Abstract
Genetic variation within a dominant riparian forest tree affects susceptibility to a leaf-galling aphid (Pemphigus betae), which induces phytochemical and structural changes in leaf tissue. Research Highlights: We show here that these changes to tree leaf tissue alter adjacent in-stream leaf litter decomposition rates and the aquatic macroinvertebrate community associated with litter in the stream for some Populus genotypes. Background and Objectives: Naturally occurring hybrid cottonwoods (Populus fremontii × Populus angustifolia) are differentially susceptible to aphid attack and vary in induced phytochemistry following attack. When leaves are galled by aphids, foliar tissue is altered structurally (through the formation of pea-sized gall structures) and phytochemically (through an increase in foliar condensed tannin concentrations). Materials and Methods: To examine the effect of aphid-galled leaves on forest stream processes, we collected both galled and un-galled leaves from five clones of three hybrid cottonwood genotypes in an experimental forest. We measured in-stream litter decomposition rates, aquatic fungal biomass and aquatic macroinvertebrate community composition. Results: Decomposition rates differed among genotypes and the galled litter treatments, with a 27% acceleration of decomposition rate for the galled litter of one genotype compared to its own un-galled litter and no differences between galled and un-galled litters for the other two genotypes. Genotype by foliar gall status interactions also occurred for measures of phytochemistry, indicating a prevalence of complex interactions. Similarly, we found variable responses in the macroinvertebrate community, where one genotype demonstrated community differences between galled and un-galled litter. Conclusions: These data suggest that plant genetics and terrestrial forest herbivory may be important in linking aquatic and terrestrial forest processes and suggest that examination of decomposition at finer scales (e.g., within species, hybrids and individuals) reveals important ecosystem patterns.
1. Introduction
Riparian forests provide a fundamental linkage between terrestrial and aquatic systems, and herbivores in terrestrial environments have the potential to alter these interactions. For example, studies have examined the effects of aboveground herbivory by arthropods and mammals on terrestrial forest nutrient cycling, decomposition rates and other ecosystem processes [1,2,3,4,5]. Herbivory has been shown to induce premature leaf abscission and reduce nutrient resorption [6,7], increase foliar fragmentation [8], and increase wound-induced carbon-to-nitrogen ratios, foliar phenolic defense compounds, lignin, and cellulose [9,10,11,12,13,14]. These effects of herbivores can decrease decomposition rates in forest ecosystems and laboratory studies [13,15,16,17], or have no effect [18,19]. Because herbivory often affects subsequent litter quality, it is likely that herbivory could alter the quantity, quality and timing of riparian forest leaf litter inputs to streams [7,20,21]. Understanding the potential interactions between herbivores and litter quality is important because the majority of plant foliage affected by herbivores is not assimilated by herbivores, but dropped as plant litter [22], so these alterations to litter inputs could potentially change organic matter linkages between forests and streams.
Leaf-galling herbivores are mainly species of invertebrates (e.g., aphids, midges, wasps, mites) that live in symbiosis with plants. Leaf galling invertebrates are often in obligate relationships with particular plant species to house and protect their offspring in gall structures. Plant species respond to herbivores in a variety of ways [1,9], but mainly respond to galling herbivores with both structural and chemical alterations to leaf, stem, or bark tissues [5]. Differential plant responses to galling herbivores may also occur due to a wide array of factors associated with the environment, individual constitutive leaf quality, and genetic differences in resistance to galling [5,17,23]. Galling herbivores have been shown to influence decomposition rates in terrestrial ecosystems [24], and galling aphids differentially influenced genotypes of Populus to alter terrestrial decomposition rates in a previous study [17]. Accordingly, we should expect that variability in plant leaf chemistry associated with galling by insects will result in differential effects on the communities of organisms that colonize and decompose leaf litter in forest streams with consequences for the process of aquatic leaf litter decomposition.
We examined whether terrestrial arthropod herbivores have the capacity to alter ecosystem processes in adjacent forest stream ecosystems in a well-studied cottonwood forest system in the western USA. Specifically, we examined the effects of a leaf-galling aphid (Pemphigus betae Doane; Homoptera: Aphididae) on in-stream leaf litter decomposition. Previous studies show that different genotypes of cottonwood (Populus angustifolia James; Salicaceae) show differential susceptibility to aphid herbivory [25,26,27,28]. Earlier work in this system reported a deceleration of terrestrial decomposition for leaves galled by the aphid compared to un-galled leaves [17]. In this case, the gall-forming aphid does not remove tissue from the tree; it induces changes in tissue structure and chemistry, causing an increase in condensed tannins, which are implicated in the slowing of decomposition. Because Populus are naturally occurring riparian trees whose litter provides essential energy to adjacent streams, we were interested in whether the effects of the galling aphid on litter chemistry would have similar “after-life” effects [13] in an aquatic ecosystem. Based on the above logic, we make the following predictions:
- (1)
- Litter chemistry will vary based on Populus genotype, galling by P. betae, and interactions between genotype and gall status. For example, while galled leaves will be characterized by elevated levels of condensed tannins relative to un-galled leaves, this response may vary by genotype.
- (2)
- Galled leaves will decompose more slowly in the stream than un-galled leaves from the same tree.
- (3)
- We predict that slower decomposition rates will be correlated with lower aquatic fungal biomass on leaves.
- (4)
- All together, differences in condensed tannins, aquatic fungal biomass, and decomposition rates will result in altered macroinvertebrate communities on galled versus un-galled leaves.
- (5)
- The effects of both tree genotype and galling aphids will decline up trophic chains [29].
