Abstract
The pinewood nematode (PWN) Bursaphelenchus xylophilus, the causal agent of the pine wilt disease (PWD), enters above-ground parts of the tree, migrates through the resin canals and feeds on plant cells causing extensive damage. In order to penetrate the cell wall and establish a parasitic relationship with host trees, the PWN needs to secretea mixture of active cell wall degrading enzymes. In maritime pine, Pinus pinaster, which is high susceptible to PWN, xyloglucan is the major hemicellulosic polysaccharide in primary cells. The xyloglucan backbone is susceptible to hydrolysis by numerous endoglucanases, some of them specific to xyloglucan. However, to completely degrade xyloglucan, all substitutions on the glucan backbones must be released, and l-fucose residues in xyloglucan branches are released by α-l-fucosidases. In the present study, the molecular characterization of two α-l-fucosidases found in PWN secretome was performed. Moreover, a novel α-l-fucosidase was identified and its cDNA and gene sequence were determined. The three-dimensional structures of these α-l-fucosidases were predicted and the transcript levels were analyzed, thus providing new insights into fundamental PWN biology and the possible role of these proteins as cell wall degrading enzymes.
1. Introduction
The pinewood nematode (PWN), Bursaphelenchus xylphilus, the causal agent of the pine wilt disease (PWD), is recognized worldwide as a major forest pest. Unlike other plant-parasitic nematodes such as cyst, root knot or root lesion nematodes, which infect the plant roots and tubers, the PWN enters above-ground parts of the tree, migrates through the resin canals and feeds on plant cells. Therefore, the pine tree cell wall, which is mainly composed of cellulose (40%–50%), hemicellulose (≈25%) and lignin (25%–35%), constitutes the first barrier that PWN must overcome. Cell wall proteins and pectins are among the other minor compounds of the cell wall [1]. The secretion of active cell wall degrading enzymes (CWDE) is essential for PWN to invade this cell wall and establish a parasitic relationship with the host tree.
Cellulose is a homopolymer of β-linked glucose units forming glucan chains linked to each other by hydrogen bonds, which form the cellulose microfibril. Its hydrolysis is achieved by the synergistic action of cellulases. In the B. xylophilus genome, the 11 genes identified as cellulases are endo-β-1,4-glucanases belonging to the glycoside hydrolase family 45 (GH45) and are not found in any other nematode genus in which the complete genome is known [2,3]. Seven of these cellulases are also found in B. xylophilus secretome [4,5].
The water-insoluble cellulose microfibrils are associated with mixtures of soluble non-cellulosic polysaccharides, the hemicelluloses, which prevent cellulose microfibrils from aggregating. These are heteropolymers with branched polysaccharides and hexose and pentose sugar monomers. Hemicelluloses in conifer cell walls are mainly xyloglucan, glucuronoarabinoxylan and galactoglucomannan [6]. In maritime pine, Pinus pinaster, which is high susceptible to PWN, xyloglucan is the major hemicellulosic polysaccharide in primary cells [7]. Xyloglucan consists of a linear β-(1-4)-linked d-glucan backbone that carries α-d-xyloxyl, β-d-galactosyl-(1-2)-α-d-xylosyl and α-l-fucosyl-(1-2)-β-d-galactosyl-(1-2)-α-d-xylosyl side chains attached to the OH-6 of β-glucosyl residues (Figure 1), depending on the plant family and specific tissue [8]. The xyloglucan backbone is susceptible to hydrolysis by numerous endoglucanases, some of which are specific to xyloglucan; however, to completely degrade xyloglucan, all substitutions on the glucan backbones must be released. This requires different enzyme activities such as xyloglucan β-1,4-endoglucanase, α-d-xylosidase, β-d-galactosidase, α-l-fucosidase and β-d-glucosidase. Among these, α-l-fucosidase is of particular importance in xyloglucan breakdown or structural modification since terminal α-l-substituents seem to be directly involved in the binding of xyloglucan to cellulose [9]. l-fucose residues in xyloglucan branches are released by α-l-fucosidases belonging to glycoside hydrolases family 29 (GH29) and GH95 [10], according to the Carbohydrate-Active Enzymes (CAZy) database classification (http://www.cazy.org/) [11].
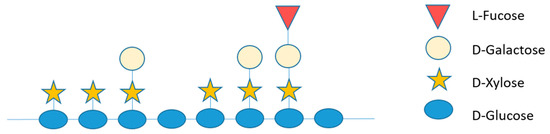
Figure 1.
Schematic representation of the hemicellulosic polysaccharide xyloglucan.
