Genome-Wide Characterization of AspATs in Populus: Gene Expression Variation and Enzyme Activities in Response to Nitrogen Perturbations
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Cultivation and N Treatments
2.2. Sequence Available and Identification of AspATs
2.3. Phylogenetic Analysis and Calculation of Ka/Ks
2.4. Genomic Distribution, Gene Structure, Protein Motif, and Cis-Regulatory Element
2.5. Transcriptomic Sequencing and Expression Analysis
2.6. In-Gel Activity of AspAT Enzymes
3. Results
3.1. Genome-Wide Identification of AspATs in Populus and Other Plant Species
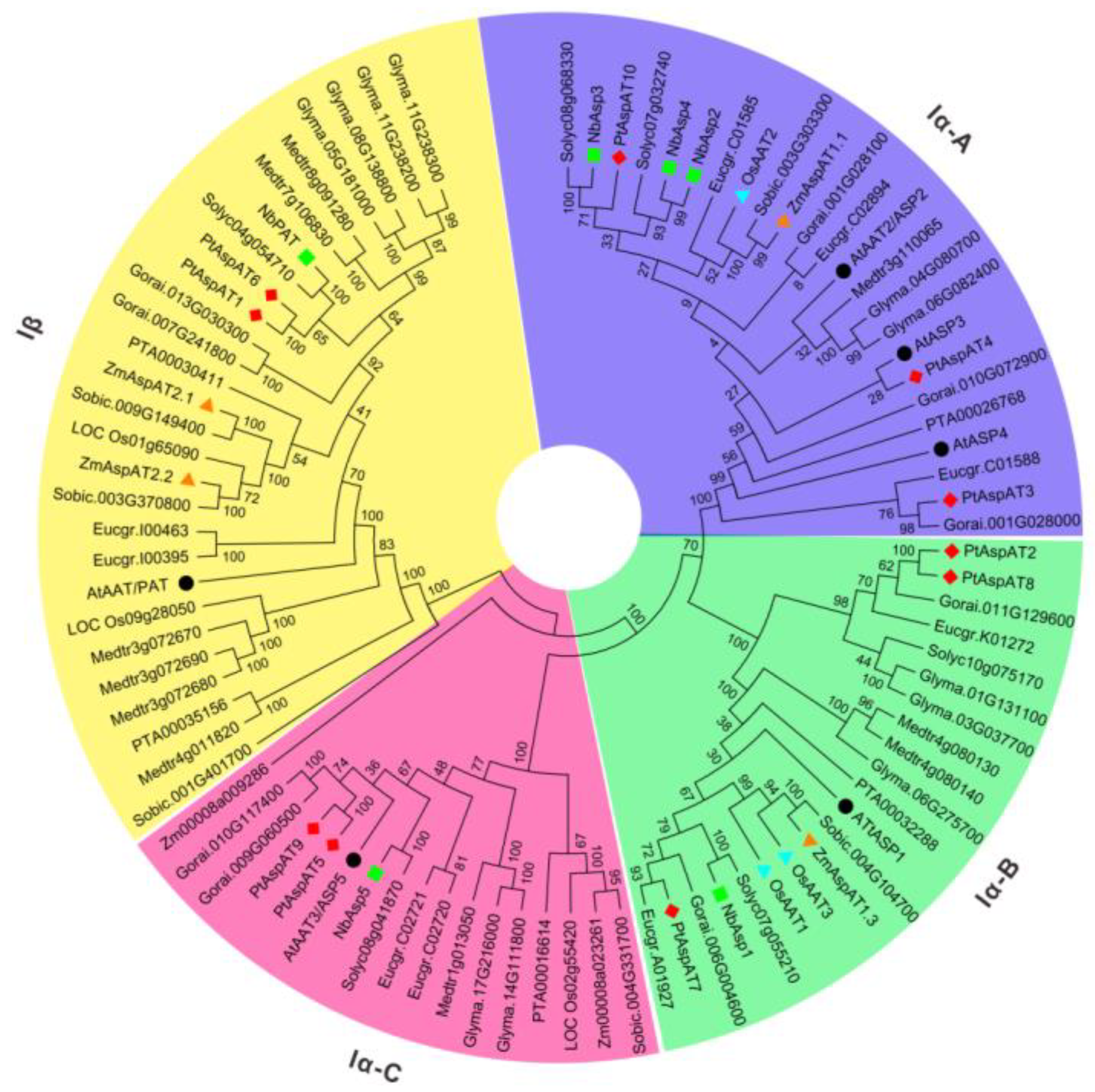
3.2. Evolutionary Relationships of AspATs between Populus and Other Species
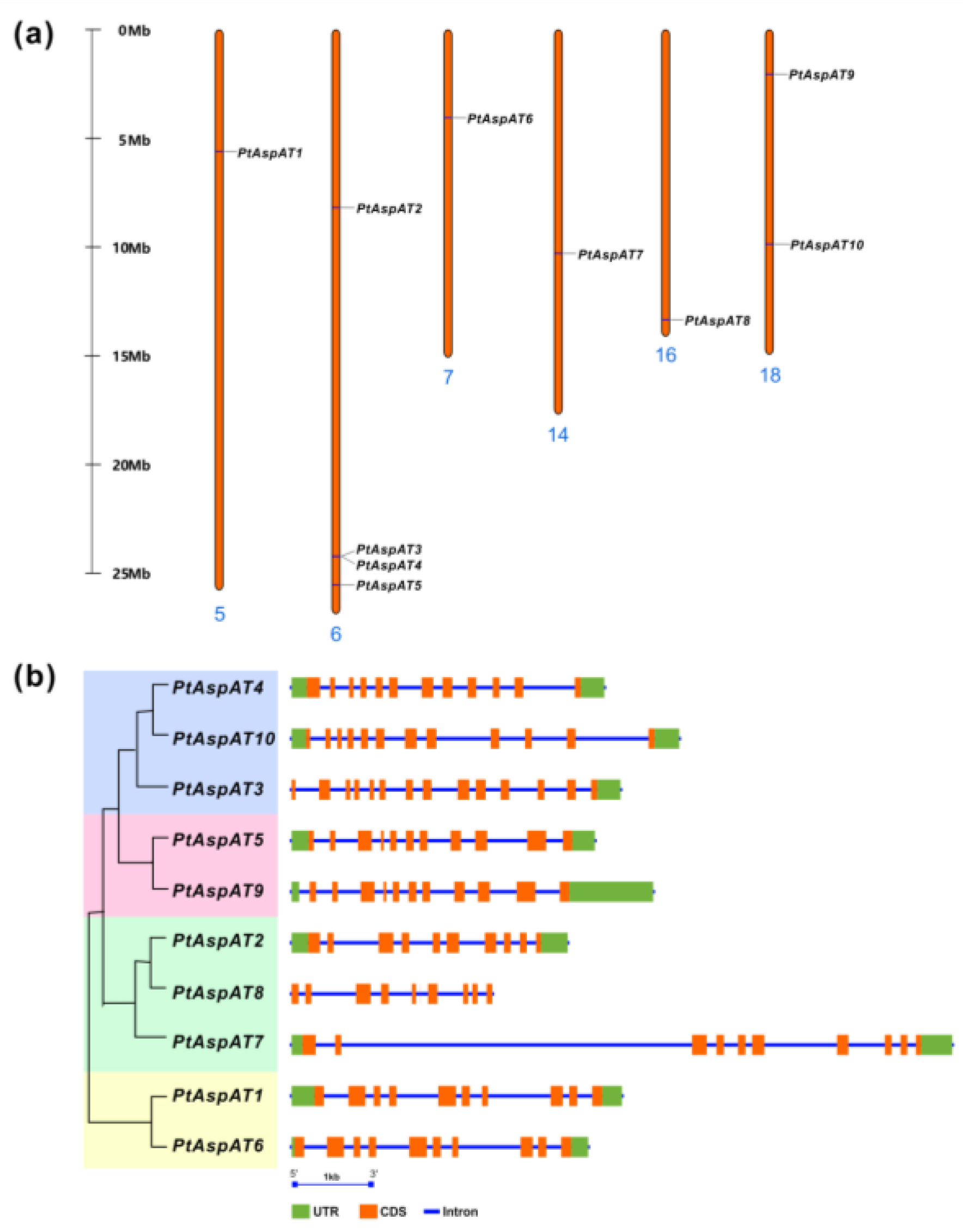
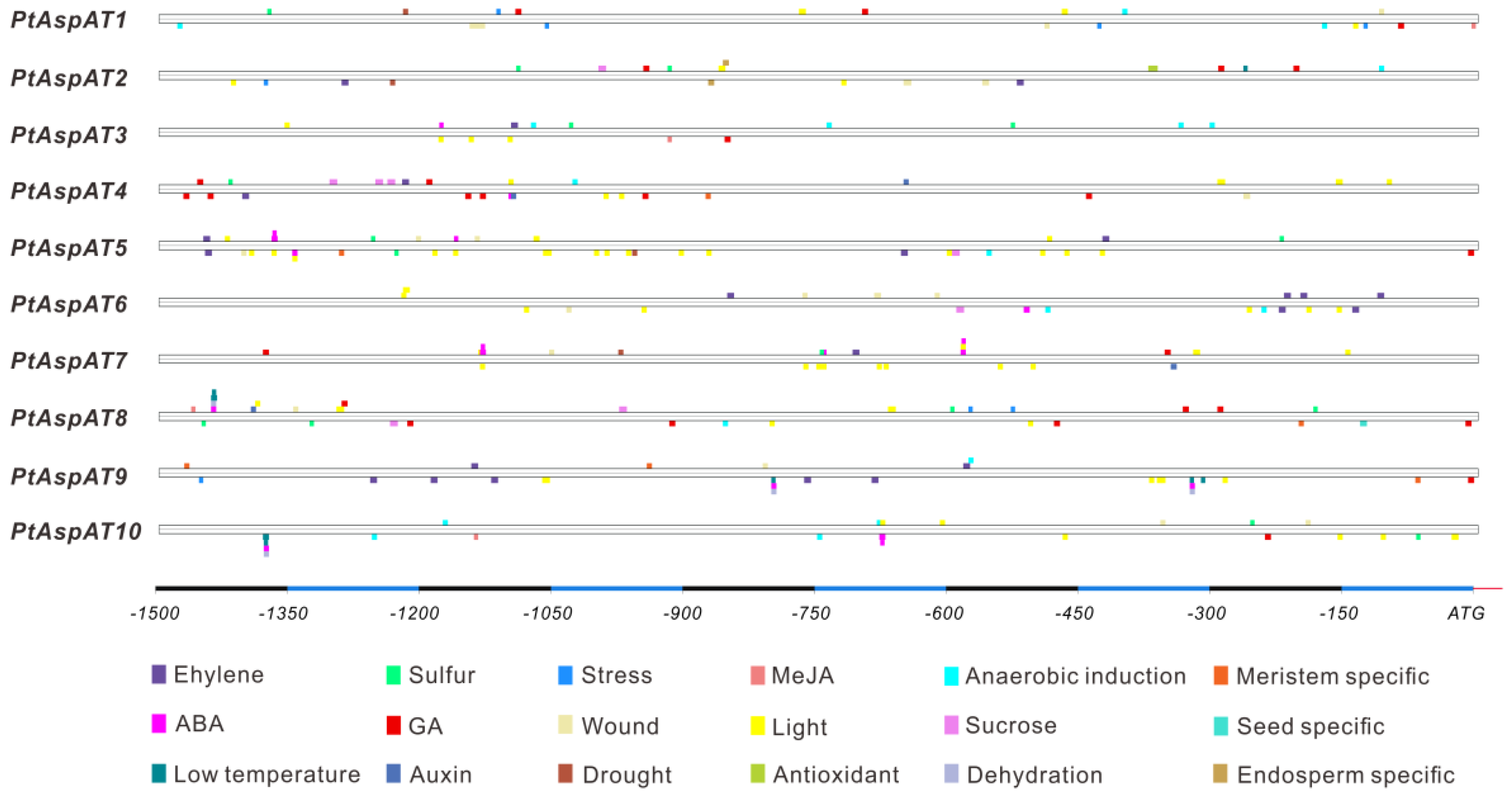
3.3. Chromosomal Location, Genomic Structure, and Cis-Elements
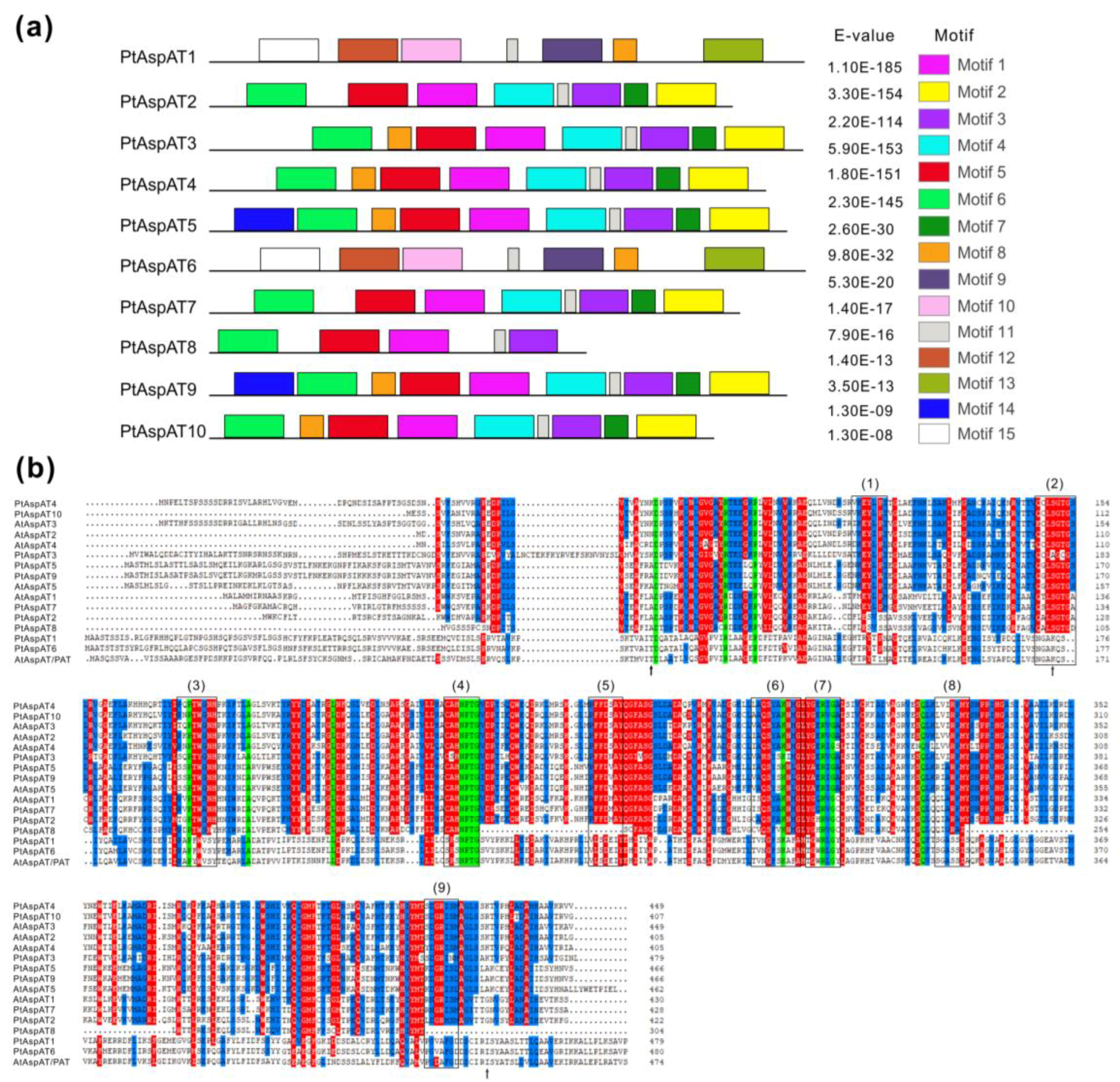
3.4. Conserved Motif and Domain of PtAspAT Proteins
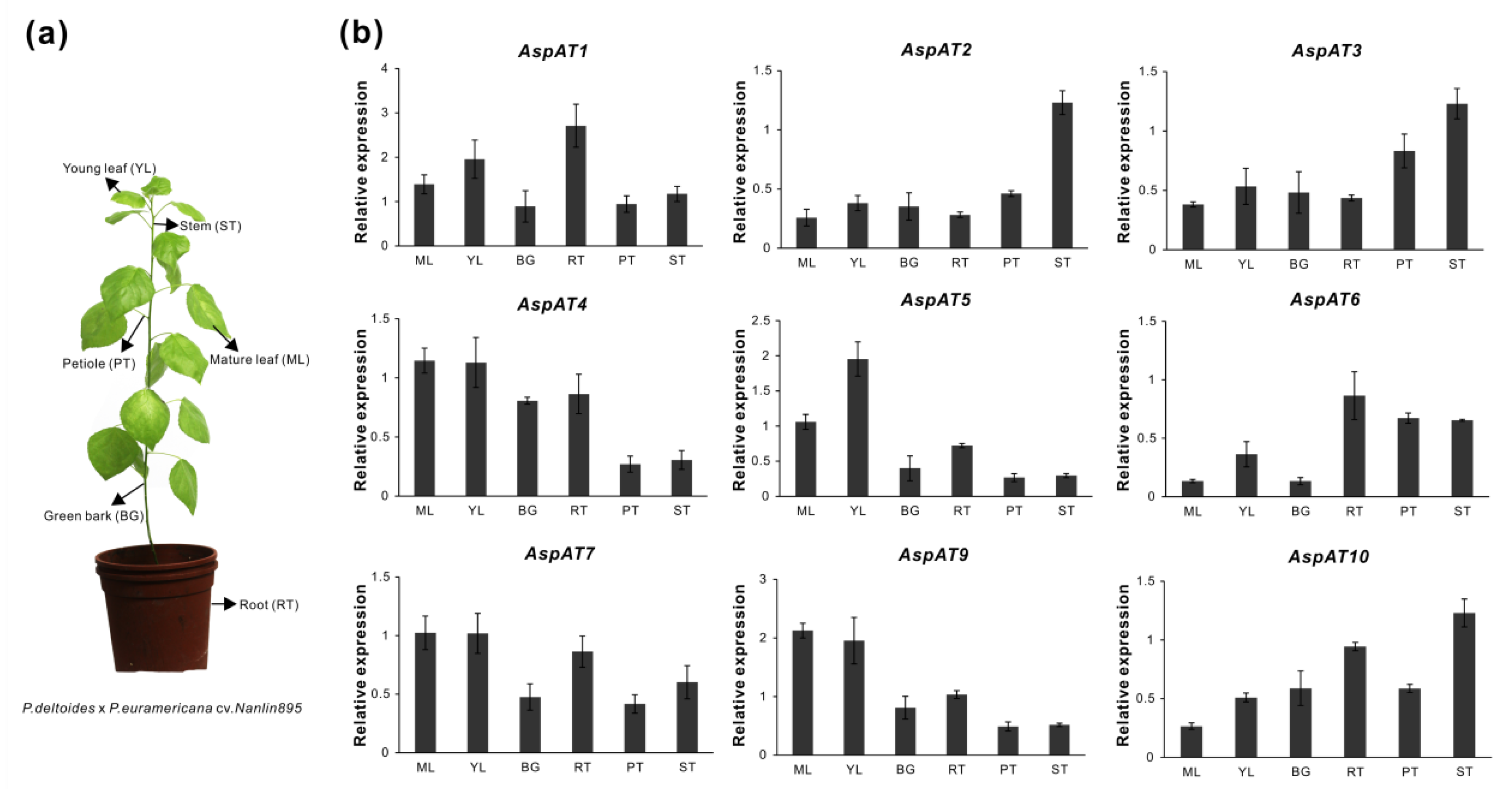
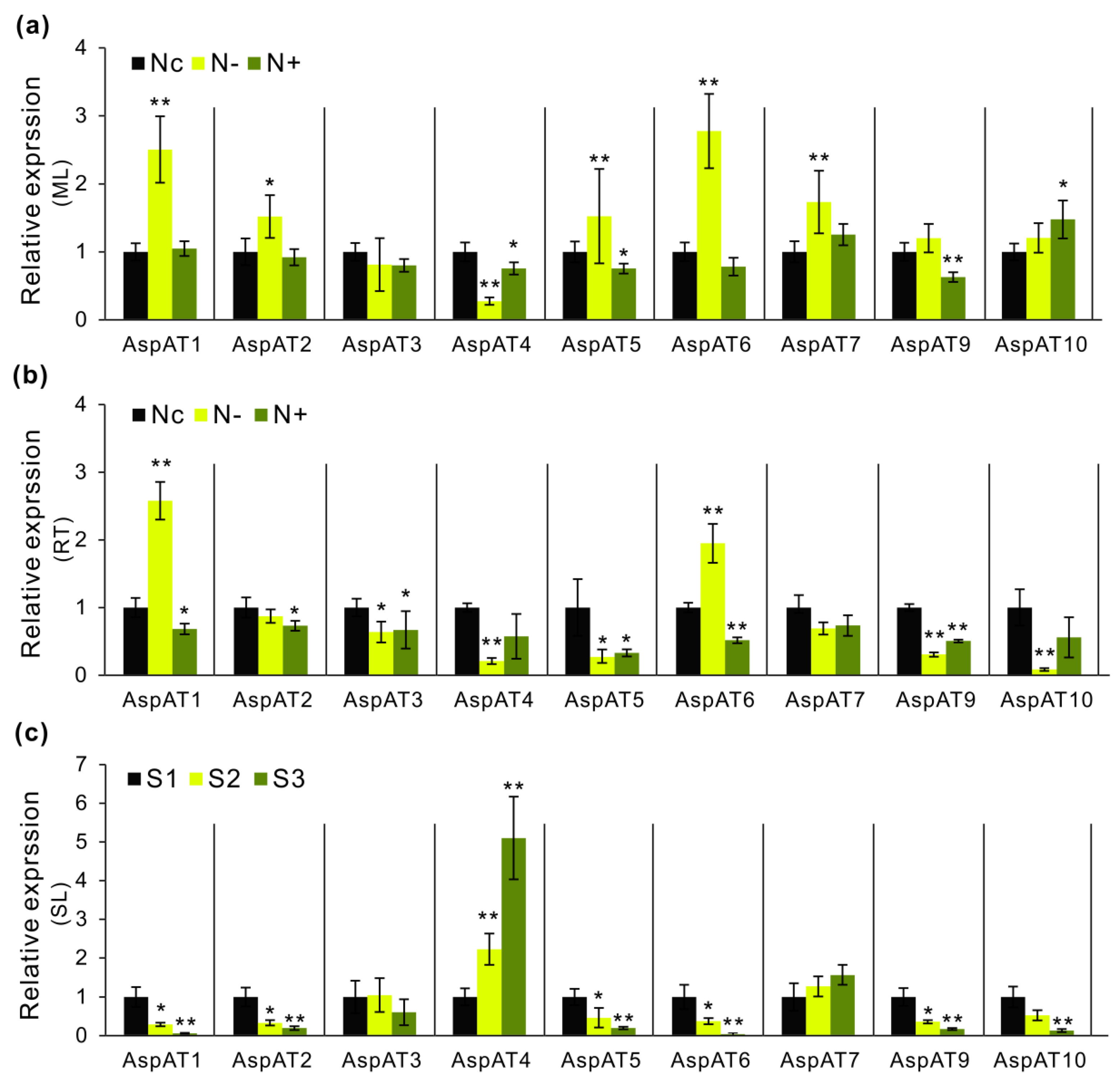
3.5. Expression Profilings of PtAspATs in Various Tissues, and in Response to N Feeding
3.6. Expression Evaluation of AspATs in Poplar Clone ‘Nanlin895’
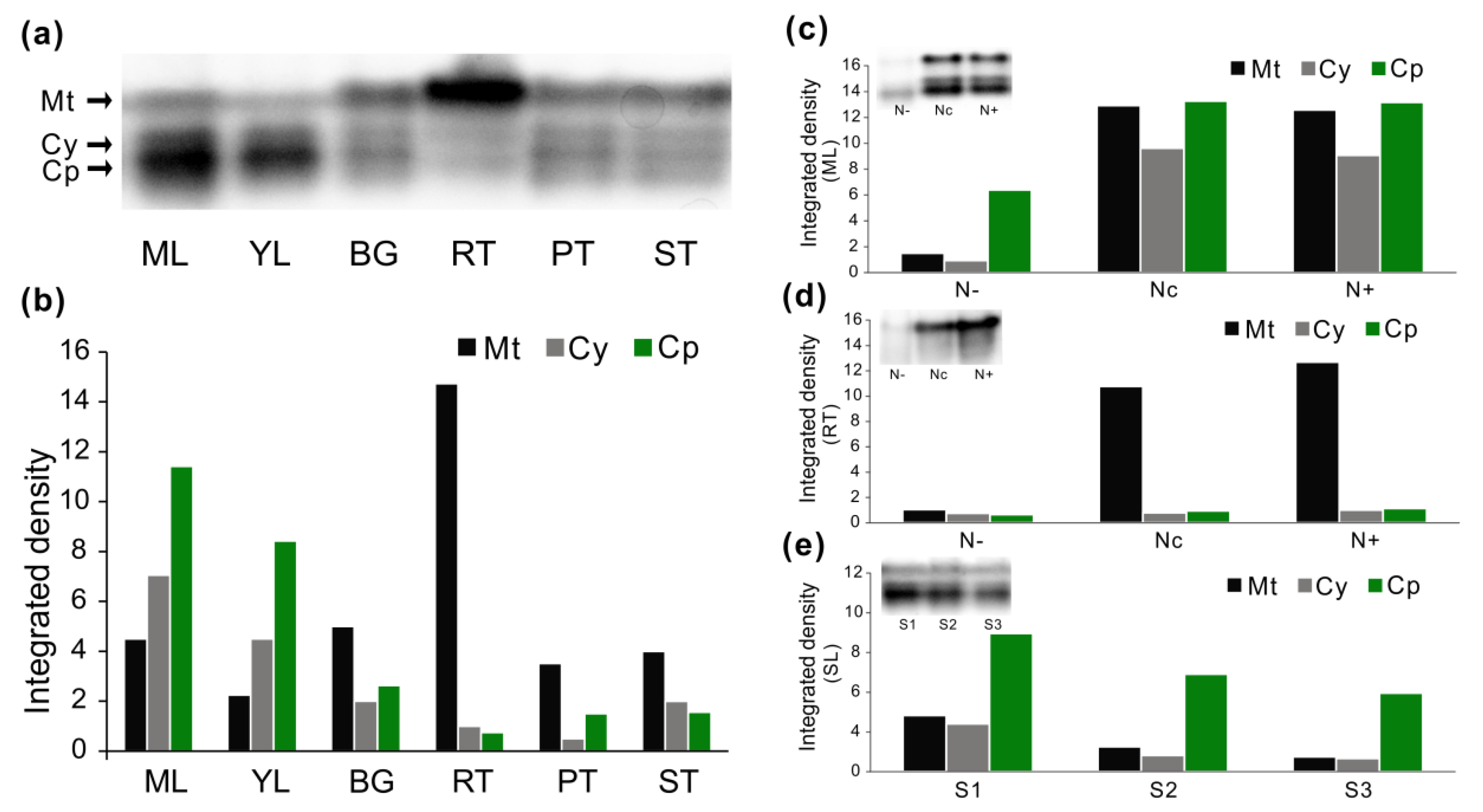
3.7. Enzyme Activities in Native Gels
4. Discussion
4.1. AspAT Gene Family in Populus
4.2. Mining Expression Profiles of AspATs in Tissues of Different Poplar Clone
4.3. Modulation of AspAT Transcripts and Activities upon N Alteration
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Schultz, C.J.; Coruzzi, G.M. The aspartate aminotransferase gene family of Arabidopsis encodes isoenzymes localized to three distinct subcellular compartments. Plant J. 1995, 7, 61–75. [Google Scholar] [CrossRef] [PubMed]
- De la Torre, F.; Cañas, R.A.; Pascual, M.B.; Avila, C.; Cánovas, F.M. Plastidic aspartate aminotransferases and the biosynthesis of essential amino acids in plants. J. Exp. Bot. 2014, 65, 5527–5534. [Google Scholar] [CrossRef]
- De la Torre, F.; El-Azaz, J.; Avila, C.; Cánovas, F.M. Deciphering the role of aspartate and prephenate aminotransferase activities in plastid nitrogen metabolism. Plant Physiol. 2014, 164, 92–104. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Xu, M.; Wang, G.; Galili, G. New insights into the metabolism of aspartate-family amino acids in plant seeds. Plant Reprod. 2018, 31, 203–211. [Google Scholar] [CrossRef]
