Soil Fungal Communities and Enzyme Activities along Local Tree Species Diversity Gradient in Subtropical Evergreen Forest
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Site and Experiment Design
2.2. Soil Sampling
2.3. Soil Physicochemical Characterization and Enzyme Activity Measurements
2.4. DNA Extraction, Amplification and Sequencing
2.5. Bioinformatic Analyses
2.6. Statistical Analysis
3. Results
3.1. The Composition of the Soil Fungal Community in the Gradient of Tree Species Diversity
3.2. Correlation between Soil Fungal Diversity and Local Tree Species Diversity
3.3. Soil Physicochemical Properties and Soil Enzymatic Activities
3.4. Relationship between Soil Properties, Enzyme Activities and Fungal Communities
4. Discussion
4.1. Soil Fungal Community Structure in the Gradient of Tree Species Diversity
4.2. Soil Fungal Richness and Diversity in the Gradient of Tree Species Diversity
4.3. Soil Fungi Trophic Modes and Functional Guilds in the Gradient of Tree Species Diversity
4.4. Different Reactions of Soil Fungal Community Structure with Soil Properties and Enzyme Activities
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Baldrian, P. Microbial activity and the dynamics of ecosystem processes in forest soils. Curr. Opin. Microbiol. 2017, 37, 128–134. [Google Scholar] [CrossRef]
- Zhu, Z.K.; Ge, T.D.; Luo, Y.; Liu, S.L.; Xu, X.L.; Tong, C.; Shibistova, O.; Guggenberger, G.; Wu, J.S. Microbial stoichiometric flexibility regulates rice straw mineralization and its priming effect in paddy soil. Soil Biol. Biochem. 2018, 121, 67–76. [Google Scholar] [CrossRef]
- Hector, A.; Schmid, B.; Beierkuhnlein, C.; Caldeira, M.C.; Diemer, M.; Dimitrakopoulos, P.G.; Finn, J.A.; Freitas, H.; Giller, P.S.; Good, J.; et al. Plant Diversity and Productivity Experiments in European Grasslands. Science 1999, 286, 1123–1127. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; Chen, Y.; Castro-Izaguirre, N.; Baruffol, M.; Brezzi, M.; Lang, A.; Li, Y.; Härdtle, W.; Von Oheimb, G.; Yang, X.; et al. Impacts of species richness on productivity in a large-scale subtropical forest experiment. Science 2018, 362, 80–83. [Google Scholar] [CrossRef] [Green Version]