The importance of fungal and bacteria α-l-fucosidases in plant polysaccharide degradation, namely, on xyloglucan has been reported previously [10,12]. A secreted α-l-fucosidase belonging to the GH29 family from the plant pathogenic fungus Fusarium graminearum was shown to be active in releasing fucose from a xyloglucan fragment [13]. Nevertheless, the importance of α-l-fucosidases from plant-parasitic nematodes and their possible role in breaking down plant cell walls have been underestimated. However, the inability of a nematode to degrade any component of the plant cell wall may result in unsuccessful infection, thus, all possible carbohydrate active enzymes that could act on plant cell wall degradation must be further explored. Recently, a α-l-fucosidase gene from B. xylophilus (Bxy-fuca) was characterized and associated with nematode development, lifespan and reproduction [14]. In our previous studies, two α-l-fucosidases were detected in B. xylophilus secretome [5] and in the present study, the molecular characterization of these two α-L-fucosidases was performed, thus providing new insights into the possible role of these proteins in degrading the plant cell walls and in PWN pathogenicity.
2. Materials and Methods
2.1. In Silico Identification of Fucosidase Sequences
Two α-l-fucosidases belonging to glycoside hydrolase family 29 (GH29) and detected in B. xylophilus secretome in our previous studies [5], were selected for further molecular characterization and named BxFUCA1 and BxFUCA2. α-l-fucosidases were also searched in B. xylophilus transcriptome BioProject PRJNA192936 [15] and genome Bioproject PRJEA64437 [2] to look for the characteristic InterPro domain IPR000933 of GH29 [16]. This domain was also searched in other plant-parasitic nematodes genomes available from WormBase Parasite [17]: Globodera rostochiensis PRJEB13504-Ro1; G. pallida PRJEB123; Meloidogyne arenaria PRJEB8714; M. floridensis PRJEB6016; M. graminicola PRJNA411966; M. hapla PRJNA29083-VW9; M. incognita PRJEB8714; M. javanica PRJEB8714, and identified predicted proteins were included for phylogenetic analysis.
2.2. Pinewood Nematodes
Nematodes from a Portuguese B. xylophilus isolate (BxPt17AS) were maintained in cultures of the fungus Botrytis cinerea grown on malt extract agar medium at 25 °C. Mixed developmental nematode stages grown for 15 days on fungal cultures were collected with distilled water using a 20 µm sieve and washed three times with sterile water or RNase free water. These nematodes were then used for DNA and RNA extraction or treatment with P. pinaster extracts. Aqueous P. pinaster extracts from the stems of non-inoculated seedlings were prepared as previously described [5] and used to incubate the nematodes, simulating pine stimulus. Nematodes were soaked in 5 mL of pine extracts for 16 h at 28 °C, washed three times in 1x M9 buffer and concentrated via centrifugation. Nematode pellets of approximately 100 µL were used for RNA extraction.
2.3. Fucosidase cDNA and Genomic Coding Sequence
Bursaphelenchus xylophilus RNA was extracted from various stages of B. xylophilus (eggs, juveniles, females and males) as previously described [5] and DNase treated with TURBO DNA-free kit (Invitrogen, Carlsbad, CA, USA). The cDNA synthesis was carried out using iScript™ cDNA Synthesis Kit (BioRad, Foster City, CA, USA). DNA extraction was performed with the DNeasy Blood and Tissue Mini kit (Qiagen, Venlo, Netherlands), following the manufacturer’s instructions. Synthetized cDNA and extracted genomic DNA were used for PCR amplification of selected fucosidase using primers BxFuF (TGCTTATCAGCTGCTGTCTT) and BxFuR (TGGCTGTTTAATGGCAAAAGGG). Primers were designed, based on transcript sequence (protein ID All_gs454_002563), to flank the predicted protein cDNA coding sequence fragment and the putative full-length protein-encoding genomic sequence, including partial sequences of 5´and 3´regions. Reactions were performed with GoTaq DNA polymerase (Promega, Fitchburg, WI, USA) in a Thermal Cycler (BioRad) with an initial denaturation step of 95 °C for 2.5 min followed by 35 reaction cycles of 95 °C for 30 s, annealing at 52 °C for 30 s and extension at 72 °C for 1.5 (cDNA template) or 2.5 min (DNA template) and a final extension at 72 °C for 5 min. Amplified products were purified using the Qiaquick PCR Purification Kit (Qiagen) and sequenced in both strands in an Automatic Sequencer 3730xl under BigDyeTM terminator cycling conditions at Macrogen, Inc. using specific primers (Supplementary Materials, Table S1).
