2020, 2020, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Hall, B.G. Building phylogenetic trees from molecular data with MEGA. Mol. Biol. Evol. 2013, 30, 1229–1235. [Google Scholar] [CrossRef]
- Yang, Y.; Jiang, X.-T.; Zhang, T. Evaluation of a hybrid approach using UBLAST and BLASTX for metagenomic sequences annotation of specific functional genes. PLoS ONE 2014, 9, e110947. [Google Scholar] [CrossRef]
- Mardika, B.A.P.; Setyorini, S.; Karimah, S.A. Implementasi sequence alignment pada lingkungan Spark—Hadoop berbasis single node. J. Tugas Akhir Fak. Inform. 2021, 8, 703–714. (In Indonesian) [Google Scholar]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K.; Mega, X. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Lestari, D.A.; Azrianingsih, R.; Hendrian, H. Filogenetik Jenis-jenis Annonaceae dari Jawa Timur Koleksi. J. Trop. Biodivers. Biotechnol. 2018, 3, 1–7. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Seidl, A.; Tremetsberger, K.; Pfanzelt, S.; Blattner, F.R.; Neuffer, B.; Friesen, N.; Hurka, H.; Shmakov, A.; Batlai, O.; Žerdoner Čalasan, A.Z.; et al. The phylogeographic history of Krascheninnikovia reflects the development of dry steppes and semi-deserts in Eurasia. Sci. Rep. 2021, 11, 6645. [Google Scholar] [CrossRef]
- Delayahe, C.; Nicolas, J. Sequencing DNA with nanopores: Troubles and biases. PLoS ONE 2021, 16, e0257521. [Google Scholar] [CrossRef]
- Tyler, A.D.; Mataseje, L.; Urfano, C.J.; Schmidt, L.; Antonation, K.S.; Mulvey, M.R.; Corbett, C.R. Evaluation of Oxford nanopore’s MinION sequencing device for microbial whole genome sequencing applications. Scientific Report. Sci. Rep. 2018, 8, 10931. [Google Scholar] [CrossRef]
- Ma, X.; Cai, Y.; Tang, L. The complete chloroplast genome of a critically endangered tree species, Hopea reticulata (Dipterocarpaceae). Mitochondrial DNA Part B Resour. 2020, 5, 3575–3576. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.-F.; Sun, Y. The complete chloroplast genome of the endangered tree Parashorea chinensis (Dipterocarpaceae). Mitochondrial DNA Part B 2019, 4, 1163–1164. [Google Scholar] [CrossRef]
- Tasma, I.M. Pemanfaatan teknologi sekuensing genom untuk mempercepat program pemuliaan tanaman. J. Litbang Pertan. 2015, 34, 159–168. (In Indonesian) [Google Scholar] [CrossRef]
- Indayati, I.; Purwanto, H. Controlling Culex quinquefasciatus Say, 1823 (Diptera: Culicidae) using several Lysinibacillus Sphaericus isolates endogenic to Indonesia. J. Biol. Trop. 2021, 21, 298–304. [Google Scholar] [CrossRef]