- Tilman, D.; Reich, P.B.; Knops, J.; Wedin, D.; Mielke, T.; Lehman, C. Diversity and Productivity in a Long-Term Grassland Experiment. Science 2001, 294, 843–845. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.L.; Xu, T.L.; Veresoglou, S.D.; Hu, H.W.; Hao, Z.P.; Hu, Y.J.; Liu, L.; Deng, Y.; Rillig, M.; Chen, B. Plant diversity represents the prevalent determinant of soil fungal community structure across temperate grasslands in northern China. Soil Biol. Biochem. 2017, 110, 12–21. [Google Scholar] [CrossRef]
- Nguyen, N.H.; Williams, L.J.; Vincent, J.B.; Stefanski, A.; Cavender-Bares, J.; Messier, C.; Paquette, A.; Gravel, D.; Reich, P.; Kennedy, P.G. Ectomycorrhizal Fungal Diversity and Saprotrophic Fungal Diversity Are Linked to Different Tree Community Attributes in a Field-Based Tree Experiment. Mol. Ecol. 2016, 25, 4032–4046. [Google Scholar] [CrossRef] [PubMed]
- Wardle, D.A.; Lindahl, B.D. Ecology. Disentangling Global Soil Fungal Diversity. Science 2014, 346, 1052–1053. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, Y.H.; Chen, X.L.; Huang, Z.Q. Meta-analysis shows positive effects of plant diversity on microbial biomass and respiration. Nat. Commun. 2019, 10, 505–511. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tedersoo, L.; Bahram, M.; Cajthaml, T.; Põlme, S.; Hiiesalu, I.; Anslan, S.; Harend, H.; Buegger, F.; Pritsch, K.; Koricheva, J.; et al. Tree Diversity and Species Identity Effects on Soil Fungi, Protists and Animals Are Context Dependent. ISME J. 2016, 10, 346–362. [Google Scholar] [CrossRef] [Green Version]
- Li, W.-Q.; Huang, Y.-X.; Chen, F.-S.; Liu, Y.-Q.; Lin, X.-F.; Zong, Y.-Y.; Wu, G.-Y.; Yu, Z.-R.; Fang, X.-M. Mixing with Broad-Leaved Trees Shapes the Rhizosphere Soil Fungal Communities of Coniferous Tree Species in Subtropical Forests. For. Ecol. Manag. 2021, 480, 118664. [Google Scholar] [CrossRef]
- Bender, S.F.; Plantenga, F.; Neftel, A.; Jocher, M.; Oberholzer, H.R.; Köhl, L.; Giles, M.; Daniell, T.J.; Heijden, M.G. Symbiotic relationships between soil fungi and plants reduce N2O emissions from soil. ISME J. 2014, 8, 1336–1345. [Google Scholar] [CrossRef]