- Zhou, Y.; Cai, H.; Xiao, J.; Li, X.; Zhang, Q.; Lian, X. Over-expression of aspartate aminotransferase genes in rice resulted in altered nitrogen metabolism and increased amino acid content in seeds. Theor. Appl. Genet. 2009, 118, 1381–1390. [Google Scholar] [CrossRef] [PubMed]
- Wadsworth, G.J. The plant aspartate aminotransferase gene family. Physiol. Plant. 1997, 100, 998–1006. [Google Scholar] [CrossRef]
- Miesak, B.; Coruzzi, G.M. Molecular and Physiological Analysis of Arabidopsis Mutants Defective in Cytosolic or Chloroplastic. Plant Physiol. 2002, 129, 650–660. [Google Scholar] [CrossRef]
- Campos, F.; Sobrino, T.; Ramos-Cabrer, P.; Castillo, J. Oxaloacetate: A novel neuroprotective for acute ischemic stroke. Int. J. Biochem. Cell Biol. 2012, 44, 262–265. [Google Scholar] [CrossRef]
- Zdzisińska, B.; Żurek, A.; Kandefer-Szerszeń, M. Alpha-Ketoglutarate as a Molecule with Pleiotropic Activity: Well-Known and Novel Possibilities of Therapeutic Use. Arch. Immunol. Ther. Exp. (Warsz). 2017, 65, 21–36. [Google Scholar] [CrossRef]
- Lin, A.P.; Abbas, S.; Kim, S.W.; Ortega, M.; Bouamar, H.; Escobedo, Y.; Varadarajan, P.; Qin, Y.; Sudderth, J.; Schulz, E.; et al. D2HGDH regulates alpha-ketoglutarate levels and dioxygenase function by modulating IDH2. Nat. Commun. 2015, 6, 1–14. [Google Scholar] [CrossRef]
- De la Torre, F.; Moya-García, A.A.; Suárez, M.-F.; Rodríguez-Caso, C.; Cañas, R.A.; Sánchez-Jiménez, F.; Cánovas, F.M. Molecular modeling and site-directed mutagenesis reveal essential residues for catalysis in a prokaryote-type aspartate aminotransferase. Plant Physiol. 2009, 149, 1648–1660. [Google Scholar] [CrossRef] [PubMed]
- De la Torre, F.; Suarez, M.F.; Santis, L.D.; Canovas, F.M. The aspartate aminotransferase family in conifers: Biochemical analysis of a prokaryotic-type enzyme from maritime pine. Tree Physiol. 2007, 27, 1283–1291. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, S.; Ito, M.; Nishida, I.; Watanabe, A. Isolation and RNA gel blot analysis of genes that could serve as potential molecular markers for leaf senescence in Arabidopsis thaliana. Plant Cell Physiol. 2001, 42, 170–178. [Google Scholar] [CrossRef] [PubMed]