2.4. Sequence Analyses
Sequence analyses were conducted using BioEdit [18]. To identify introns in the genomic sequence, Softberry FGENESH [19] was used and the gene scheme was generated using Exon-Intron Graphic maker [20] available from WormWeb.org. Molecular mass values and isoelectric points of deduced amino acid sequence were predicted using the deduced protein sequence by analysis with ProtParam tool software available at the ExPASy website [21]. InterPro [16] was used for the protein family search, predicting the presence of domains and important sites, and SignalP 4.1 Server [22] was used to predict the presence of the signal peptides. Deduced amino acid sequences were BLASTp against the NCBI non-redundant protein database [23] and UniProt Knowledgebase (reviewed) database (UniProtKB/Swiss-Prot) [24], where records were manually annotated with information extracted from literature and curator evaluated computational analysis. Sequence alignments were obtained using BioEdit between deduced amino acid sequences and predicted fucosidases found after BLASp searches and identified in plant-parasitic nematodes’ available genomes, as described above. This alignment was used to estimate phylogenetic relationships in MEGA 6 [25] by the maximum likelihood method based on the Jones-Taylor-Thornton (JTT) matrix-based model.
2.5. Protein Homology Modeling
The 3-D structures were modeled in silico with the protein structure homology-modeling server SWISS-MODEL [26]. The best template for protein 3-D structure prediction was selected with the SWISS-MODEL template library search. The optimality of the predicted structures was estimated with global model quality estimation (GMQE) and qualitative model energy analysis (QMEAN) [27], both implemented in SWISS-MODEL. The resulting GMQE score was expressed as a number between 0 and 1, reflecting the expected accuracy of a model built with that alignment and template and the coverage of the target. Higher numbers indicate higher reliability, whereas for QMEAN, scores of −4.0 or below indicate that the predicted structures are very low quality. Additionally, the obtained structures were validated with a Ramachandran plot analysis implemented on the RAMPAGE server [28]. The structures with a higher number of residues in favored regions (>90%) and lower number of residues in outlier region were selected. The binding of possible ligands to predicted protein structures was previewed using the protein-small molecule docking web service SwissDock [29] and visualized using UCSF Chimera 1.4 software [30].
2.6. Fucosidases’ Transcript Levels
The relative transcript abundance of fucosidases in B. xylophilus grown on fungus and after P. pinaster extract stimulus was assessed by reverse transcription quantitative real time PCR (RT-qPCR) with SsoAdvance Universal SYBR Green supermix (BioRad), according to standard protocols, using the CFX96 Touch™ Real-Time PCR Detection System (BioRad). The cDNAs of extracted RNAs were synthesized as previously described and used for qPCR. The amplification kinetics of each transcript was normalized with the amplification kinetics of the actin and 18S genes, chosen as endogenous controls and amplified with primers as previously described [5]. Other primers used in qPCR were designed based on B. xylophilus transcript sequences (Supplementary Table S1). qPCRs were done at 98 °C for 30 s, followed by 40 cycles of 98 °C for 5 s and 60 °C for 30 s. Melting curve analyses were performed and validation experiments were first carried out to ensure equivalent amplification efficiency for all transcripts. The RT-qPCRs were conducted for three biological repetitions, with three technical replicates for each qPCR. Amplification efficiencies and Ct values were determined by the CFX ManagerTM Software 2.1 (BioRad) and the mean Ct values were used for relative transcript level analysis (ΔΔCt method) with statistically significant differences tested using the pair-wise fixed reallocation randomization test© in REST software [31].
3. Results
3.1. Sequence Analyses
Analyzing B. xylophilus genome [2], three genes (BXY_1120100.1, BXY_0325100.1 and BXY_0325000.1) were identified as coding putative GH29 proteins. After analyzing B. xylophilus transcriptome data from B. xylophilus BioProject PRJNA192936 [15], only two proteins were identified with possible α-l-fucosidase activity belonging to GH29, one of them (All_gs454_001152) was 100% identical to the genome predicted BXY_0325100.1 gene product, named BxFUCA1, which corresponds to the α-l-fucosidase (Bxy-FUCA) recently described and characterized by Tang et al. [14]. The other fucosidase (All_gs454_002563) presented 57%, 67% and 52% sequence identity to the genome predicted BXY_1120100.1, BXY_0325100.1 and BXY_0325000.1 gene products, respectively. This protein, named BxFUCA2, was selected for further gene and cDNA coding sequencing.
The BxFUCA1 protein sequence deduced from transcriptome and genome data, is 462 amino acids long with a calculated molecular mass of 52.9 KDa and a theoretical isoelectric point (pI) of 6.23. The protein sequence includes a N-terminal signal peptide of 15 amino acids with no predicted transmembrane domain. After removing this signal peptide sequence, a molecular mass of 51.3 KDa and pI of 6.25 were estimated.
Nucleotide sequences of 2332 bp long and 1491 bp long were determined for BxFUCA2, and submitted to Genbank under the accession numbers MN935185 and MN935184 for genomic amplicon and cDNA amplicon, respectively. The genomic sequence revealed the presence of eight introns (Figure 2).
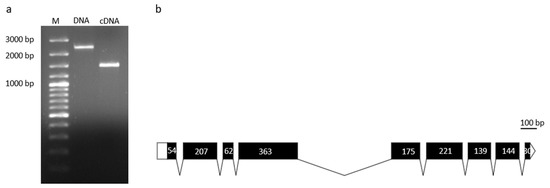
Figure 2.