- Hiiesalu, I.; Bahram, M.; Tedersoo, L. Plant species richness and productivity determine the diversity of soil fungal guilds in temperate coniferous forest and bog habitats. Mol. Ecol. 2017, 26, 4846–4858. [Google Scholar] [CrossRef]
- Schlatter, D.C.; Kahl, K.; Carlson, B.; Huggins, D.R.; Paulitz, T. Fungal Community Composition and Diversity Vary with Soil Depth and Landscape Position in a No-till Wheat-Based Cropping System. FEMS Microbiol. Ecol. 2018, 94, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Kooch, Y.; Razie, S.J.; Masoud, T. Increasing Tree Diversity Enhances Microbial and Enzyme Activities in Temperate Iranian Forests. Trees 2018, 32, 809–822. [Google Scholar] [CrossRef]
- Cui, Y.; Fang, L.; Guo, X.; Wang, X.; Wang, Y.; Zhang, Y.; Zhang, X. Responses of soil bacterial communities, enzyme activities, and nutrients to agricultural-to-natural ecosystem conversion in the Loess Plateau, China. J. Soil Sediment. 2019, 19, 1427–1440. [Google Scholar] [CrossRef]
- Adamczyk, B.; Kilpeläinen, P.; Kitunen, V.; Smolander, A. Potential activities of enzymes involved in n, c, p and s cycling in boreal forest soil under different tree species. Pedobiologia 2014, 57, 97–102. [Google Scholar] [CrossRef]
- Cui, Y.; Fang, L.; Guo, X.; Wang, X.; Zhang, Y.; Li, P.; Zhang, X. Ecoenzymatic stoichiometry and microbial nutrient limitation in rhizosphere soil in the arid area of the northern Loess Plateau, China. Soil Biol. Biochem. 2018, 116, 11–21. [Google Scholar] [CrossRef]
- Ding, J.; Zhang, Y.; Wang, M.; Sun, X.; Cong, J.; Deng, Y.; Lu, H.; Yuan, T.; Van Nostrand, J.D.; Li, D.; et al. Soil organic matter quantity and quality shape microbial community compositions of subtropical broadleaved forests. Mol. Ecol. 2015, 24, 5175–5185. [Google Scholar] [CrossRef]
- Stark, S.; Männistö, M.K.; Eskelinen, A. Nutrient Availability and PH Jointly Constrain Microbial Extracellular Enzyme Activities in Nutrient-Poor Tundra Soils. Plant Soil 2014, 383, 373–385. [Google Scholar] [CrossRef]