- Ding, S.; Dong, X.; Yang, J.; Guo, C.; Cao, B.; Guo, Y.; Hu, G. Complete chloroplast genome of Clethra fargesii Franch., an original Sympetalous plant from Central China: Comparative analysis, adaptive evolution, and phylogenetic relationships. Forests 2021, 12, 441. [Google Scholar] [CrossRef]
- Mursyidin, D.H.; Makruf, M.I. Keanekaragaman dan kekerabatan genetik Artocarpus berdasarkan penanda DNA kloroplas matK & rbcL: Kajian in silico. Floribunda 2020, 6, 195–204. (In Indonesian) [Google Scholar] [CrossRef]
- CBOL Plant Working Group A DNA barcode for land plants. Proc. Natl. Acad. Sci. USA 2009, 106, 12794–12797. [CrossRef]
- Dobrogojski, J.; Adamiec, M.; Luciński, R. The chloroplast genome: A review. Acta Physiol. Plant. 2020, 42, 1–13. [Google Scholar] [CrossRef]
- Papuangan, N. Amplification and analysis of rbcL gene (ribulose-1,5-bisphosphate carboxylase) of clove in ternate island. In IOP Conference Series: Earth and Environmental Science; International Conference on Life Sciences and Technology; IOP Publishing: Bristol, UK, 2019; Volume 276, pp. 1–7. [Google Scholar] [CrossRef]
- Heckenhauer, J.; Samuel, R.; Ashton, P.S.; Turner, B.; Barfuss, M.H.J.; Jang, T.; Temsch, E.M.; McCann, J.; Salim, K.A.; Attanayake, A.M.A.S.; et al. Phylogenetic analyses of plastid DNA suggest a different interpretation of morphological evolution than those used as the basis for previous classifications of Dipterocarpaceae (Malvales). Bot. J. Linn. Soc. 2017, 185, 1–26. [Google Scholar] [CrossRef]
- Heckenhauer, J.; Samuel, R.; Ashton, P.S.; Abu Salim, K.A.; Paun, O. Phylogenomics resolves evolutionary relationships and provides insights into floral evolution in the tribe Shoreeae (Dipterocarpaceae). Mol. Phylogenet. Evol. 2018, 127, 1–13. [Google Scholar] [CrossRef]
- Hasibuan, F.E.; Mantiri, F.R.; Rumendel, R.R.H. Kajian variasi sekuens intraspesies dan filogenetik monyet hitam Sulawesi (Macaca nigra) dengan menggunakan gen coi. J. Ilm. Sains 2017, 17, 60–66. (In Indonesian) [Google Scholar]
- Gong, Y.N.; Yang, S.L.; Shih, S.R.; Huang, Y.C.; Chang, P.Y.; Huang, C.G.; Kao, K.C.; Hu, H.C.; Liu, Y.C.; Tsao, K.C. Molecular evolution and the global reemergence of enterovirus D68 by genome-wide analysis. Medicine 2016, 95, e4416. [Google Scholar] [CrossRef]
- Cabelin, V.L.D.; Alejandro, G.J.D. Efficiency of matK, rbcL, trnH–psbA, and trnL-F (cpDNA) to molecularly authenticate Philippine ethnomedicinal Apocynaceae through DNA barcoding. Pharmacogn. Mag. 2016, 12, S384–S388. [Google Scholar] [CrossRef] [PubMed]