- De la Torre, F.; De Santis, L.; Suárez, M.F.; Crespillo, R.; Cánovas, F.M. Identification and functional analysis of a prokaryotic-type aspartate aminotransferase: Implications for plant amino acid metabolism. Plant J. 2006, 46, 414–425. [Google Scholar] [CrossRef] [PubMed]
- Cañas, R.A.; Quilleré, I.; Christ, A.; Hirel, B. Nitrogen metabolism in the developing ear of maize (Zea mays): Analysis of two lines contrasting in their mode of nitrogen management. New Phytol. 2009, 184, 340–352. [Google Scholar] [CrossRef]
- Plett, D.; Holtham, L.; Baumann, U.; Kalashyan, E.; Francis, K.; Enju, A.; Toubia, J.; Roessner, U.; Bacic, A.; Rafalski, A.; et al. Nitrogen assimilation system in maize is regulated by developmental and tissue-specific mechanisms. Plant Mol. Biol. 2016, 92, 293–312. [Google Scholar] [CrossRef] [PubMed]
- Pick, T.R.; Brautigam, A.; Schluter, U.; Denton, A.K.; Colmsee, C.; Scholz, U.; Fahnenstich, H.; Pieruschka, R.; Rascher, U.; Sonnewald, U.; et al. Systems Analysis of a Maize Leaf Developmental Gradient Redefines the Current C4 Model and Provides Candidates for Regulation. Plant Cell 2011, 23, 4208–4220. [Google Scholar] [CrossRef] [PubMed]
- Nomura, M.; Higuchi, T.; Katayama, K.; Taniguchi, M.; Miyao-tokutomi, M.; Matsuoka, M.; Tajima, S. The Promoter for C 4 -type Mitochondrial Aspartate Aminotransferase Does not Direct Bundle Sheath-specific Expression in Transgenic Rice Plants. Plant Cell Physiol. 2005, 46, 743–753. [Google Scholar] [CrossRef]
- Seebauer, J.R.; Moose, S.P.; Fabbri, B.J.; Crossland, L.D.; Below, F.E. Amino acid metabolism in maize earshoots. Implications for assimilate preconditioning and nitrogen signaling. Plant Physiol. 2004, 136, 4326–4334. [Google Scholar] [CrossRef] [PubMed]
- Babst, B.A.; Sjödin, A.; Jansson, S.; Orians, C.M. Local and systemic transcriptome responses to herbivory and jasmonic acid in Populus. Tree Genet. Genomes 2009, 5, 459–474. [Google Scholar] [CrossRef]