- Chen, L.; Xiang, W.H.; Wu, H.L.; Ouyang, S.; Lei, P.F.; Hu, Y.J.; Ge, T.D.; Ye, J.; Kuzyakov, Y. Contrasting patterns and drivers of soil fungal communities in subtropical deciduous and evergreen broadleaved forests. Appl. Microbiol. Biotechnol. 2019, 103, 5421–5433. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.R.; Chen, Z.; Piao, S.L.; Peng, C.H.; Ciais, P.; Wang, Q.F.; Li, X.R.; Zhu, X.J. High Carbon Dioxide Uptake by Subtropical Forest Ecosystems in the East Asian Monsoon Region. Proc. Nati. Acad. Sci. USA 2014, 111, 4910–4915. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Xiang, W.H.; Lei, P.F.; Deng, X.W.; Tian, D.L.; Fang, X.; Peng, C.H. Standing fine root mass and production in four Chinese subtropical forests along succession and species diversity gradient. Plant Soil 2014, 376, 445–459. [Google Scholar] [CrossRef]
- Zhao, L.J.; Xiang, W.H.; Li, J.X.; Lei, P.F.; Deng, X.W.; Fang, X.; Peng, C.H. Effects of topographic and soil factors on woody species assembly in a Chinese subtropical evergreen broadleaved forest. Forests 2015, 6, 650–669. [Google Scholar] [CrossRef] [Green Version]
- Ouyang, S.; Xiang, W.; Wang, X.; Zeng, Y.; Lei, P.; Deng, X.; Peng, C. Significant Effects of Biodiversity on Forest Biomass during the Succession of Subtropical Forest in South China. For. Ecol. Manag. 2016, 372, 291–302. [Google Scholar] [CrossRef]
- Chen, L.; Xiang, W.H.; Wu, H.L.; Ouyang, S.; Zhou, B.; Zeng, Y.L.; Chen, Y.L.; Kuzyakov, Y. Tree species identity surpasses richness in affecting soil microbial richness and community composition in subtropical forests. Soil Biol. Biochem. 2019, 130, 113–121. [Google Scholar] [CrossRef]
- Jacob, A.; Hertel, D.; Leuschner, C. Diversity and Species Identity Effects on Fine Root Productivity and Turnover in a Species-Rich Temperate Broad-Leaved Forest. Funct. Plant Biol. 2014, 41, 678–689. [Google Scholar] [CrossRef]
- Xiang, W.H.; Fan, G.W.; Lei, P.F.; Zeng, Y.L.; Tong, J.; Fang, X.; Deng, X.W.; Peng, C.H. Fine Root Interactions in Subtropical Mixed Forests in China Depend on Tree Species Composition. Plant Soil 2015, 395, 335–349. [Google Scholar] [CrossRef]
- Walkley, A. A critical examination of a rapid method for determining organic carbon in soils-effect of variations in digestion conditions and of inorganic soil constituents. Soil Sci. 1947, 63, 251–264. [Google Scholar] [CrossRef]