2. Materials and Methods
2.1. Common Garden Experimental Forest
For litter collection, we use an experimental forest designed to isolate the effects of genetic differences from environmental differences in the field. This experimental forest represents a common garden of naturally occurring hybrid cottonwood genotypes and was planted from wild cuttings in the spring of 1991 (Ogden, Utah, USA). The hybridizing system includes Fremont cottonwood (Populus fremontii), narrowleaf cottonwood (Populus angustifolia), F1 hybrids (P. fremontii × P. angustifolia) and backcross hybrids to the narrowleaf parent (Figure 1). In a previous study, the proportion of 35 species-specific (P. fremontii) markers was determined for each of three genotypes (11, 1017, 3200) using restriction fragment length polymorphisms (RFLP; see [30,31]). In the fall of 2003, after aphids had vacated their galls, naturally abscised leaf litter was collected weekly from five clones of each of three backcrosses to P. angustifolia hybrid genotypes in the garden using a mixture of whole-tree mesh enclosures and mesh branch bags. Litter from these five clonal replicates was air-dried and mixed to produce a genotype-level mixture. Litter was then sorted between un-galled litter and litter galled by the aphid Pemphigus betae for each genotype. This gall is conspicuous and easily distinguished from the galls of other species of Pemphigus and Chaetophorus aphids (Figure 1).
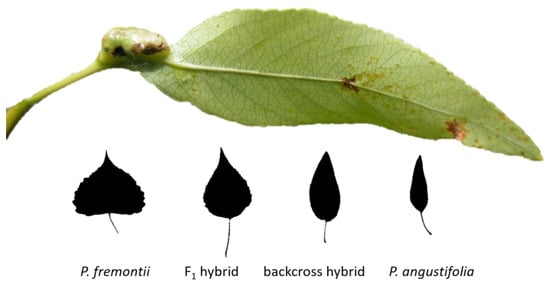
Figure 1.
Cottonwood species hybridize between two parents (Populus fremontii and Populus angustifolia) to create an F1 hybrid and a series of backcrosses to the P. angustifolia parent (backcross hybrids). Shown is a Pemphigus betae gall on a narrowleaf cottonwood. Photo by Rick Lindroth, 2005.
2.2. Site Description
Litterbags were incubated nearby in the Weber River (Utah, USA) to determine litter decomposition rates. The Weber River is a fourth-order river at the location used in this study (41°8′15.94′′ N, 111°56′44.78′′ W), but overall a fifth order river [32] with its terminus at the Great Salt Lake. The riparian zone is dominated by P. fremontii, P. angustifolia and both F1 and backcross hybrids. Water quality parameters were measured throughout the study period using a Hydrolab minisonde (Hydrolab-Hach Corporation, Loveland, CO, USA). Temperature, dissolved oxygen, pH, total dissolved solids, specific conductivity, and salinity were measured at each harvest date. All water quality measures are presented in Table 1.

Table 1.
Water quality and site measurements in the Weber River (UT, USA) for the study period from December 2003 to June 2004 (n = 3). Values represent means ± 1 S.E.
2.3. Initial Litter Chemistry
Leaf litter was ground for initial chemical analyses in a Wiley Mill (Thomas Scientific, Swedesboro, NJ, USA) to pass mesh size 40. Subsamples (25–50 mg) were extracted for condensed tannins with 70% acetone and 10 mM ascorbic acid. We used the butanol-HCl method to determine condensed tannin (CT) concentrations [33], with standards purified from P. angustifolia following the methods of Hagerman and Butler [34]. We quantified absorbance on a Spectramax-Plus 384 spectrophotometer (Molecular Devices, Sunnyvale, CA, USA). We also determined total litter %N and %P by modified micro-Kjeldahl digestion [35] followed by analysis on a Lachat AE Flow Injection Analyzer (Lachat Instruments, Inc., Loveland, CO, USA), using the salicylate and molybdate-ascorbic acid methods, respectively (Lachat instruments, Inc. 1992).
2.4. Litter Decomposition
Six leaf litter treatments included galled and un-galled litter from each of three backcross hybrid genotypes (3200, 11 and 1017). Two g of air-dried litter was placed into 1 mm mesh litterbags. We used five replicate litterbags (n = 5) for each treatment at three harvest dates for a total of 90 litterbags. Litterbags were randomly assigned a harvest date and a location within the stream. Bags were anchored in the stream along 2 m lengths of steel rebar and wedged into place in active depositional areas near the shore. Litterbags were color-coded by harvest date to assist harvesting and avoid disturbing neighboring bags. Litterbags were harvested from the stream on three harvest dates, after 17, 108 and 189 days, and placed into individual polyethylene zipper bags for transport on ice to the laboratory.
Litterbags were processed within 12 h of harvesting. Leaf material was rinsed with stream water to remove and collect all sediment, organic debris, and invertebrates. Ten leaf punches were taken from leaf material for ergosterol analysis (see below). Remaining leaf material was flash frozen on dry ice in the field and lyophilized for three days. Sediment and invertebrates were sieved through 250 μm nets for preservation in 70% ethanol. Freeze-dried leaf material was weighed and ground in a Wiley Mill to pass mesh size 40. Ground material was combusted at 500 °C in a muffle furnace (Barnstead International, Dubuque, IA, USA) for 1 h to determine percent organic material and ash-free dry mass (AFDM).
2.5. Aquatic Fungal Biomass
Aquatic fungal biomass was estimated using an ergosterol assay by High-Performance Liquid Chromatography (HPLC; sensu Suberkropp [36]). Ten (11 mm diameter) leaf discs were punched from remaining leaf material in each litterbag at harvest date 17, which approximated peak fungal biomass on decomposing tissue. Five of these leaf discs were flash-frozen and lyophilized for three days to determine the dry weight of the leaf material. The other five leaf discs were preserved in 5 mL HPLC-grade (99.99%) methanol for ergosterol extraction. Leaf discs were refluxed at 80 °C for 30 min in a mixture of methanol (25 mL), and alcoholic KOH (5 mL). The leaf particles were removed and discarded, and the extractant was added to 20 mL of pentane. The pentane was evaporated under a stream of N2, and the residue was re-dissolved into 1 mL of HPLC-grade methanol and filtered through HPLC-certified, 13 mm syringe filters with 0.2 μm PTFE membranes, for processing on the HPLC. With methanol as the mobile phase and a 1.0 mL/min flow rate, we passed samples through a C-18 column and found an ergosterol standard retention time of 6.35 min. Ergosterol concentration was converted to fungal biomass (mg/g leaf litter) using a 5.5 μg ergosterol mg−1 fungal biomass conversion factor (sensu Gessner and Chauvet [37]).