- Batushansky, A.; Kirma, M.; Grillich, N.; Toubiana, D.; Pham, P.A.; Balbo, I.; Fromm, H.; Galili, G.; Fernie, A.R.; Fait, A. Combined Transcriptomics and Metabolomics of Arabidopsis thaliana Seedlings Exposed to Exogenous GABA Suggest Its Role in Plants Is Predominantly Metabolic. Mol. Plant 2014, 7, 1065–1068. [Google Scholar] [CrossRef]
- Luo, J.; Zhou, J.; Li, H.; Shi, W.; Polle, A.; Lu, M.; Sun, X.; Luo, Z. Bin Global poplar root and leaf transcriptomes reveal links between growth and stress responses under nitrogen starvation and excess. Tree Physiol. 2015, 35, 1283–1302. [Google Scholar] [CrossRef] [PubMed]
- Silvente, S.; Camas, A.; Lara, M. Molecular cloning of the cDNA encoding aspartate aminotransferase from bean root nodules and determination of its role in nodule nitrogen metabolism. J. Exp. Bot. 2003, 54, 1545–1551. [Google Scholar] [CrossRef] [PubMed]
- Barghini, E.; Cossu, R.M.; Cavallini, A.; Giordani, T. Transcriptome analysis of response to drought in poplar interspecific hybrids. Genomics Data 2015, 3, 143–145. [Google Scholar] [CrossRef] [PubMed]
- Gaufichon, L.; Rothstein, S.J.; Suzuki, A. Asparagine metabolic pathways in Arabidopsis. Plant Cell Physiol. 2016, 57, 675–689. [Google Scholar] [CrossRef] [PubMed]
- Schultz, C.J.; Hsu, M.; Miesak, B.; Coruzzi, G.M. Arabidopsis mutants define an in vivo role for isoenzymes of aspartate aminotransferase in plant nitrogen assimilation. Genetics 1998, 149, 491–499. [Google Scholar] [PubMed]
- Brauc, S.; De Vooght, E.; Claeys, M.; Höfte, M.; Angenon, G. Influence of over-expression of cytosolic aspartate aminotransferase on amino acid metabolism and defence responses against Botrytis cinerea infection in Arabidopsis thaliana. J. Plant Physiol. 2011, 168, 1813–1819. [Google Scholar] [CrossRef]
- Graindorge, M.; Giustini, C.; Jacomin, A.C.; Kraut, A.; Curien, G.; Matringe, M. Identification of a plant gene encoding glutamate/aspartate-prephenate aminotransferase: The last homeless enzyme of aromatic amino acids biosynthesis. FEBS Lett. 2010, 584, 4357–4360. [Google Scholar] [CrossRef]