- Guan, S.Y. Soil Enzymes and Their Research Methods; Agriculture Press: Beijing, China, 1986. [Google Scholar]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef] [PubMed]
- Chao, A. Nonparametric Estimation of the Number of Classes in a Population. Scand. J. Stat. 1984, 11, 265–270. [Google Scholar]
- Prescott, C.E.; Grayston, S.J. Tree Species Influence on Microbial Communities in Litter and Soil: Current Knowledge and Research Needs. For. Ecol. Manag. 2013, 309, 19–27. [Google Scholar] [CrossRef]
- Aponte, C.; García, L.V.; Marañón, T.; Gardes, M. Indirect host effect on ectomycorrhizal fungi: Leaf fall and litter quality explain changes in fungal communities on the roots of co-occurring Mediterranean oaks. Soil Biol. Biochem. 2010, 42, 788–796. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.H.; Ni, J.; Tang, F.P.; Jiang, L.F.; Guo, T.R.; Pei, K.Q.; Sun, L.F.; Liang, Y. The effects of different human disturbance regimes on root fungal diversity of Rhododendron ovatum in subtropical forests of China. Can. J. For. Res. 2017, 47, 659–666. [Google Scholar] [CrossRef]
- Hooper, D.U.; Chapin, F.S., III; Ewel, J.J.; Wardle, D.A. Effects of biodiversity on ecosystem functioning: A consensus of current knowledge. Ecol. Monogr. 2005, 75, 3–35. [Google Scholar] [CrossRef]
- Wardle, D.A. The Influence of Biotic Interactions on Soil Biodiversity. Ecol. Lett. 2006, 9, 870–886. [Google Scholar] [CrossRef] [PubMed]
- Urbanová, M.; Šnajdr, J.; Baldrian, P. Composition of Fungal and Bacterial Communities in Forest Litter and Soil Is Largely Determined by Dominant Trees. Soil Biol. Biochem. 2015, 84, 53–64. [Google Scholar] [CrossRef]
- Yang, H.; Guo, Z.L.; Chu, X.L.; Man, R.Z.; Chen, J.X.; Liu, C.J.; Tao, J.; Jiang, Y. Comment on Impacts of Species Richness on Productivity in a Large-Scale Subtropical Forest Experiment. Science 2019, 363, 80–83. [Google Scholar] [CrossRef]
- Peay, K.G.; Baraloto, C.; Fine, P.V. Strong Coupling of Plant and Fungal Community Structure across Western Amazonian Rainforests. ISME J. 2013, 7, 1852–1861. [Google Scholar] [CrossRef]
- Schmidt, R.; Mitchell, J.; Scow, K. Cover Cropping and No-till Increase Diversity and Symbiotroph: Saprotroph Ratios of Soil Fungal Communities. Soil Biol. Biochem. 2019, 129, 99–109. [Google Scholar] [CrossRef] [Green Version]