2.6. Aquatic Invertebrates
Preserved invertebrate samples were sieved through 1 mm mesh to separate micro- from macroinvertebrates. Macroinvertebrate samples from all harvest dates were sorted under 2× magnification and counted and identified under a dissecting scope (40×) to the lowest taxonomic level using several keys [38,39,40]. Reference specimens are maintained in the LeRoy Leaf Litter (L3) Lab at The Evergreen State College. In total, we identified 35 genera from a total of 25 families and 16 orders.
2.7. Statistical Analyses
Initial leaf chemistry and fungal biomass measurements were compared with standard two-way analysis of variance (ANOVA), with gall status and effect of genotype treated as fixed factors. Pairwise comparisons were conducted using Tukey’s HSD tests following significant ANOVA results. Analysis of leaf litter decay rate constants (k) required a natural log-transformation of AFDM remaining for two reasons, to meet normality and equal variance assumptions and to determine the exponential decay rate (k) [41,42]. Decay constants (k) were compared using an equality of slopes test with Hommel’s corrected p-values in SAS 8.01 (SAS Institute, Inc. 1999–2000, sensu Swan & Palmer [43]). Patterns of %N, %P, %C, and AFDM of remaining leaf litter were analyzed throughout the decomposition process and by genotype, gall status, and all possible interactions with days in stream using three-way ANOVAs.
Invertebrate community data were analyzed using a variety of community analysis techniques. Taxa abundance, taxa richness, taxa evenness and Shannon’s (H’) and Simpson’s (D) diversity indices were calculated for each litterbag at all harvest dates. Values were compared using ANOVA and post-hoc comparisons (Tukey’s HSD). To visualize community-wide responses to leaf litter treatments, we used a non-metric multidimensional scaling (NMDS) ordination method and a Bray-Curtis similarity measure in PC-ORD 4.02 (MJM Software, McCune, B. and Mefford, M.J. 1999. Gleneden Beach, OR, USA). To test for differences among groups we used a multi-response permutation procedure (MRPP) in the same program. In all analyses, a p-value less than 0.05 was used to denote statistical significance.
3. Results
3.1. Leaf Chemistry and Aquatic Decomposition
Gall status and genotype were both significant interacting factors influencing initial leaf chemistry (Table 2). Both condensed tannins and %N were significantly influenced by Populus genotype, gall status, and genotype*gall interactions. Genotypes 11 and 3200 showed significantly higher condensed tannins for galled leaves compared to un-galled leaves (Table 3), but genotype 1017 did not differ by gall status. Genotypes 11 and 3200 also showed significantly higher %N for un-galled leaves compared to galled leaves, but genotype 1017 was invariable. Initial %P only varied by gall status, and only for genotype 3200 which had higher %P in galled leaves (Table 3).

Table 2.
Two-way ANOVA results for initial leaf chemistry and fungal biomass measurements at Harvest 1. Values represent overall F(df1,df2)-ratios and p-values as well as effect test F-ratios and p-values for each main effect (Genotype, Gall status), and the Genotype*Gall interaction. Significant effects are shown in bold.

Table 3.
Initial leaf litter chemistry (% condensed tannins, % nitrogen, % phosphorus) values, fungal biomass (mg/g) at day 17, and decomposition rate constants (k day−1; across entire 189 day study) for galled and un-galled litter of three cottonwood backcross hybrid genotypes. Chemistry values represent means ± 1 S.E. (n = 2), fungal biomass values represent means. ± 1 S.E. (n = 5), decomposition rate constants (k) represent the slopes of the exponential regression lines ± S.E. for the slope (n = 5). Lower case letters denote significant differences at an alpha of 0.05 following a Tukey’s HSD test, or an equality of slopes test in the case of k.
For the two genotypes that showed induced condensed tannin concentrations in galled tissue (11 and 3200), galled leaves decomposed at slightly slower rates, although these data only show a trend toward deceleration, not a statistically significant difference (Figure 2). However, genotype 1017, which did not induce increased condensed tannins in litter (Table 3), showed the opposite pattern in decomposition, where galled leaves decomposed 27% faster in the stream than un-galled leaves (Figure 2). Additionally, galled litter from genotype 1017 decomposed 17% faster than all other litter types used in this study, regardless of their gall status. Decomposition rate constant comparisons are conservative measures of differences among treatments. We can also compare remaining AFDM across treatments throughout the decomposition process. These results show that AFDM was significantly influenced by Populus genotype, the genotype*gall interaction, day in the stream, and the genotype*day interaction (Table 4).
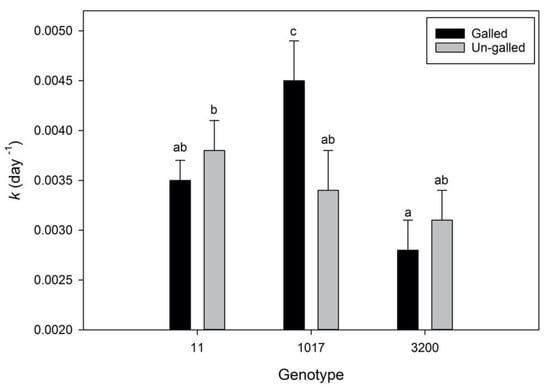
Figure 2.
Decomposition rate constants (k day−1) for three backcross hybrid cottonwood genotypes comparing galled to un-galled leaf litter. Values represent mean slopes ± 1 S.E. and lower case letters denote significantly different slopes based on contrasts at a Hommel’s corrected alpha level of 0.05.