- Murooka, Y.; Mori, Y.; Hayashi, M. Variation of the Amino Acid Content of Arabidopsis Seeds by Expressing Soybean Aspartate Aminotransferase Gene. J. Biosci. Bioeng. 2002, 94, 225–230. [Google Scholar] [CrossRef]
- Polle, A.; Douglas, C. The molecular physiology of poplars: Paving the way for knowledge-based biomass production. Plant Biol. 2010. [Google Scholar] [CrossRef]
- Jansson, S.; Douglas, C.J. Populus: A Model System for Plant Biology. Annu. Rev. Plant Biol. 2007, 58, 435–458. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zhen, C.; Xu, W.; Wang, C.; Cheng, Y. Simple, rapid and efficient transformation of genotype Nisqually-1: A basic tool for the first sequenced model tree. Sci. Rep. 2017, 7, 2638. [Google Scholar] [CrossRef] [PubMed]
- Wojciechowska, N.; Marzec-Schmidt, K.; Kalemba, E.M.; Zarzyńska-Nowak, A.; Jagodziński, A.M.; Bagniewska-Zadworna, A. Autophagy counteracts instantaneous cell death during seasonal senescence of the fine roots and leaves in Populus trichocarpa. BMC Plant Biol. 2018, 18, 260. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Li, J.; Zhao, X.Q.; Wang, J.; Wong, G.K.S.; Yu, J. KaKs_Calculator: Calculating Ka and Ks Through Model Selection and Model Averaging. Genomics Proteomics Bioinforma. 2006, 4, 259–263. [Google Scholar] [CrossRef]
- Lynch, M.; Conery, J.S. The Evolutionary Fate and Consequences of Duplicate Genes. Science (80-). 2000, 290, 1151–1155. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.-Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Bailey, T.L.; Williams, N.; Misleh, C.; Li, W.W. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006, 34, W369–W373. [Google Scholar] [CrossRef]
- De Castro, E.; Sigrist, C.J.A.; Gattiker, A.; Bulliard, V.; Langendijk-Genevaux, P.S.; Gasteiger, E.; Bairoch, A.; Hulo, N. ScanProsite: Detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 2006, 34, W362–W365. [Google Scholar] [CrossRef]
- Shi, R.; Wang, J.P.; Lin, Y.C.; Li, Q.; Sun, Y.H.; Chen, H.; Sederoff, R.R.; Chiang, V.L. Tissue and cell-type co-expression networks of transcription factors and wood component genes in Populus trichocarpa. Planta 2017, 245, 927–938. [Google Scholar] [CrossRef]