- Waldrop, M.P.; Zak, D.R.; Blackwood, C.B.; Curtis, C.D.; Tilman, D. Resource Availability Controls Fungal Diversity across a Plant Diversity Gradient. Ecol. Lett. 2006, 9, 1127–1135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raza, W.; Ling, N.; Zhang, R.F.; Huang, Q.W.; Xu, Y.C.; Shen, Q.R. Success Evaluation of the Biological Control of Fusarium Wilts of Cucumber, Banana, and Tomato since 2000 and Future Research Strategies. Crit. Rev. Biotechnol. 2017, 37, 202–212. [Google Scholar] [CrossRef] [PubMed]
- Gonthier, P.; Giordano, L.; Zampieri, E.; Lione, G.; Vizzini, A.; Colpaert, J.V.; Balestrini, R. An ectomycorrhizal symbiosis differently affects host susceptibilityto two congeneric fungal pathogens. Fungal Ecol. 2019, 39, 250–256. [Google Scholar] [CrossRef]
- Chen, L.; Xiang, W.; Ouyang, S.; Wu, H.; Xia, Q.; Ma, J.; Zeng, Y.; Lei, P.; Xiao, W.; Li, S.; et al. Tight coupling of fungal community composition with soil quality in a Chinese fir plantation chronosequence. Land Degrad. Dev. 2020, 32, 1164–1178. [Google Scholar] [CrossRef]
- Duan, C.J.; Fang, L.C.; Yang, C.L.; Chen, W.B.; Cui, Y.X.; Li, S.Q. Reveal the response of enzyme activities to heavy metals through in situ zymography. Ecotox. Environ. Safe 2018, 156, 106–115. [Google Scholar] [CrossRef] [PubMed]
- Bandick, A.K.; Dick, R.P. Field management effects on soil enzyme activities. Soil Biol. Biochem. 1999, 31, 1471–1479. [Google Scholar] [CrossRef]
- Wang, Q.; Wang, C.; Yu, W.W.; Turak, A.; Chen, D.W.; Huang, Y.; Ao, J.H.; Jiang, Y.; Huang, Z.R. Effects of nitrogen and phosphorus inputs on soil bacterial abundance, diversity, and community composition in Chinese fir plantations. Front. Microbiol. 2018, 9, 1543. [Google Scholar] [CrossRef]
- Riley, R.; Salamov, A.A.; Brown, D.W.; Nagy, L.G.; Floudas, D.; Held, B.W.; Levasseur, A.; Lombard, V.; Morin, E.; Otillar, R.; et al. Extensive Sampling of Basidiomycete Genomes Demonstrates Inadequacy of the White-Rot/Brown-Rot Paradigm for Wood Decay Fungi. Proc. Nati. Acad. Sci. USA 2014, 111, 9923–9928. [Google Scholar] [CrossRef] [Green Version]


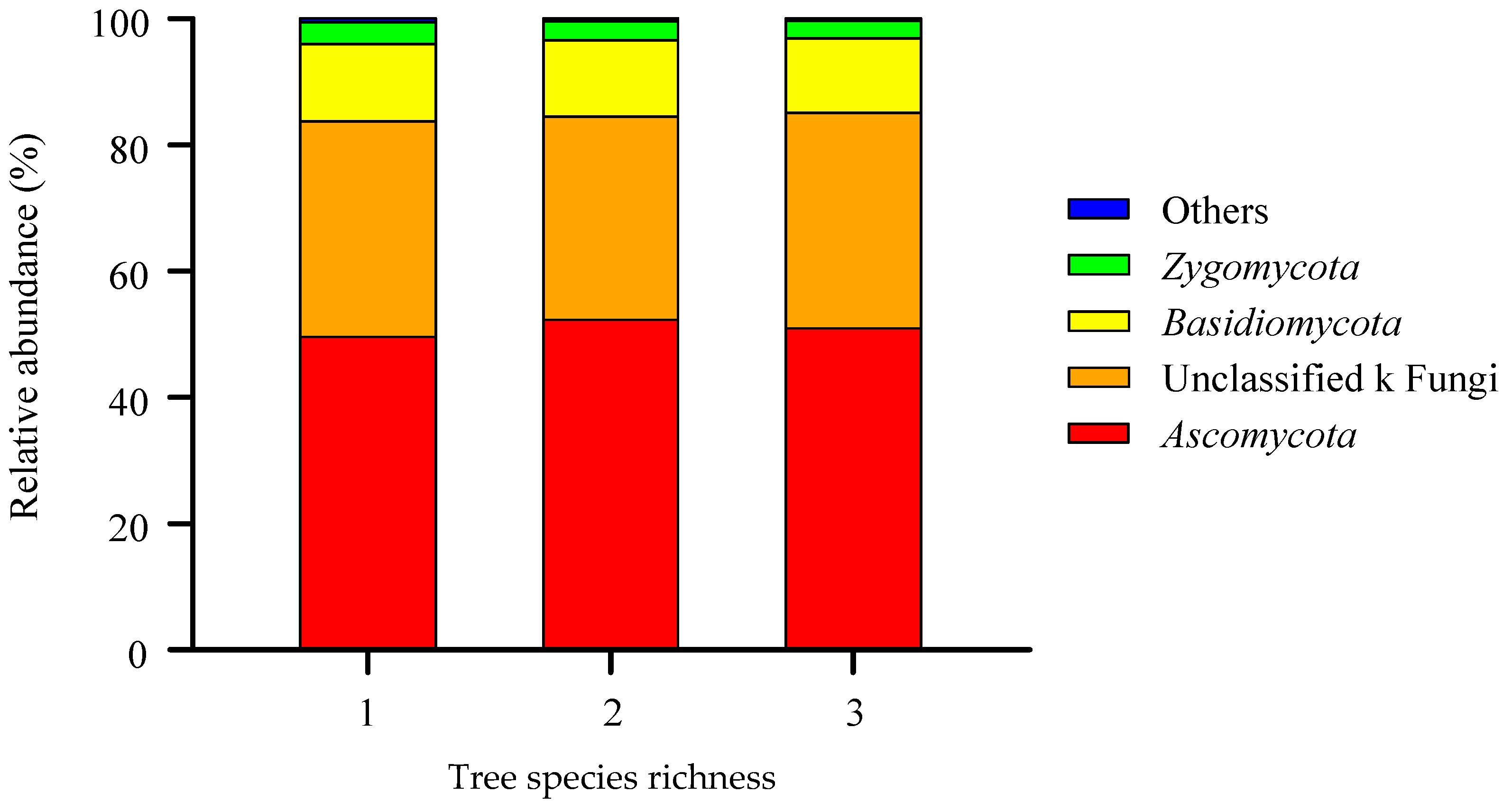