- Suka, I.M.G. Isolasi dan amplifikasi DNA Genomik Kamper (Dryobalanops aromatica) Asal Sumatera Utara. Bachelor’s. Thesis, Sumatera Utara University, Medan, Indonesian, 2018. (In Indonesian). [Google Scholar]
- Ismail, M.; Ahmad, A.; Nadeem, M.; Javed, M.A.; Khan, S.H.; Khawaish, I.; Sthanadar, A.A.; Qari, S.H.; Alghanem, S.M.; Khan, K.A.; et al. Development of DNA barcodes for selected Acacia species by using rbcL and matK DNA markers. Saudi J. Biol. Sci. 2020, 27, 3735–3742. [Google Scholar] [CrossRef] [PubMed]
- Siregar, I.Z.; Dwiyanti, F.G.; Pratama, R.; Matra, D.D.; Majiidu, M. Generating long-read sequences using Oxford Nanopore Technology from Diospyros celebica genomic DNA. BMC Res. Notes 2021, 14, 75. [Google Scholar] [CrossRef]
- Lannoy, C.D.; Ridder, D.D.; Risse, J. The long reads ahead: De novo genome assembly using the MinION. F1000Research 2017, 6, 1083. [Google Scholar] [CrossRef] [PubMed]
- Yasodha, R.; Vasudeva, R.; Balakrishnan, S.; Sakthi, A.R.; Abel, N.; Binai, N.; Rajashekar, B.; Bachpai, V.K.W.; Pillai, C.; Dev, S.A. Draft genome of a high value tropical timber tree, Teak (Tectona grandis L. f): Insights into SSR diversity, phylogeny and conservation. DNA Res. 2018, 25, 409–419. [Google Scholar] [CrossRef]
- Fleischmann, A.; Michael, T.P.; Rivadavia, F.; Sousa, A.; Wang, W.; Temsch, E.M.; Greilhuber, J.; Müller, K.F.; Heubl, G. Evolution of genome size and chromosome number in the carnivorous plant genus Genlisea (Lentibulariaceae), with a new estimate of the minimum genome size in angiosperms. Ann. Bot. 2014, 114, 1651–1663. [Google Scholar] [CrossRef]
- Holliday, J.A.; Aitken, S.N.; Cooke, J.E.K.; Fady, B.; Gonzalez-Martinez, S.C.; Heuertz, M.; Jaramillo-Correa, J.-P.; Lexer, C.; Staton, M.; Whetten, R.W.; et al. Advances in ecological genomics in forest trees and applications to genetic resources conservation and breeding. Mol. Ecol. 2017, 26, 706–717. [Google Scholar] [CrossRef]
- Pellicer, J.; Fay, M.F.; Leitch, I.J. The largest eukaryotic genome of them all? Bot. J. Linn. Soc. 2010, 164, 10–15. [Google Scholar] [CrossRef]
- [ONT] Oxford Nanopore Technologies. Whole Genome Sequencing: Large Genomes. Available online: http://nanoporetech.com/publications (accessed on 22 December 2020).
- Smith, D.R. Does cell size impact chloroplast genome size? Front. Plant Sci. 2017, 8, 2116. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Huang, W.; Cai, S.; Zhao, J. The complete chloroplast genome sequence of Dryobalanops aromatica. Mitochondrial DNA Part B Resour. 2021, 6, 1687–1688. [Google Scholar] [CrossRef] [PubMed]