- Han, M.; Heppel, S.C.; Su, T.; Bogs, J.; Zu, Y.; An, Z.; Rausch, T. Enzyme Inhibitor Studies Reveal Complex Control of Methyl-D-Erythritol 4-Phosphate (MEP) Pathway Enzyme Expression in Catharanthus roseus. PLoS ONE 2013, 8, e62467. [Google Scholar] [CrossRef] [PubMed]
- Vandesompele, J.; de Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; de Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, 1–12. [Google Scholar] [CrossRef]
- Swanson, W.J.; Yang, Z.; Wolfner, M.F.; Aquadro, C.F. Positive Darwinian selection drives the evolution of several female reproductive proteins in mammals. Proc. Natl. Acad. Sci. USA 2001, 98, 2509–2514. [Google Scholar] [CrossRef] [PubMed]
- Maciaga, M.; Szkop Michałand Paszkowski, A. Biochemical characterization of aspartate aminotransferase allozymes from common wheat. Cent. Eur. J. Biol. 2013, 8, 1183–1193. [Google Scholar] [CrossRef][Green Version]
- Holland, C.K.; Berkovich, D.A.; Kohn, M.L.; Maeda, H.; Jez, J.M. Structural basis for substrate recognition and inhibition of prephenate aminotransferase from Arabidopsis. Plant J. 2018, 94, 304–314. [Google Scholar] [CrossRef] [PubMed]
- Chao, Q.; Gao, Z.-F.; Zhang, D.; Zhao, B.-G.; Dong, F.-Q.; Fu, C.-X.; Liu, L.-J.; Wang, B.-C. The developmental dynamics of the Populusstem transcriptome. Plant Biotechnol. J. 2019, 17, 206–219. [Google Scholar] [CrossRef] [PubMed]
- Dornfeld, C.; Weisberg, A.J.; Ritesh, K.C.; Dudareva, N.; Jelesko, J.G.; Maeda, H.A. Phylobiochemical Characterization of Class-Ib Aspartate/Prephenate Aminotransferases Reveals Evolution of the Plant Arogenate Phenylalanine Pathway. Plant Cell 2014, 26, 3101–3114. [Google Scholar] [CrossRef] [PubMed]
- Han, M.; Wong, J.; Su, T.; Beatty, P.H.; Good, A.G. Identification of Nitrogen Use Efficiency Genes in Barley: Searching for QTLs Controlling Complex Physiological Traits. Front. Plant Sci. 2016, 7, 1587. [Google Scholar] [CrossRef]
- Bao, H.; Li, E.; Mansfield, S.D.; Cronk, Q.C.B.; El-Kassaby, Y.A.; Douglas, C.J. The developing xylem transcriptome and genome-wide analysis of alternative splicing in Populus trichocarpa (black cottonwood) populations. BMC Genom. 2013, 14, 359. [Google Scholar] [CrossRef]