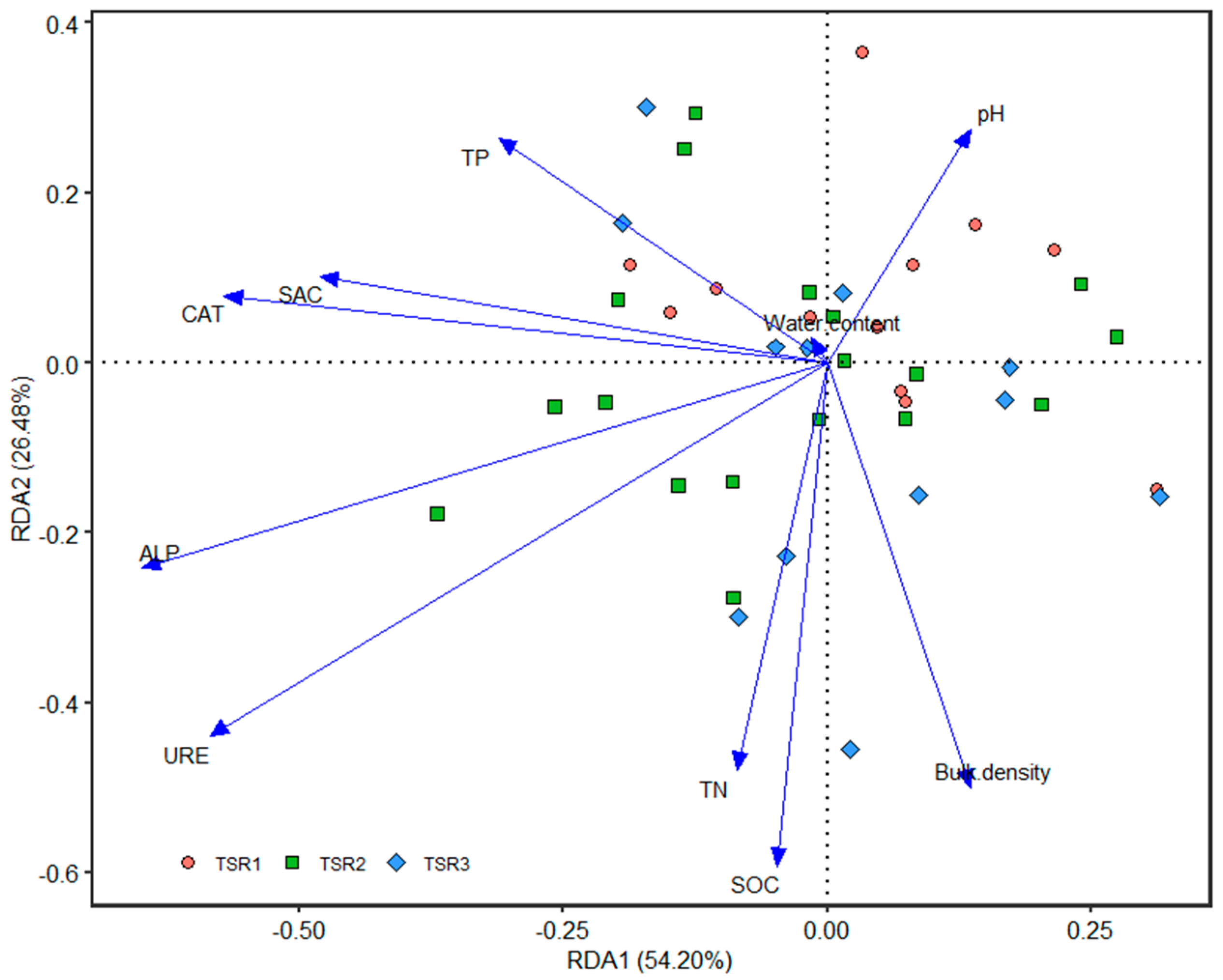

| Tree Species Richness | AP | FP | PP | DS-PS | SS | US | WS | Ec | EO-MRAB | En |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 0.03 ± 0.01 b | 0.002 ± 0.001 b | 0.04 ± 0.01 b | 0.97 ± 0.96 a | 0.37 ± 0.18 a | 15.70 ± 6.02 a | 0.13 ± 0.05 a | 82.38 ± 5.81 a | 0.36 ± 0.28 a | 0.03 ± 0.01 a |
| 2 | 0.04 ± 0.01 b | 0.002 ± 0.001 b | 0.05 ± 0.01 b | 6.58 ± 5.61 a | 0.10 ± 0.08 a | 11.89 ± 2.64 a | 1.27 ± 1.08 a | 78.20 ± 8.83 a | 1.82 ± 1.39 a | 0.04 ± 0.01 a |
| 3 | 0.12 ± 0.03 a | 0.000 ± 0.001 a | 0.22 ± 0.03 a | 0.63 ± 0.62 a | 0.24 ± 0.19 a | 32.24 ± 10.23 a | 1.67 ± 0.78 a | 63.15 ± 10.32 a | 1.59 ± 1.58 a | 0.13 ± 0.05 a |
| Tree Species Richness | Bulk Density (g/cm3) | Water Content (%) | pH | SOC (g/kg) | TN (g/kg) | TP (g/kg) |
|---|---|---|---|---|---|---|
| 1 | 1.05 ± 0.04 a | 14.20 ± 0.63 a | 3.99 ± 0.06 a | 18.68 ± 1.78 a | 1.46 ± 0.34 b | 0.27 ± 0.01 a |
| 2 | 1.12 ± 0.03 a | 14.32 ± 0.51 a | 3.99 ± 0.03 a | 28.06 ± 2.54 b | 1.32 ± 0.23 b | 0.30 ± 0.01 a |
| 3 | 1.06 ± 0.04 a | 13.57 ± 1.29 a | 3.96 ± 0.06 a | 28.55 ± 3.20 b | 2.58 ± 0.29 a | 0.24 ± 0.01 a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fu, Z.; Chen, Q.; Lei, P.; Xiang, W.; Ouyang, S.; Chen, L. Soil Fungal Communities and Enzyme Activities along Local Tree Species Diversity Gradient in Subtropical Evergreen Forest. Forests 2021, 12, 1321. https://doi.org/10.3390/f12101321
Fu Z, Chen Q, Lei P, Xiang W, Ouyang S, Chen L. Soil Fungal Communities and Enzyme Activities along Local Tree Species Diversity Gradient in Subtropical Evergreen Forest. Forests. 2021; 12(10):1321. https://doi.org/10.3390/f12101321
Chicago/Turabian StyleFu, Ziqi, Qin Chen, Pifeng Lei, Wenhua Xiang, Shuai Ouyang, and Liang Chen. 2021. "Soil Fungal Communities and Enzyme Activities along Local Tree Species Diversity Gradient in Subtropical Evergreen Forest" Forests 12, no. 10: 1321. https://doi.org/10.3390/f12101321
APA StyleFu, Z., Chen, Q., Lei, P., Xiang, W., Ouyang, S., & Chen, L. (2021). Soil Fungal Communities and Enzyme Activities along Local Tree Species Diversity Gradient in Subtropical Evergreen Forest. Forests, 12(10), 1321. https://doi.org/10.3390/f12101321