- Ashton, P.S. Dipterocarpaceae. In Tree Flora of Sabah and Sarawak; Soepadmo, E., Saw, L.G., Chung, R.C.K., Eds.; Forest Research Institute: Kuala Lumpur, Malaysia, 2004; Volume 5, p. 388. [Google Scholar]
- Symington, C.F. Foresters’ Manual of Dipterocarps, 2nd ed.; Ashton, P.S., Appanah, S., Eds.; Malayan Forest Records; Forest Research Institute Malaysia: Kuala Lumpur, Malaysia, 2004; Volume 16, p. 519. [Google Scholar]
- Newman, M.F.; Burgess, P.F.; Whitemore, T.C. Pedoman Identifikasi Pohon-Pohon Dipterocarpaceae: Pulau Kalimantan; Prosea Indonesia: Bogor, Indonesia, 1999; p. 151. (In Indonesian) [Google Scholar]
- Clegg, M.T. Chloroplast gene sequences and the study of plant evolution. Proc. Natl. Acad. Sci. USA 1993, 90, 363–367. [Google Scholar] [CrossRef]
- Clegg, M.T.; Gaut, B.S.; Learn, G.H.; Morton, B.R. Rates and patterns of chloroplast DNA evolution. Proc. Natl. Acad. Sci. USA 1994, 91, 6795–6801. [Google Scholar] [CrossRef]
- Xie, D.F.; Yu, Y.; Deng, Y.Q.; Li, J.; Liu, H.Y.; Zhou, S.D.; He, X.J. Comparative analysis of the chloroplast genomes of the Chinese endemic genus Urophysa and their contribution to choroplast phylogeny and adaptive evolution. Int. J. Mol. Sci. 2018, 19, 1847. [Google Scholar] [CrossRef]
- Zhang, F.; Wang, N.; Cheng, G.; Shu, X.; Wang, T.; Zhuang, W.; Lu, R.; Wang, Z. Comparative chloroplast genomes of four Lycoris species (Amaryllidaceae) provides new insight into interspecific relationship and phylogeny. Biology 2021, 10, 715. [Google Scholar] [CrossRef]
- Zhao, J.; Xu, Y.; Xi, L.; Yang, J.; Chen, H.; Zhang, J. Characterization of the chloroplast genome sequence of Acer miaotaiense: Comparative and phylogenetic analyses. Molecules 2018, 23, 1740. [Google Scholar] [CrossRef]
- Bowman, S.M.; Patel, M.; Yerramsetty, P.; Mure, C.M.; Zielinski, A.M.; Bruenn, J.A.; Berry, J.O. A novel RNA binding protein affects rbcL gene expression and is specific to bundle sheat chloroplasts in C4 plants. BMC Plant Biol. 2013, 13, 138. [Google Scholar] [CrossRef]
- Yao, X.; Tang, P.; Li, Z.; Li, D.; Liu, Y.; Huang, H. The first complete chloroplast genome sequences in Actinidiaceae: Genome structure and comparative analysis. PLoS ONE 2015, 10, e0129347. [Google Scholar] [CrossRef] [PubMed]
- Ramadhani, R. Bar Code DNA Untuk Identifikasi Jenis—Jenis kemenyan (Styrax sp.) di Sumatera Utara. Bachelor’s Thesis, Sumatera Utara University, Medan, Indonesian, 2017. (In Indonesian). [Google Scholar]