- Srivastava, V.; Srivastava, M.K.; Chibani, K.; Nilsson, R.; Rouhier, N.; Melzer, M.; Wingsle, G. Alternative splicing studies of the reactive oxygen species gene network in Populus reveal two isoforms of high-isoelectric-point superoxide dismutase. Plant Physiol. 2009, 149, 1848–1859. [Google Scholar] [CrossRef]
- Song, D.; Xi, W.; Shen, J.; Bi, T.; Li, L. Characterization of the plasma membrane proteins and receptor-like kinases associated with secondary vascular differentiation in poplar. Plant Mol. Biol. 2011, 76, 97–115. [Google Scholar] [CrossRef] [PubMed]
- Han, M.; Okamoto, M.; Beatty, P.H.; Rothstein, S.J.; Good, A.G. The Genetics of Nitrogen Use Efficiency in Crop Plants. Annu. Rev. Genet 2015, 49, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Tegeder, M.; Masclaux-Daubresse, C. Source and sink mechanisms of nitrogen transport and use. New Phytol. 2018, 217, 35–53. [Google Scholar] [CrossRef] [PubMed]
- Wullschleger, S.D.; Weston, D.J.; Difazio, S.P.; Tuskan, G.A. Revisiting the sequencing of the first tree genome: Populus trichocarpa. Tree Physiol. 2013, 33, 357–364. [Google Scholar] [CrossRef]
- Rai, V.; Muthuraj, M.; Gandhi, M.N.; Das, D.; Srivastava, S. Real-time iTRAQ-based proteome profiling revealed the central metabolism involved in nitrogen starvation induced lipid accumulation in microalgae. Sci. Rep. 2017, 7, 1–16. [Google Scholar] [CrossRef]
- Luo, J.; Li, H.; Liu, T.; Polle, A.; Peng, C.; Luo, Z. Bin Nitrogen metabolism of two contrasting poplar species during acclimation to limiting nitrogen availability. J. Exp. Bot. 2013, 64, 4207–4224. [Google Scholar] [CrossRef]
- Stitt, M.; Gibon, Y. Why measure enzyme activities in the era of systems biology? Trends Plant Sci. 2014, 19, 256–265. [Google Scholar] [CrossRef]
- Su, T.; Han, M.; Min, J.; Chen, P.; Mao, Y.; Huang, Q.; Tong, Q.; Liu, Q.; Fang, Y. Genome-Wide Survey of Invertase Encoding Genes and Functional Characterization of an Extracellular Fungal Pathogen-Responsive Invertase in Glycine max. Int. J. Mol. Sci. 2018, 19, 2395. [Google Scholar] [CrossRef]
- Albacete, A.; Cantero-Navarro, E.; Großkinsky, D.K.; Arias, C.L.; Balibrea, M.E.; Bru, R.; Fragner, L.; Ghanem, M.E.; de La Cruz González, M.; Hernández, J.A.; et al. Ectopic overexpression of the cell wall invertase gene CIN1 leads to dehydration avoidance in tomato. J. Exp. Bot. 2015, 66, 863–878. [Google Scholar] [CrossRef]