Molecular characterization of BxFUCA2. Amplicons of genomic DNA (2332 bp) and cDNA (1491 bp) coding sequences (a). Schematic representation of Bxfuca2 gene structure (b). Relative positions and respective sizes of exons are indicated as dark boxes and introns as lines.
The deduced BxFUCA2 protein sequence is 464 amino acids long with a calculated molecular mass of 53.4 KDa and a theoretical pI of 5.54. The protein sequence includes a N-terminal signal peptide of 16 amino acids with no predicted transmembrane domain. Removing this signal peptide sequence, a molecular mass of 51.7 KDa and pI of 5.54 were estimated.
The typical domain of the glycoside hydrolase family 29 (IPR000933), the conserved α-l-fucosidase metazoa-type domain (IPR016286) and the glycoside hydrolase family 29 conserved site (IPR018526), identifying the putative active site with the conserved pattern P-x(2)-L-x(3)-K-W-E-x-C, were mapped for both predicted proteins BxFUCA1 and BxFUCA2, as well as the sequence WxDx containing the catalytic nucleophile aspartate (D) [32] (Figure 3).
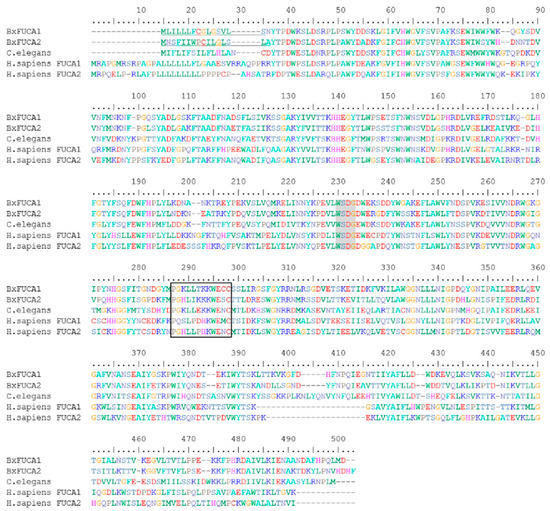
Figure 3.
Multiple sequence alignment of the predicted BxFUCA1 and BxFUCA2 proteins from Bursaphelenchus xylophilus with described α-l-fucosidases from other organisms. BxFUCA1 and BxFUCA2 sequences were aligned with α-l-fucosidase from Caenorhabditis elegans (NP_510020.2), Homo sapiens plasma FUCA2 (NP_114409.2) and H. sapiens tissue FUCA1 (NP_00138.2). The predicted BxFUCA1 and BxFUCA2 signal peptides are underlined, the sequence WxDx containing the catalytic nucleophile aspartate (D) is shaded in grey and the putative active site with the conserved pattern P-x(2)-L-x(3)-K-W-E-x-C (IPR018526) is marked with the black box.
BxFUCA1 and BxFUCA2 predicted proteins share 67.4% of amino acid sequence identity. The top 10 BLASTp hits of each predicted protein against NCBI non-redundant protein database were hypothetical proteins and predicted α-l-fucosidases from different nematode species (Caenorhabditis elegans, C. nigoni, C. briggsae, Steinernema carpocapsae, Ancylostoma caninum, A. duodenale, A. ceylanicum, Nippostrongylus brasiliensis and Haemonchus contornus) with sequence identities of 49%–54%. The BLASTp search against UniProt reviewed database revealed BxFUCA1 and BxFUCA2 sequence identities of 46%–53% with 12 reviewed α-l-fucosidases, included in the phylogenetic analysis as reference α-l-fucosidases. Predicted α-l-fucosidases from plant-parasitic nematodes genome sequencing data, available on WormBase Parasite database, were also included in this study. Phylogenetic analysis exhibited a clear separation of fusosidases from animal origin with those of bacteria or fungi origin and a nematode-specific divergence inside animal origin fucosidases is denoted. Additionally, all predicted plant-parasitic nematode fucosidases, excluding B. xylophiulus fucosidases, form a separate cluster from other animal-parasitic and free-living nematodes. Considering B. xylophiulus fucosidases, phylogenetic analysis revealed that BxFUCA1 is identical to BXY_0325100.1 gene product and that BxFUCA2 differs from the three predicted proteins deduced from B. xylophilus genome, thus forming a separate cluster from all the other nematode α-l-fucosidases (Figure 4).