- Carneiro de Melo Moura, C.; Brambach, F.; Jair Hernandez Bado, K.; Krutovsky, K.V.; Kreft, H.; Tjitrosoedirdjo, S.S.; Siregar, I.Z.; Gailing, O. Integrating DNA barcoding and traditional taxonomy for the identification of dipterocarps in remnant lowland forests of Sumatra. Plants 2019, 8, 461. [Google Scholar] [CrossRef] [PubMed]
- Soltis, P.S.; Soltis, D.E. Applying the bootstrap in phylogeny reconstruction. Stat. Sci. 2003, 18, 256–267. [Google Scholar] [CrossRef]
- Kang, Y.; Deng, Z.; Zang, R.; Long, W. DNA barcoding analysis and phylogenetic relationships of tree species in tropical cloud forests. Sci. Rep. 2017, 7, 12564. [Google Scholar] [CrossRef]
- Tsumura, Y.; Kawahara, T.; Wickneswari, R.; Yoshimura, K. Molecular phylogeny of Dipterocarpaceae in Southeast Asia using RFLP of PCR-amplified chloroplast genes. TAG Theor. Appl. Genet. Theor. Angew. Genet. 1996, 93, 22–29. [Google Scholar] [CrossRef]
- Kajita, T.; Kamiya, K.; Nakamura, N.; Tachida, H.; Wickneswari, R.; Tsumura, Y.; Yoshimaru, H.; Yamazaki, T. Molecular phylogeny of Dipetrocarpaceae in Southeast Asia based on nucleotide sequences of matK, trnL Intron, and trnL-trnF intergenic Spacer Region in chloroplast DNA. Mol. Phylogenet. Evol. 1998, 10, 2. [Google Scholar] [CrossRef]
- Dayanandan, N.; Ashton, P.A.; William, S.M.; Primack, R.B. Phylogeny of the tropical tree family Dipterocarpaceae based on nucleotide sequences of chloroplast rbcL gene. Am. J. Bot. 1999, 86, 1182–1190. [Google Scholar] [CrossRef]
- Yulita, K.S.; Bayer, R.J.; West, J.G. Molecular phylogenetic study of Hopea and Shorea (Dipterocarpaceae): Evidence from the trnL-trnF and internal transcribed spacer regions. Plant Species Biol. 2005, 20, 167–182. [Google Scholar] [CrossRef]
- Indrioko, S.; Gailing, O.; Finkeldey, R. Molecular phylogeny of Dipterocarpaceae in Indonesia based on chloroplast DNA. Plant Syst. Evol. 2006, 261, 99–115. [Google Scholar] [CrossRef]
- Al-Juhani, W.S.; Khalik, K.N.A. Identification and phylogenetics study of Arabis alpina L. From the Kingdom of Saudi Arabia. Pak. J. Bot. 2021, 53, 1057–1064. [Google Scholar] [CrossRef]
- Manshoor, N.; Aizam, E.A.; Qamarusy, S.K.; Shafarin, M.; Ahmat, N. Mass fragmentation patterns as fingerprints in identification of known oligostilbenes in Dryobalanops spp. Extracts. Int. J. Pharmacogn. Phytochem. Res. 2015, 7, 1147–1152. [Google Scholar]
- Hattimare, R. Crown shyness in various tree species. Int. J. Sci. Dev. Res. 2018, 3, 322–324. [Google Scholar]