Table 4.
Three-way ANOVA results for litter chemistry throughout the decomposition process. Values represent overall F(df1,df2)-ratios and p-values as well as effect test p-values for each main effect (Genotype, Gall status, Harvest Day), and all possible interactions. Significant effects are shown in bold.
We predicted that initial litter chemistry might be correlated with decomposition rate (k), but we found non-significant correlations between condensed tannins and decomposition for galled litter (Pearson’s r = −0.9956, p = 0.0597), and for un-galled litter (Pearson’s r = −0.7868, p = 0.4235). We also hypothesized that genetic differences among the three genotypes might explain differences in chemistry or decomposition rate. For each backcross genotype we used the number of species-specific P. fremontii markers determined in a previous study [31] (Genotype 11 = 0.1818, 1017 = 0.0286, 3200 = 0.0571), and found that the proportion of Fremont markers was positively correlated with condensed tannin concentration (%CT: r = 0.9976, p = 0.0441) but not decomposition rate for un-galled litter (k: r = 0.8276, p = 0.3794). For galled litter, the proportion of Fremont markers did not correlate with either condensed tannin concentration or decomposition rate (%CT: r = 0.1566, p = 0.8999; k: r = 0.2483, p = 0.8402).
Throughout the decomposition process, litter %N and %P changed through time differentially for the different genotypes and based on a genotype*gall interaction (Table 4). Percent N was significantly influenced by genotype, day, and the genotype*gall interaction, but gall status and all other interactions were insignificant. (Table 4). Nitrogen concentrations increased in all litter types throughout the decomposition process, and %N tended to be higher in un-galled litter from genotypes 11 and 3200, with no difference between galled and un-galled litter for genotype 1017. A similar result was found for %P with a significant effect of genotype, day, and the genotype*gall interaction, but gall status and all other interactions were insignificant. Phosphorus also increased throughout the decomposition process, and %P also tended to be higher in un-galled litter from genotypes 11 and 3200. A slightly different pattern was shown for %C, which was influenced by genotype, gall status, day, and both the genotype*day and gall*day interactions. Carbon concentrations are higher in un-galled litter for genotypes 11 and 1017, but lower for un-galled litter from genotype 3200. Carbon concentrations decrease throughout the decomposition process, but %C decreases faster for genotype 1017 than the other two genotypes and %C also decreases faster for galled tissue than un-galled tissue.
3.2. Fungal Response to Galled Litter
Fungal biomass on leaf tissue post-incubation did not differ significantly by genotype, gall status, or their interaction (Table 2). Galled litter from genotype 1017, although it decomposed the fastest, did not show the highest concentrations of ergosterol on leaf tissue. Patterns in decomposition rates were not associated with patterns in fungal biomass accumulation. Fungal biomass was not correlated with decomposition rate constant (k) (Pearson’s r = −0.0699, p = 0.8954), or CT concentration (Pearson’s r = −0.4237, p = 0.4025). Aquatic fungal biomass was correlated with proportion of Fremont markers for galled tissue (r = −0.9951, p = 0.0166), but not un-galled tissue (r = 0.7645, p = 0.4460).
3.3. Macroinvertebrate Response to Galled Litter
Macroinvertebrate communities were significantly influenced by the gall and genotype litter treatments, but not necessarily in a manner consistent with chemistry and decomposition differences above. Macroinvertebrate communities on litter were significantly different between genotypes 11 and 3200 on harvest date 108 (Overall MRPP: A = 0.0937, p = 0.0458; 11 ≠ 3200: A = 0.1751, p = 0.0097; Figure 3), but did not differ on harvest dates 17 and 189 (results not shown). For one of these genotypes (11) macroinvertebrate communities also differed significantly between galled and un-galled litter at day 108 (11: A = 0.0486, p = 0.0405; Figure 4), even though the decomposition rates of galled and un-galled litter were indistinguishable for this genotype. When we compared galled to un-galled litter for genotypes 1017 and 3200, we found no significant differences in macroinvertebrate community structure (1017: A = −0.0335, p = 0.8058; 3200: A = −0.0391, p = 0.9198; Figure 4), even though the galled litter for genotype 1017 decomposed 27% faster than the un-galled litter.
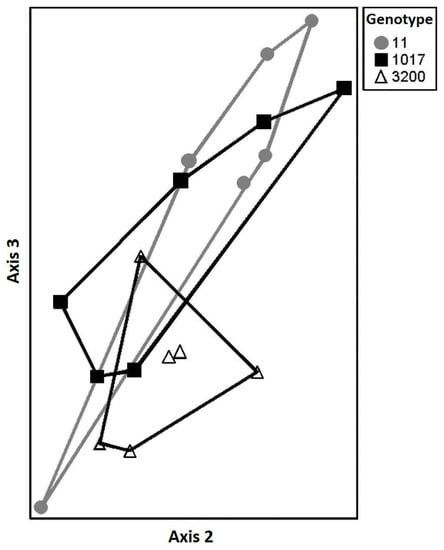
Figure 3.
Non-metric multidimensional scaling (NMDS) ordination showing two distinct macroinvertebrate communities among three backcross hybrid cottonwood genotypes on harvest date 107 (Overall MRPP: A = 0.0937, p = 0.0458; 11 ≠ 3200: A = 0.1751, p = 0.0097). Symbols represent entire macroinvertebrate communities on a given litterbag. Distance between symbols is a graphical representation of community similarity with points closer in space more similar than points further apart for genotype 11 (•), genotype 1017 (■) and genotype 3200 (∆).
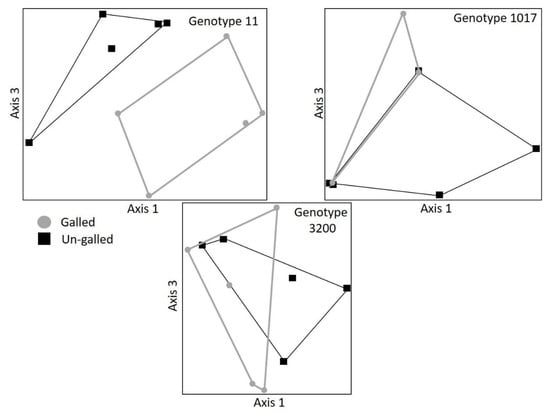
Figure 4.