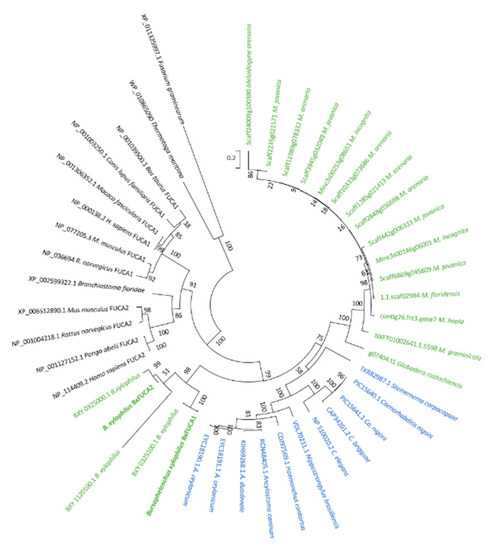
Figure 4.
Phylogenetic analysis of predicted Bursaphelenchus xylophilus α-l-fucosidases. The maximum likelihood method based on amino acid sequence alignment of the two α-l-fucosidases characterized in this study (BxFUCA1 and BxFUCA2) and other homologous sequences found after Blastp searches on the NCBI non-redundant protein database: Caenorhabditis elegans, C. nigoni, C. briggsae, Steinernema carpocapsae, Ancylostoma caninum, A. duodenale, A. ceylanicum, Nippostrongylus brasiliensis and Haemonchus contornus; on the UniProt reviewed database: Homo sapiens FUCA1, H. sapiens FUCA2, Rattus norvegicus FUCA1, R. norvegicus FUCA2, Canis lupus familiaris FUCA1, Mus musculus FUCA1, M. musculus FUCA2, Bos taurus FUCA1, Pongo abelii FUCA2, Macaca fascicularis FUCA1, Branchiostoma floridae, C. elegans; and predicted from WormBase Parasite available plant-parasitic genomes data: Bursaphelenchus xylophilus, Meloidogyne incognita, M. hapla, M. floridensis, M. arenaria, M. graminicola, M. jacanica and G. rostochiensis. Predicted α-l-fucosidases from the bacteria Thermotoga maritima and the fungus Fusarium graminearum are also included. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Initial tree(s) for the heuristic search were obtained by applying the neighbor-joining method to a matrix of pair-wise distances estimated using a Jones-Taylor-Thornton model. The tree is drawn to scale with branch lengths measured in the number of substitutions per site. The analysis involved 44 amino acid sequences. All positions with less than 50% site coverage were eliminated.
3.2. Structural Analyses
To better understand the structure and function of these proteins, 3-D structures were generated by homology modeling. BxFUCA1 and BxFUCA2 structures were predicted using the crystal structure of the GH29 family α-l-fucosidase from Fusarium graminearum (PDB ID: 4ni3.1.A) as a template. Both proteins presented two domains. For BxFUCA1, a C-terminal domain composed of seven anti-parallel β-sheets and a N-terminal domain formed by five parallel β-sheets arranged around the central axis and surrounded by eight α-helices forming the catalytic pocket, were predicted. The BxFUCA1 3-D structure is similar to the Bxy-FUCA structure predicted by Tang et al. [14]. For BxFUCA2, the C-terminal domain was composed of eight anti-parallel β-sheets and the N-terminal catalytic domain was formed by five parallel β-sheets arranged around the central axis and surrounded by eight α-helices (Figure 4). In addition to the GMQE and QMEAN values of 0.64 and −3.84 for BxFUCA1 and 0.64 and −3.63 for BxFUCA2 predicted structures, the Ramachandran plots validated these structural models with more than 90% of the residues located in the most favored regions (Supplementary Materials, Figure S1). Docking of a α-l-fucose molecule (CHEBI ID: 42548) to the catalytic pocket of BxFUCA1 structure was previewed with a full fitness of −2164.72 Kcal/mol and an estimated ΔG of −6.36 kcal/mol. For BxFUCA2, the docking of this fucose molecule to the catalytic pocket was also previewed with a full fitness of −2135.23 Kcal/mol and an estimated ΔG of −6.41 kcal/mol (Figure 5).
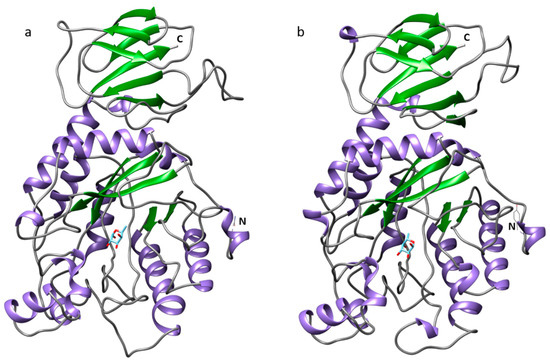
Figure 5.
Predicted three-dimensional structure of two α-l-fucosidases from Bursaphelenchus xylophilus and docking of a α-l-fucose molecule. Structures were obtained using the crystal structure of α-l-fucosidase from Fusarium graminearum (PDB ID: 4ni3.1.A) as a template for BxFUCA1 (a) and BxFUCA2 (b) in SwissModel and docking of the fucose molecule was previewed in SwissDock and visualized on Chimera 1.14.