- Harada, K.; Dwiyanti, F.G.; Siregar, I.Z.; Subiakto, A.; Chong, L.; Diway, B.; Lee, Y.-F.; Ninomiya, I.; Kamiya, K. Genetic variation and genetic structure of two closely related Dipterocarp Species, Dryobalanops aromatica C.F. Gaertn. and D. beccarii Dyer. J. Bot. Gard. Hortic. 2018, 16, 179–196. [Google Scholar] [CrossRef]
- Putri Fiqa, A.; Fauziah, F.; Ayu Lestari, D.; Budiharta, S. The importance of in-situ conservation area in mining concession in preserving diversity, threatened and potential floras in East Kalimantan, Indonesia. Biodiversitas J. Biol. Divers. 2019, 20, 198–210. [Google Scholar] [CrossRef][Green Version]

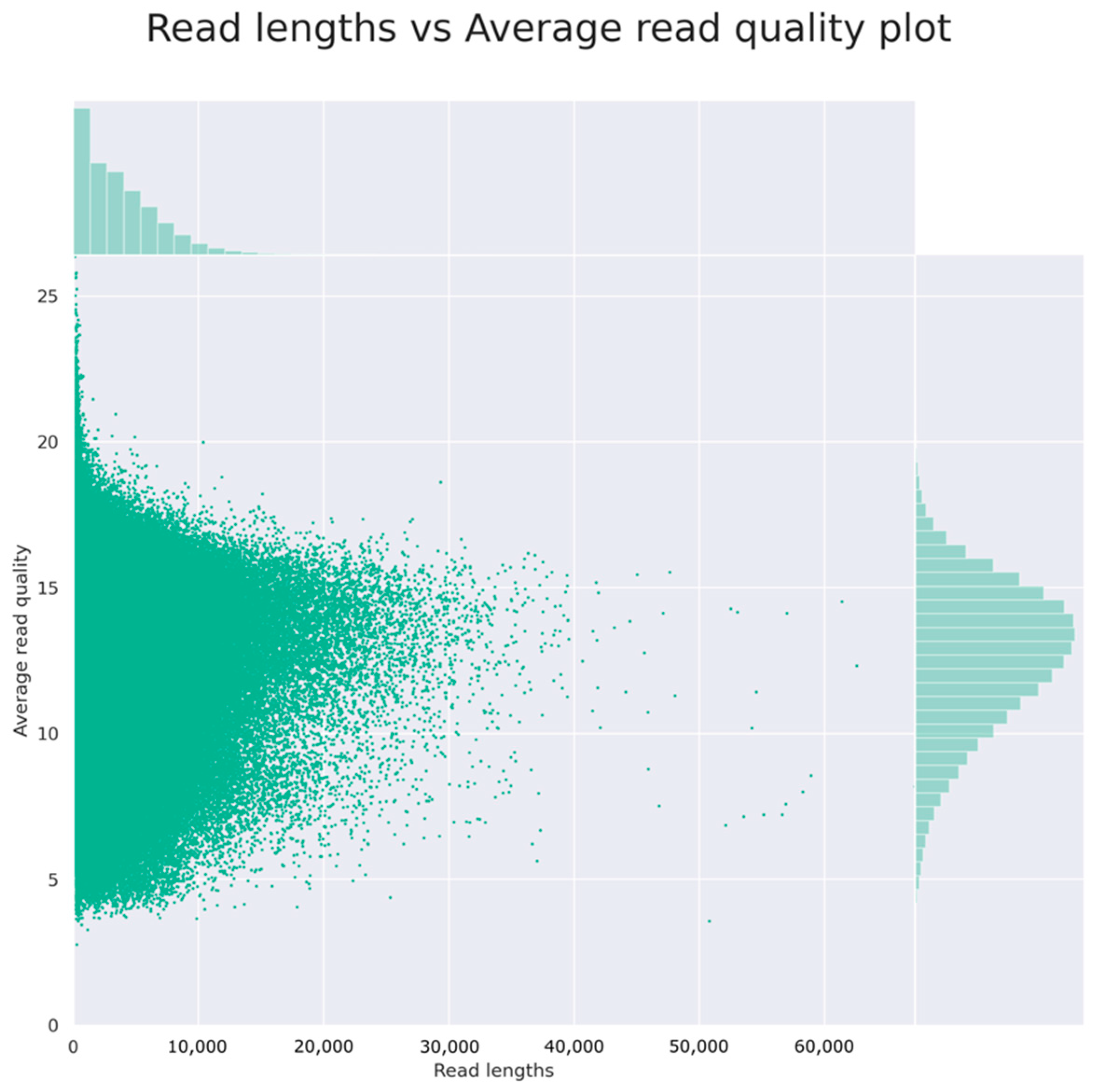
| Raw Reads | Filtered Reads | Assembled Reads | |
|---|---|---|---|
| Mean read length/contig length (bp) | 3862 | 4438 | 148,856 |
| Mean read quality | 12,6 | 12,7 | - |
| Number of reads/contig | 418,943 | 351,411 | 1 |
| Read length N50 (bp) | 6061 | 6114 | - |
| Total bases (bp) | 1,617,953,241 | 1,559,878,347 | - |
| Average coverage | - | - | 186.804 |
| Assembly Size (contig, bp) | 148,856 |
| GC content (%) | 36.92 |
| Genome fraction (%) | 89.99 |
| Number of indels | 411 |
| Alignment to reference | 135,411 |
| Total number of gene features | 349 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wahyuni, D.; Dwiyanti, F.G.; Pratama, R.; Majiidu, M.; Rachmat, H.H.; Siregar, I.Z. Chloroplast Genome Draft of Dryobalanops aromatica Generated Using Oxford Nanopore Technology and Its Potential Application for Phylogenetic Study. Forests 2021, 12, 1515. https://doi.org/10.3390/f12111515
Wahyuni D, Dwiyanti FG, Pratama R, Majiidu M, Rachmat HH, Siregar IZ. Chloroplast Genome Draft of Dryobalanops aromatica Generated Using Oxford Nanopore Technology and Its Potential Application for Phylogenetic Study. Forests. 2021; 12(11):1515. https://doi.org/10.3390/f12111515
Chicago/Turabian StyleWahyuni, Dwi, Fifi Gus Dwiyanti, Rahadian Pratama, Muhammad Majiidu, Henti Hendalastuti Rachmat, and Iskandar Zulkarnaen Siregar. 2021. "Chloroplast Genome Draft of Dryobalanops aromatica Generated Using Oxford Nanopore Technology and Its Potential Application for Phylogenetic Study" Forests 12, no. 11: 1515. https://doi.org/10.3390/f12111515
APA StyleWahyuni, D., Dwiyanti, F. G., Pratama, R., Majiidu, M., Rachmat, H. H., & Siregar, I. Z. (2021). Chloroplast Genome Draft of Dryobalanops aromatica Generated Using Oxford Nanopore Technology and Its Potential Application for Phylogenetic Study. Forests, 12(11), 1515. https://doi.org/10.3390/f12111515