NMDS ordinations showing a distinct difference macroinvertebrate community structure between galled and un-galled litter for one of three backcross hybrid cottonwood genotypes (MRPP results 11: A = 0.0486, p = 0.0405; 1017: A = −0.0335, p = 0.8058; 3200: A = −0.0391, p = 0.9198). Symbols represent entire macroinvertebrate communities on galled (•) versus un-galled (■) litterbags.
We found no differences in invertebrate species richness, evenness or diversity indices (H’ or D) among genotypes, nor between galled and un-galled litter at any harvest date, and there were also no significant genotype by gall status interactions (whole model p > 0.05). However, we did find significantly higher overall invertebrate abundances on litter from genotype 11 than that of genotype 3200 at harvest date 189 (F(2,26) = 3.49, p = 0.045). Examining these macroinvertebrate data at an individual taxon level, litter from genotype 11 on harvest date 108 was colonized by greater abundances of Chelifera sp. (Empididae: Diptera), Chironomidae (Diptera), Fossaria sp. (Gastropoda), and Oligochaeta than either genotype 1017 or 3200. Galled litter of all three genotypes was colonized by greater abundances of Nectopsyche sp. (Leptoceridae: Trichoptera) and Oecetis sp. (Leptoceridae: Trichoptera) than un-galled litter (Table S1). Although these taxon-specific differences were apparent, none of these taxa were determined to be indicator species for any one cottonwood genotype, nor for galled or un-galled litter overall.
4. Discussion
In forest ecosystems, tree genetics can have strong and complex effects on communities and ecosystems [29,44,45,46,47]. Our study furthers our understanding of these interactions by showing that differences among tree genotypes in terms of herbivory (galling) can have extended effects on communities and ecosystems [17] in a riparian forest stream system.
4.1. Herbivory and Ecosystem Function
Our data suggest gall-induced leaf condensed tannin concentrations may not be the only factor that leads to altered leaf decomposition. We found significant induction of condensed tannins for two genotypes, and although a third genotype did not induce tannins in galled litter, there was a significant acceleration of the decomposition process for galled tissue. The mechanism behind the accelerated decomposition of galled tissue is, as yet, unclear, but may be either structural or chemical.
Can herbivory or galling accelerate decomposition? A previous study on terrestrial herbivory and stream function found that second-flush leaves (after intense herbivory by the gypsy moth (Lymantria dispar L.)) decomposed faster than first-flush leaves [48] due to lower levels of percent fiber and lower fiber:protein ratios in second flush leaves of Chestnut oak (Quercus prinus L.). A similar study found that moose browsing of birch (Betula papyrifera Marsh.) resulted in higher quality second-flush litter and subsequent acceleration of decomposition [49]. Both of these previous studies confirm the herbivory-acceleration hypothesis [21,50]. Our results also partially confirm the herbivory-acceleration hypothesis [21,50], but only for one genotype (1017). In both previous studies discussed above, the trees produced a second flush of litter that was of higher quality than initial flushes [48,49]. In our case, the litter was modified by a leaf-galling aphid, and both chemical and structural changes were made in situ. There may be fundamental differences in the manner by which herbivores can affect ecosystem functions due to the pathways through which they alter organic matter inputs.
4.2. Genotype-Specific Responses
Previous work in this cottonwood system shows high levels of variability in both terrestrial and aquatic decomposition rates (k) among backcross cottonwood individuals [17,29,44,45], and in general, deceleration of terrestrial decomposition rates due to the effects of the galling aphid, P. betae, on specific backcross genotypes [17]. Pemphigus betae galling tended to induce condensed tannins, lignin and phosphorus concentrations which was associated with slower overall decomposition for four of five Populus genotypes in a previous terrestrial study [17]. For a different species of Pemphigus (P. spirotheceae), galls tended to increase phenolic content and C:N ratio, leading to slower terrestrial mass loss for Populus nigra var. italica [51]. Our results are also variable and show chemical induction (genotypes 11 and 3200), altered decomposition rates (genotype 1017), and responses in the decomposer community (genotype 11). In concert with previous studies, variability in our results demonstrates a lack of predictability for gall influences on in-stream communities and ecosystem processes, at this point in time.
Considering galling herbivores to be part of the biotic environment of a plant allows us to place our results in a G × E (genotype × environment) interaction context. Previous studies have found strong G × E effects on both terrestrial and aquatic ecosystems. For example, in previous work with Populus tremuloides under low and high nutrient treatments, there were significant influences of genotype, nutrient status, and their interactions on terrestrial mass remaining, nitrogen, and condensed tannins in leaves [52] and both C:N and aquatic decomposition rates [53]. Our results provide evidence for another genotype-mediated ecological interaction with herbivores that crosses ecosystem boundaries.
4.3. Herbivory and Higher Trophic Levels
Aquatic fungi are thought to be generalist detritivores in stream systems that sometimes discriminate among litter species [54,55], but we only found a tight correlation between aquatic fungal biomass and the proportion of Fremont markers in galled leaf litter. Because we did not find strong correlations between fungal biomass and any measured phytochemicals, we assume that other genetically-controlled traits may be affecting fungal biomass in tandem with phytochemicals. Although the mechanisms are unclear, we hope this study provides a foundation upon which further examination of this interesting interaction between galling-herbivores and fungal detritivores can build.