3.3. Transcript Level Analysis
The relative transcript abundance of BxFUCA1 and BxFUCA2 in B. xylophilus grown on fungus and after P. pinaster extract stimulus was assessed by RT-qPCR. Both genes were found to be expressed in fungus and after pine extract stimulus, with no significant differences found between the two conditions after statistical analysis (p > 0.1). The transcript level of BxFUCA2 in B. xylophilus under pine tree extract was higher than BxFUCA1 (Figure 6).
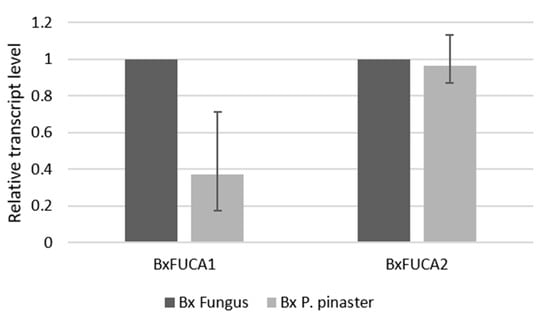
Figure 6.
Relative transcript levels of BxFUCA1 and BxFUCA2 measured by RT-qPCR. Differences in BxFUCA1 and BXFUCA2 transcript levels between Bursaphelenchus xylophilus grown on fungus (Bx Fungus) and after Pinus pinaster pine extract stimulus (Bx P. pinaster). Bars represent the standard error range of three biological replicates.
4. Discussion
Several studies have shown that plant-parasitic nematodes possess a set of synergistically active CWDE to allow their invasion and migration through the plant host tissues. The study of nematode CWDE not only enhance our understanding on the nematode-host interaction but also provides information on novel sources of enzymes with potential use in the biomass industry. There is a sustained focus on the discovery and application of carbohydrate-active enzymes for the selective deconstruction of plant biomass for the production of food, chemicals, biomaterials and biofuels [12].
A genome wide analysis of the CWDE from the genome sequenced phyto-parasitic nematode species identified 119 putative CWDE in B. xylophilus genome, most of them glycoside hydrolases. The number of GH genes and their family varies between analyzed species, indicating a possible evolution of plant-parasitic nematodes according to their feeding behavior [3]. Although not mentioned by the authors as predicted CWDE in published B. xylophilus genome [2,3] or even first published secretome [4], three genes and proteins with possible fucosidase activity belonging to GH29 were identified in B. xylophilus genome and secretome and were found listed in the supplementary tables of the respective publications. These proteins were identified as GH29 and from these, only the one coded by the BXY_0325100.1 gene (Bxy-fuca) was further characterized and described as playing a crucial role in the development, lifespan and reproduction of B. xylophilus [14]. The two proteins identified as α-l-fucosidases in our previous study on B. xylophilus secretome [5] were further characterized in this study, and a new α-l-fucosidase of B. xylophilus (BxFUCA2) was described.
The characterization of the complete cDNA coding sequences of BxFUCA1 and BxFUCA2 and analysis of their deduced amino acid sequences confirmed that these proteins are α-L-fucosidases belonging to the GH29 CAZy family, and they contain the typical domains and conserved sites of these proteins. A signal peptide was detected in predicted amino acid sequences, suggesting that these proteins are secreted, which is in accordance with previous data on B. xylophilus secretome [5]. Phylogenetic analysis of these proteins showed that fucosidases from B. xylophilus form a separated cluster, which is distant from the other plant-parasitic nematodes fucosidases, indicating a possible evolution of these proteins according to nematodes’ host and feeding behavior. Unlike other plant-parasitic nematodes, such as Globodera sp. or Meloidogyne sp. that infect the plant roots and/or tubers, B. xylophilus enters the above-ground parts of the pine tree, migrates through the resin canals and feeds on cells.
To date, GH29 crystal structures have been determined only for bacterial and fungi species and not yet for any eukaryotic species. The 3-D structure of BxFUCA1 and BxFUCA2 were predicted by homology-modeling with the available structure of the secreted α-l-fucosidase from the plant pathogenic fungus Fusarium graminearum. This fungal fucosidase was found to be active in releasing fucose from xyloglucan [13], the major hemicellulosic polysaccharide in pine cell walls [7]. A similar role for B. xylophilus-secreted BxFUCA1 and BxFUCA2 could be inferred, acting on the fucosylated side groups of xyloglucan, breaking down the self-aggregation of xyloglucan chains, the binding of xyloglucan to cellulose and exposing the xyloglucan backbone for further degradation. These proteins may also act on other fucosylated substrates like pectin or glycoproteins, contributing to deconstruction of the plant cell wall. The possible involvement of BxFUCA1 in hemicellulose digestion along with its crucial role in the development, lifespan and reproduction of B. xylophilus, has recently been suggested in a work describing the Bxy-fuca gene [14].