- Franklin, O.; Cambui, C.A.; Gruffman, L.; Palmroth, S.; Oren, R.; Näsholm, T. The carbon bonus of organic nitrogen enhances nitrogen use efficiency of plants. Plant Cell Environ. 2017, 40, 25–35. [Google Scholar] [CrossRef]

| Plant Species | Numbers of AspATs in Subfamilies | Total | |||
|---|---|---|---|---|---|
| Iα-A | Iα-B | Iα-C | Iβ | ||
| Populus trichocarpa | 3 | 3 | 2 | 2 | 10 |
| Arabidopsis thaliana | 3 | 1 | 1 | 1 | 6 |
| Eucalyptus grandis | 3 | 2 | 2 | 2 | 9 |
| Glycine max | 2 | 3 | 2 | 4 | 11 |
| Grossypium raimondii | 3 | 2 | 2 | 2 | 9 |
| Medicago truncatula | 1 | 2 | 1 | 7 | 11 |
| Nicotiana benthamiana | 2 | 1 | 1 | 1 | 6 |
| Oryza sativa | 1 | 2 | 1 | 2 | 6 |
| Pinus taeda | 1 | 1 | 1 | 2 | 5 |
| Sorghum bicolor | 1 | 1 | 1 | 3 | 6 |
| Solanum lycopersicum | 2 | 2 | 1 | 1 | 6 |
| Zea mays | 1 | 1 | 2 | 2 | 6 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Su, T.; Han, M.; Min, J.; Cao, D.; Zhai, G.; Zhou, H.; Li, N.; Li, M. Genome-Wide Characterization of AspATs in Populus: Gene Expression Variation and Enzyme Activities in Response to Nitrogen Perturbations. Forests 2019, 10, 449. https://doi.org/10.3390/f10050449
Su T, Han M, Min J, Cao D, Zhai G, Zhou H, Li N, Li M. Genome-Wide Characterization of AspATs in Populus: Gene Expression Variation and Enzyme Activities in Response to Nitrogen Perturbations. Forests. 2019; 10(5):449. https://doi.org/10.3390/f10050449
Chicago/Turabian StyleSu, Tao, Mei Han, Jie Min, Dan Cao, Guangqing Zhai, Huaiye Zhou, Nanyue Li, and Mingzhi Li. 2019. "Genome-Wide Characterization of AspATs in Populus: Gene Expression Variation and Enzyme Activities in Response to Nitrogen Perturbations" Forests 10, no. 5: 449. https://doi.org/10.3390/f10050449
APA StyleSu, T., Han, M., Min, J., Cao, D., Zhai, G., Zhou, H., Li, N., & Li, M. (2019). Genome-Wide Characterization of AspATs in Populus: Gene Expression Variation and Enzyme Activities in Response to Nitrogen Perturbations. Forests, 10(5), 449. https://doi.org/10.3390/f10050449