Macroinvertebrate communities, which have shown weak discriminatory ability among genotypes of cottonwoods in previous studies [29,45], seem influenced by both genotype and gall-status, to a small degree in this study. For one genotype (11) that induces condensed tannins in galled tissue, macroinvertebrate communities on galled litter differed when compared to communities found on un-galled litter. A third genotype (1017), which showed no difference in macroinvertebrate colonization between galled versus un-galled tissue, showed a significant difference in decomposition rate, with galled tissue decomposing 27% faster than un-galled tissue. Sampling a greater number of genotypes and an ability to focus on clear mechanisms for macroinvertebrate discrimination would improve this study. Regardless, it appears that macroinvertebrate communities are influenced to some degree by these litter treatments, and the multiple trophic links between leaf litter and these decomposers does not result in an attenuation of the genetic or galling effects as we predicted. However, none of the single invertebrate taxa that were associated with a specific genotype or galled litter are in the shredder feeding guild, nor hypothesized to be tightly associated with leaf litter inputs. Accordingly, these species-level statistical results may not be biologically relevant, should be interpreted with caution.
Overall, experiment-wide correlations between induced chemistry, decomposition, fungal biomass and proportion of Fremont markers were not detectably different from zero. When galled and un-galled litters were separated for analysis, our sample sizes (for Populus genotypes) were often too small to see significant correlations, even when our correlations were strong. Future work should include more clonal replicates of each Populus genotype, more gall-susceptible genotypes, larger litter collections, and a separation of litter collections by clonal individual to increase replication at the genotype level, which would better determine whether galls accelerate or decelerate litter decomposition. The main difficulty facing increased sample sizes is in collecting enough galled leaf litter for these studies. We chose three Populus genotypes that were heavily influenced by galling aphids, but the majority of leaves collected were still un-galled. Future studies would benefit from increased sample sizes for all analyses, and our results should be interpreted in this light.
5. Conclusions
This study demonstrates the potential cross-ecosystem effects gall-forming herbivores may have on adjacent stream ecosystems. Overall, our results show that the potential effects of galling aphids on aquatic ecosystem and community processes may be genotype specific, just as susceptibility to aphid herbivory is genotype specific. Extrapolating these results to a landscape level could result in a genotypic-geographic mosaic [56] of genetically based interactions that could provide dynamic inputs to both terrestrial and aquatic systems. Future work could also examine the effects of other interacting arthropod herbivores that may alter leaf structure and chemistry through different pathways in order to determine the effects of chronic or outbreak herbivory events on linked forests and streams.
Supplementary Materials
The following are available online at https://www.mdpi.com/1999-4907/11/2/182/s1, Table S1: Total aquatic macroinvertebrate abundance, taxa richness, and abundances by taxon for three hybrid Populus genotypes (3200, 11, 1017) and for galled (Pemphigus betae aphid) and ungalled leaf tissues for each.
Author Contributions
Conceptualization, C.J.L. and J.A.S.; methodology, C.J.L. and J.A.S.; fieldwork, C.J.L., J.A.S., J.K.B., and D.F.; data curation, C.J.L.; statistical analysis, C.J.L., J.K.B., and D.F..; writing—original draft preparation, C.J.L.; writing—review and editing, J.A.S., J.K.B., D.F., and C.J.L.; visualization, C.J.L. and D.F. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Science Foundation, grant numbers DEB-1836387, DEB-0130487, IRCEB-0078280, and FIBR-0425908.
Acknowledgments
We would like to thank the Ogden Nature Center (UT) for cooperation with this research and for help maintaining our common gardens. Members of the Marks, Whitham, Hart, and Hungate labs at NAU provided crucial comment on this research at early stages: specifically, S. C. Hart, D. Guido, R. K. Bangert, G. M. Wimp, G. A. Haden, E. C. Dinger and Z. G. Compson. Finally, this work would not have been possible without a host of graduate and undergraduate researchers who assisted with field and lab work: K. Kolanoski, K. Pearson, A. Martinez, J. Moan, E. Mester, E. Yazzie, D. Jamieson, C. Bartlett, M. Stritar, and A. Posey.
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Holland, E.A.; Detling, J.K. Plant response to herbivory and belowground nitrogen cycling. Ecology 1990, 71, 1040–1049. [Google Scholar] [CrossRef]
- Pastor, J.; Dewey, B.; Naiman, R.J.; McInnes, P.F.; Cohen, Y. Moose browsing and soil fertility in the boreal forests of Isle Royale National Park. Ecology 1993, 74, 467–480. [Google Scholar] [CrossRef]
- Sirotnak, J.M.; Huntly, N.J. Direct and indirect effects of herbivores on nitrogen dynamics: Voles in riparian areas. Ecology 2000, 81, 78–87. [Google Scholar] [CrossRef]