Our findings suggest that α-l-fucosidases belonging to the GH29 family, from B. xylophilus and other plant-parasitic nematodes, should be included in CWDE studies as this family of proteins is involved in many crucial biological processes that include fucosylated glycoconjugates, and it has been recognized as being involved in xyloglucan degradation. Xyloglucan constitutes a significant reserve of metabolically accessible monosaccharides for diverse phytopathogenic, saprophytic and gut symbiotic micro-organisms. To overcome the intrinsic stability of the diverse glycosidic bonds in this plant polysaccharide, bacteria and fungi have evolved extensive repertoires of xyloglucan-active enzymes [10] and plant-parasitic nematodes, like B. xylophilus, also had to do it.
The molecular characterization of the two α-l-fucosidases identified in B. xylophilus secretome increases our knowledge on this important group of enzymes, and provides new insights about the importance of these proteins in B. xylophilus pathogenicity. Moreover, the limited homology of these proteins with other known fucosidases makes them promising targets for the further development of target-specific strategies for PWN management.
Supplementary Materials
The following are available online at https://www.mdpi.com/1999-4907/11/3/265/s1, Figure S1: Ramachandran analysis of predicted three-dimensional structure of two α-l-fucosidase from Bursaphelenchus xylophilus. Ramachandran plot of BxFUCA1 (a) and BxFUCA2 (b) predicted structures, Table S1: Primers used for BxFUCA2 cDNA and DNA coding sequence amplification, sequencing and qPCR and for BxFUCA1 qPCR.
Author Contributions
Conceptualization, J.M.S.C., I.A. and L.F.; Formal analysis, J.M.S.C.; Funding acquisition, J.M.S.C., I.A. and L.F.; Investigation, J.M.S.C.; Methodology, J.M.S.C.; Project administration, L.F.; Supervision, I.A.; Validation, J.M.S.C.; Writing—original draft, J.M.S.C.; Writing—review & editing, J.M.S.C., I.A. and L.F. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Portuguese Foundation for Science and Technology (FCT) through national funds and the co-funding by FEDER, PT2020 and COMPETE 2020 under the projects POINTERS- PTDC/ASP-SIL/31999/2017 (POCI-01-145-FEDER-031999) and UID/BIA/04004/2019; Project ReNATURE—Valorization of the Natural Endogenous Resources of the Centro Region (Centro 2020, Centro-01-0145-FEDER-000007) and Instituto do Ambiente, Tecnologia e Vida.
Acknowledgments
The authors would like to thank Joana Duarte by her technical assistance on Bursaphelenchus xylophilus cultures maintenance.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Plomion, C.; Leprovost, G.; Stokes, A. Wood Formation in Trees. Plant Physiol. 2001, 127, 1513–1523. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, T.; Cotton, J.A.; Dalzell, J.J.; Hasegawa, K.; Kanzaki, N.; McVeigh, P.; Takanashi, T.; Tsai, I.J.; Assefa, S.A.; Cock, P.J.A.; et al. Genomic insights into the origin of parasitism in the emerging plant pathogen Bursaphelenchus xylophilus. PLoS Pathog. 2011, 7, e1002219. [Google Scholar] [CrossRef] [PubMed]
- Rai, K.M.; Balasubramanian, V.K.; Welker, C.M.; Pang, M.; Hii, M.M.; Mendu, V. Genome wide comprehensive analysis and web resource development on cell wall degrading enzymes from phyto-parasitic nematodes. BMC Plant Biol. 2015, 15, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Shinya, R.; Morisaka, H.; Kikuchi, T.; Takeuchi, Y.; Ueda, M.; Futai, K. Secretome analysis of the pine wood nematode Bursaphelenchus xylophilus reveals the tangled roots of parasitism and its potential for molecular mimicry. PLoS ONE 2013, 8, e67377. [Google Scholar] [CrossRef]
- Cardoso, J.M.S.; Anjo, S.I.; Fonseca, L.; Egas, C.; Manadas, B.; Abrantes, I. Bursaphelenchus xylophilus and B. mucronatus secretomes: A comparative proteomic analysis. Sci. Rep. 2016, 6, 39007. [Google Scholar] [CrossRef]
- Scheller, H.V.; Ulvskov, P. Hemicelluloses. Annu. Rev. Plant Biol. 2010, 61, 263–289. [Google Scholar] [CrossRef]
- Acebes, J.L.; Lorences, E.P.; Revilla, G.; Zarra, I. Pine xyloglucan. Occurrence, localization and interaction with cellulose. Physiol. Plant. 1993, 89, 417–422. [Google Scholar] [CrossRef]