- Stark, S.; Wardle, D.A.; Ohtonen, R.; Helle, T.; Yeates, G.W. The effect of reindeer grazing on decomposition, mineralization and soil biota in a dry oligotrophic Scots pine forest. Oikos 2000, 90, 301–310. [Google Scholar] [CrossRef]
- Wardle, D.A.; Bonner, K.I.; Barker, G.M. Linkages between plant litter decomposition, litter quality, and vegetation responses to herbivores. Funct. Ecol. 2002, 16, 585–595. [Google Scholar] [CrossRef]
- Faeth, S.H.; Connor, E.F.; Simberloff, D.S. Early leaf abscission: A neglected source of mortality for folivores. Am. Nat. 1981, 117, 409–415. [Google Scholar] [CrossRef]
- Hunter, M.D. Insect population dynamics meets ecosystem ecology: Effects of herbivory on soil nutrient dynamics. Agric. Forest Entomol. 2001, 3, 77–84. [Google Scholar] [CrossRef]
- Risley, L.S. The influence of herbivores on seasonal leaf-fall: Premature leaf abscission and petiole clipping. J. Agric. Entomol. 1986, 3, 152–162. [Google Scholar] [CrossRef]
- Tuomi, J.; Niemelae, P.; Haukioja, E.; Siren, S.; Neuvonen, S. Nutrient stress: An explanation for plant anti-herbivore responses to defoliation. Oecologia 1984, 61, 208–210. [Google Scholar] [CrossRef]
- Seastedt, T.R.; Ramundo, R.A.; Hayes, D.C. Maximization of densities of soil animals by foliage herbivory: Empirical evidence, graphical and conceptual models. Oikos 1988, 51, 243–248. [Google Scholar] [CrossRef]
- Tuomi, J.; Niemelae, P.; Siren, S. The Panglossian paradigm and delayed inducible accumulation of foliar phenolics in mountain birch. Oikos 1990, 59, 399–410. [Google Scholar] [CrossRef]
- Merrill, E.H.; Stanton, N.L.; Hak, J.C. Responses of bluebunch wheatgrass, Idaho fescue, and nematodes to ungulate grazing in Yellowstone National Park. Oikos 1994, 69, 231–240. [Google Scholar] [CrossRef]
- Findlay, S.; Carreiro, M.; Krischic, V.; Jones, C.J. Effects of damage to living plants on leaf litter quality. Ecol. Appl. 1996, 6, 269–275. [Google Scholar] [CrossRef]
- Bardgett, R.D.; Wardle, D.A.; Yeates, G.W. Linking above-ground and below-ground interactions: How plant responses to foliar herbivory influence soil organisms. Soil Biol. Biochem. 1998, 30, 1867–1878. [Google Scholar] [CrossRef]
- Grime, J.P.; Cornelissen, J.H.C.; Thompson, K.; Hodgson, J.G. Evidence of a causal connection between anti-herbivore defense and the decomposition rate of leaves. Oikos 1996, 77, 489–494. [Google Scholar] [CrossRef]
- Cornelissen, J.H.C.; Pérez-Harguindeguy, N.; Díaz, S.; Grime, J.P.; Marzano, B.; Cabido, M.; Vendramini, F.; Cerabolini, B. Leaf structure and defence control litter decomposition rate across species and life forms in regional floras on two continents. New Phytol. 1999, 143, 191–200. [Google Scholar] [CrossRef]
- Schweitzer, J.A.; Bailey, J.K.; Hart, S.C.; Wimp, G.M.; Chapman, S.K.; Whitham, T.G. The interaction of plant genotype and herbivory decelerate leaf litter decomposition and alter nutrient dynamics. Oikos 2005, 110, 133–145. [Google Scholar] [CrossRef]
- Schowalter, T.D.; Sabin, T.E.; Stafford, S.G.; Sexton, J.M. Phytophage effects on primary production, nutrient turnover, and litter decomposition of young Douglas-fir in western Oregon. Forest Ecol. Manag. 1991, 42, 229–243. [Google Scholar] [CrossRef]
- Reynolds, B.C.; Hunter, M.D. Responses of soil respiration, soil nutrients, and litter decomposition to inputs from canopy herbivores. Soil Biol. Biochem. 2001, 33, 1641–1652. [Google Scholar] [CrossRef]
- Choudhury, D. Herbivore induced changes in leaf-litter resource quality: A neglected aspect of herbivory in ecosystem nutrient dynamics. Oikos 1988, 51, 389–393. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Tilman, D.; Knops, J.M.H. Herbivore effects on plant and nitrogen dynamics in oak savanna. Ecology 1998, 79, 165–177. [Google Scholar] [CrossRef]
- McNaughton, S.J.; Oesterheld, M.; Frank, D.A.; Williams, K.J. Ecosystem-level patterns of primary productivity and herbivory in terrestrial habitats. Nature 1989, 341, 142–144. [Google Scholar] [CrossRef]
- Belovsky, G.E.; Slade, J.B. Insect herbivory accelerates nutrient cycling and increases plant production. Proc. Natl. Acad. Sci. USA 2000, 97, 14412–14417. [Google Scholar] [CrossRef] [PubMed]
- Frost, C.J.; Dean, J.M.; Smyers, E.C.; Mescher, M.C.; Carlson, J.E.; De Moraes, C.M.; Tooker, J.F. A petiole-galling insect herbivore decelerates leaf lamina decomposition rates. Funct. Ecol. 2012, 26, 628–636. [Google Scholar] [CrossRef]
- Moran, N.A.; Whitham, T.G. Differential colonization of resistant and susceptible host plants: Pemphigus and Populus. Ecology 1990, 71, 1059–1067. [Google Scholar] [CrossRef]
- Larson, K.C.; Whitham, T.G. Competition between gall aphids and natural plant sinks: Plant architecture affects resistance to galling. Oecologia 1997, 109, 575–582. [Google Scholar] [CrossRef] [PubMed]
- Bailey, J.K.; Wooley, S.C.; Lindroth, R.L.; Whitham, T.G. Importance of species interactions to community heritability: A genetic basis to trophic-level interactions. Ecol. Lett. 2006, 9, 78–85. [Google Scholar] [CrossRef]
- Keith, A.R.; Bailey, J.K.; Lau, M.K.; Whitham, T.G. Genetics-based interactions of foundation species affect community diversity, stability and network structure. Proc. Biol. Sci. 2017, 284, 20162703. [Google Scholar] [CrossRef]
- LeRoy, C.J.; Whitham, T.G.; Keim, P.; Marks, J.C. Plant genes link forests and streams. Ecology 2006, 87, 255–261. [Google Scholar] [CrossRef] [PubMed]
- Keim, P.; Paige, K.N.; Whitham, T.G.; Lark, K.G. Genetic analysis of an interspecific hybrid swarm of Populus: Occurrence of unidirectional introgression. Genetics 1989, 123, 557–565. [Google Scholar]
- Martinsen, G.D.; Whitham, T.G.; Turek, R.J.; Keim, P. Hybrid populations selectively filter gene introgression between species. Evolution 2001, 55, 1325–1335. [Google Scholar] [CrossRef]
- Strahler, A.N. Quantitative analysis of watershed geomorphology. Trans. Am. Geophys. Union 1957, 38, 913–920. [Google Scholar] [CrossRef]
- Porter, L.J.; Hrstich, L.N.; Chan, B.C. The conversion of procyanidins and prodelphinidins to cyanidin and delphinidin. Phytochemistry 1986, 25, 223–230. [Google Scholar] [CrossRef]
- Hagerman, A.E.; Butler, L.G. Choosing appropriate methods and standards for assaying tannin. J. Chem. Ecol. 1989, 15, 1795–1810. [Google Scholar] [CrossRef] [PubMed]
- Parkinson, J.A.; Allen, S.E. A wet oxidation procedure suitable for the determination of nitrogen and mineral nutrients in biological material. Commun. Soil Sci. Plan. 1975, 6, 1–11. [Google Scholar] [CrossRef]
- Suberkropp, K. Fungal growth, production, and sporulation during leaf decomposition in two streams. Appl. Environ. Microb. 2001, 67, 5063–5068. [Google Scholar] [CrossRef]
- Gessner, M.O.; Chauvet, E. Ergosterol-to-biomass conversion factors for aquatic hyphomycetes. Appl. Environ. Microbiol. 1993, 59, 502–507. [Google Scholar] [CrossRef]
- Merritt, R.W.; Cummins, K.W. An Introduction to the Aquatic Insects of North America, 3rd ed.; Kendall-Hunt Publishing Company: Dubuque, IA, USA, 1996. [Google Scholar]
- Wiggins, G.B. Larvae of the North American caddisfly genera (Trichoptera), 2nd ed.; University of Toronto Press: Toronto, ON, Canada, 1996. [Google Scholar]
- Thorpe, J.H.; Covich, A.P. Ecology and Classification of North American Freshwater Invertebrates, 2nd ed.; Academic Press: San Diego, CA, USA, 2001. [Google Scholar]
- Jenny, H.; Gessel, S.P.; Bingham, F.T. Comparative study of decomposition rates of organic matter in temperate and tropical regions. Soil Sci. 1949, 68, 419–432. [Google Scholar] [CrossRef]
- Webster, J.R.; Benfield, E.F. Vascular plant breakdown in freshwater ecosystems. Annu. Rev. Ecol. Syst. 1986, 17, 567–594. [Google Scholar] [CrossRef]
- Swan, C.M.; Palmer, M.A. Leaf diversity alters litter breakdown in a Piedmont stream. J. N. Am. Benthol. Soc. 2004, 23, 15–28. [Google Scholar] [CrossRef]
- Schweitzer, J.A.; Bailey, J.K.; Rehill, B.J.; Martinsen, G.D.; Hart, S.C.; Lindroth, R.L.; Keim, P.; Whitham, T.G. Genetically based trait in a dominant tree affects ecosystem processes. Ecol. Lett. 2004, 7, 127–134. [Google Scholar] [CrossRef]
- LeRoy, C.J.; Whitham, T.G.; Keim, P.; Marks, J.C. Within-species variation in foliar chemistry influences leaf-litter decomposition in a Utah river. J. N. Am. Benthol. Soc. 2007, 26, 426–438. [Google Scholar] [CrossRef]
- Whitham, T.G.; Bailey, J.K.; Schweitzer, J.A.; Shuster, S.M.; Bangert, R.K.; LeRoy, C.J.; Lonsdorf, E.V.; Allan, G.J.; DiFazio, S.P.; Potts, B.M.; et al. A framework for community and ecosystem genetics: From genes to ecosystems. Nat. Rev. Genet. 2006, 7, 510–523. [Google Scholar] [CrossRef] [PubMed]
- Jackrel, S.L.; Morton, T.C.; Woottan, J.T. Intraspecific leaf chemistry drives locally accelerated ecosystem function in aquatic and terrestrial communities. Ecology 2016, 97, 2125–2135. [Google Scholar] [CrossRef]
- Hutchens, J.J.; Benfield, E.F. Effects of forest defoliation by the gypsy moth on detritus processing in southern Appalachian streams. Am. Midl. Nat. 2000, 143, 397–404. [Google Scholar] [CrossRef]
- Irons, J.G., III; Bryant, J.P.; Oswood, M.W. Effects of moose browsing on decomposition rates of birch leaf litter in a subarctic stream. Can. J. Fish. Aquat. Sci. 1991, 48, 442–444. [Google Scholar] [CrossRef]
- Chapman, S.K.; Hart, S.C.; Cobb, N.S.; Whitham, T.G.; Koch, G.W. Insect herbivory increases litter quality and decomposition: An extension of the acceleration hypothesis. Ecology 2003, 84, 2867–2876. [Google Scholar] [CrossRef]
- Kunkler, N.; Brandl, R.; Brandle, M. Changes in clonal poplar leaf chemistry caused by stem galls alter herbivory and leaf litter decomposition. PLoS ONE 2013, 8, e79994. [Google Scholar] [CrossRef]
- Madritch, M.J.; Donaldson, J.R.; Lindroth, R.L. Genetic identity of Populus tremuloides litter influences decomposition and nutrient release in a mixed forest stand. Ecosystems 2006, 9, 528–537. [Google Scholar] [CrossRef]
- LeRoy, C.J.; Wooley, S.C.; Lindroth, R.L. Genotype and soil nutrient environment influence aspen litter chemistry and in-stream decomposition. Freshw. Sci. 2012, 31, 1244–1253. [Google Scholar] [CrossRef]
- Bärlocher, F.; Kendrick, B. Dynamics of the fungal population on leaves in a stream. J. Ecol. 1974, 62, 761–791. [Google Scholar] [CrossRef]
- Bärlocher, F.; Graça, M.A.S. Exotic riparian vegetation lowers fungal diversity but not leaf decomposition in Portuguese streams. Freshwat. Biol. 2002, 47, 1123–1135. [Google Scholar] [CrossRef]
- Thompson, J.N. Specific hypotheses on the geographic mosaic of coevolution. Am. Nat. 1999, 153, S1–S14. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).