- Pauly, M.; Keegstra, K. Biosynthesis of the plant cell wall matrix polysaccharide xyloglucan. Annu. Rev. Plant Biol. 2016, 67, 235–259. [Google Scholar] [CrossRef]
- Levy, S.; York, W.S.; Stuike-Prill, R.; Meyer, B.; Staehelin, L.A. Simulations of the static and dynamic molecular conformations of xyloglucan. The role of the fucosylated sidechain in surface-specific sidechain folding. Plant J. 1991, 1, 195–215. [Google Scholar] [CrossRef]
- van den, B.J.; de Vries, R.P. Fungal enzyme sets for plant polysaccharide degradation. Appl. Microbiol. Biotechnol. 2011, 91, 1477–1492. [Google Scholar] [CrossRef]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, 490–495. [Google Scholar] [CrossRef] [PubMed]
- Attia, M.A.; Brumer, H. Recent structural insights into the enzymology of the ubiquitous plant cell wall glycan xyloglucan. Curr. Opin. Struct. Biol. 2016, 40, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Walton, J.D.; Brumm, P.; Phillips, G.N. Structure and substrate specificity of a eukaryotic fucosidase from Fusarium graminearum. J. Biol. Chem. 2014, 289, 25624–25638. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Ma, R.; Zhu, N.; Guo, K.; Guo, Y.; Bai, L.; Yu, H.; Hu, J.; Zhang, X. Bxy-fuca encoding α-L-fucosidase plays crucial roles in development and reproduction of the pathogenic pinewood nematode, Bursaphelenchus xylophilus. Pest Manag. Sci. 2019, 76, 205–214. [Google Scholar] [CrossRef] [PubMed]
- Figueiredo, J.; Simões, M.J.; Gomes, P.; Barroso, C.; Pinho, D.; Conceição, L.; Fonseca, L.; Abrantes, I.; Pinheiro, M.; Egas, C. Assessment of the geographic origins of pinewood nematode isolates via single nucleotide polymorphism in effector genes. PLoS ONE 2013, 8, e83542. [Google Scholar] [CrossRef]
- Mitchell, A.L.; Sangrador-Vegas, A.; Luciani, A.; Madeira, F.; Nuka, G.; Salazar, G.A.; Chang, H.Y.; Richardson, L.J.; Qureshi, M.A.; Fraser, M.I.; et al. InterPro in 2019: Improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res. 2019, 47, 351–360. [Google Scholar] [CrossRef]
- Howe, K.L.; Bolt, B.J.; Shafie, M.; Kersey, P.; Berriman, M. WormBase ParaSite—A comprehensive resource for helminth genomics. Mol. Biochem. Parasitol. 2017, 215, 2–10. [Google Scholar] [CrossRef]
- Hall, T. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Softberry FGENESH. Available online: http://www.softberry.com/ (accessed on 1 December 2019).
- Exon-Intron Graphic Maker. Available online: http://wormweb.org/exonintron (accessed on 1 December 2019).
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar]
- Petersen, T.N.; Brunak, S.S.; von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef]
- NCBI- National Center for Biothecnology Information. Available online: http://www.ncbi.nlm.nih.gov (accessed on 1 December 2019).
- UniProtKB/Swiss-Prot. Available online: https://www.uniprot.org/ (accessed on 1 December 2019).
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Biasini, M.; Bienert, S.; Waterhouse, A.; Arnold, K.; Studer, G.; Schmidt, T.; Kiefer, F.; Cassarino, T.G.; Bertoni, M.; Bordoli, L.; et al. SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014, 42, 252–258. [Google Scholar] [CrossRef] [PubMed]
- Benkert, P.; Künzli, M.; Schwede, T. QMEAN server for protein model quality estimation. Nucleic Acids Res. 2009, 37, 510–514. [Google Scholar] [CrossRef] [PubMed]
- Lovell, S.C.; Davis, I.W.; Adrendall, W.B.; de Bakker, P.I.W.; Word, J.M.; Prisant, M.G.; Richardson, J.S.; Richardson, D.C. Structure validation by C alpha geometry: Phi, psi and C beta deviation. Proteins Struct. Funct. Genet. 2003, 50, 437–450. [Google Scholar] [CrossRef] [PubMed]
- Grosdidier, A.; Zoete, V.; Michielin, O. SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res. 2011, 39, 270–277. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. Relative expression software tool (REST©) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002, 30, 1–10. [Google Scholar] [CrossRef]
- Tarling, C.A.; He, S.; Sulzenbacher, G.; Bignon, C.; Bourne, Y.; Henrissat, B.; Withers, S.G. Identification of the catalytic nucleophile of the family 29 α-L-Fucosidase from Thermotoga maritima through trapping of a covalent glycosyl-enzyme intermediate and mutagenesis. J. Biol. Chem. 2003, 278, 47394–47399. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).