Structural and Comparative Analysis of the Complete Chloroplast Genome of a Mangrove Plant: Scyphiphora hydrophyllacea Gaertn. f. and Related Rubiaceae Species
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material, DNA Extraction, and Sequencing
2.2. Genomic Assembly, Annotation and Validation
2.3. Simple Sequence Repeat Analysis
2.4. Codon Usage Analysis
2.5. Genome Comparison
2.6. Phylogenetic Analysis
3. Results and Discussion
3.1. Basic Characteristics of cp GENOME of S. hydrophyllacea
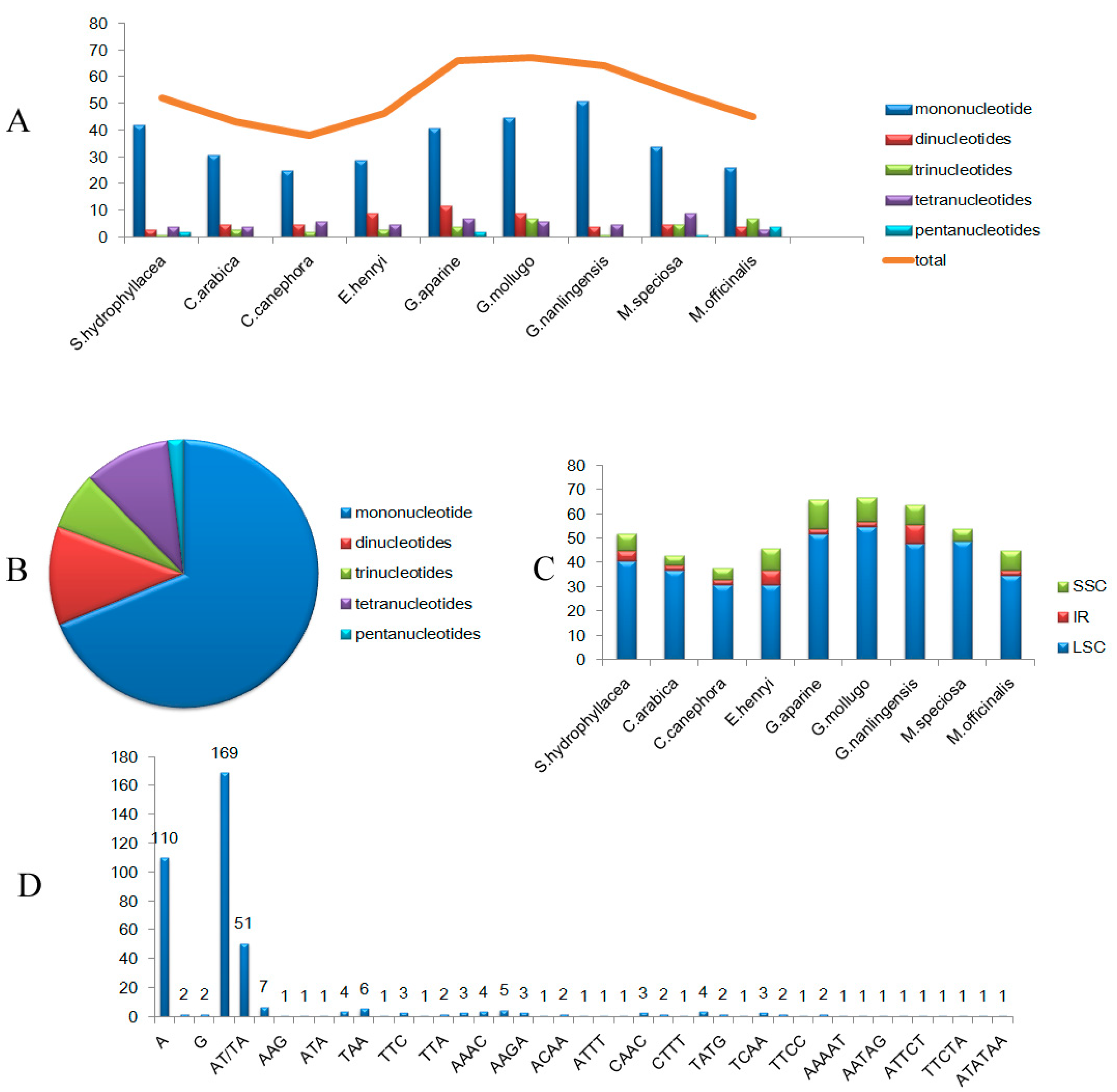
3.2. SSR Analysis
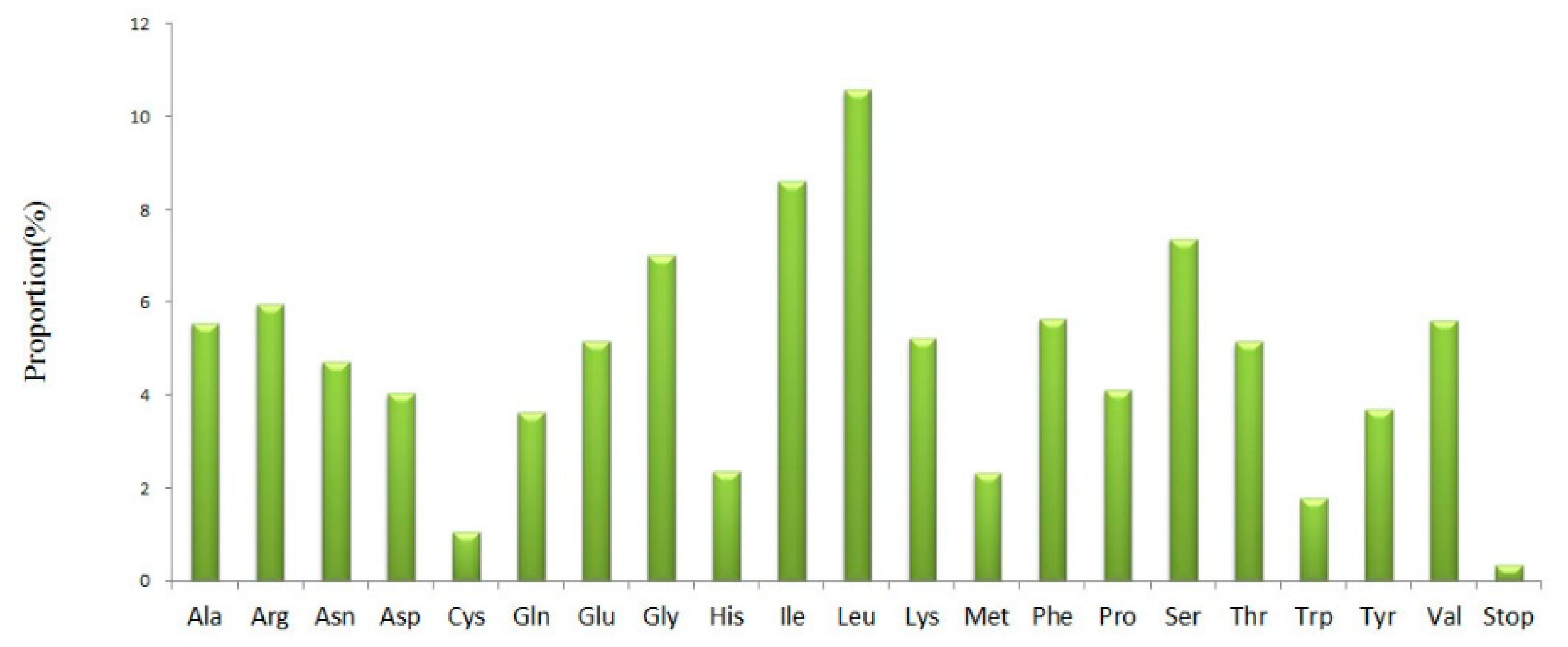
3.3. Codon Usage and Putative RNA Editing Sites in cp Genes of S. hydrophyllacea
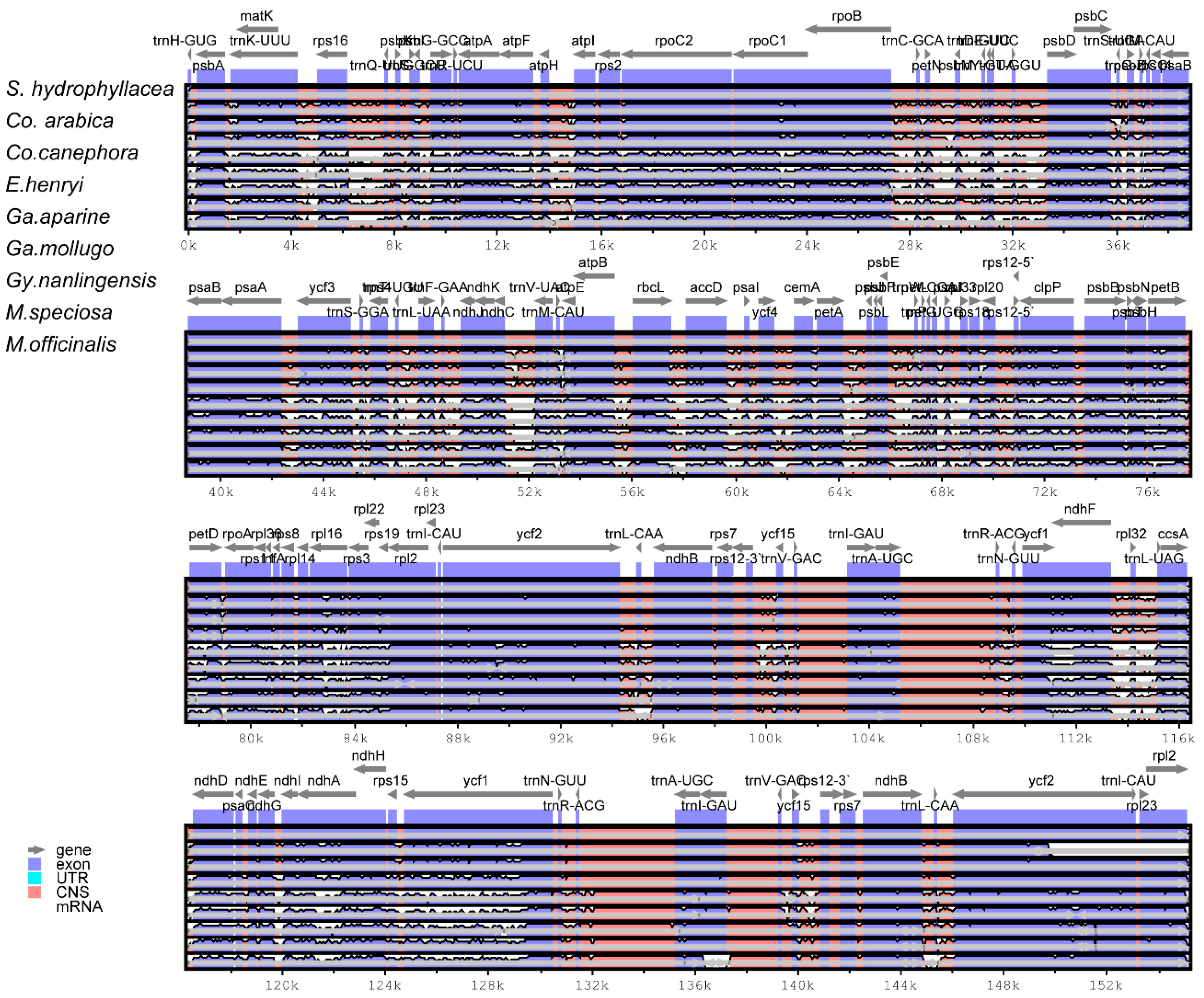
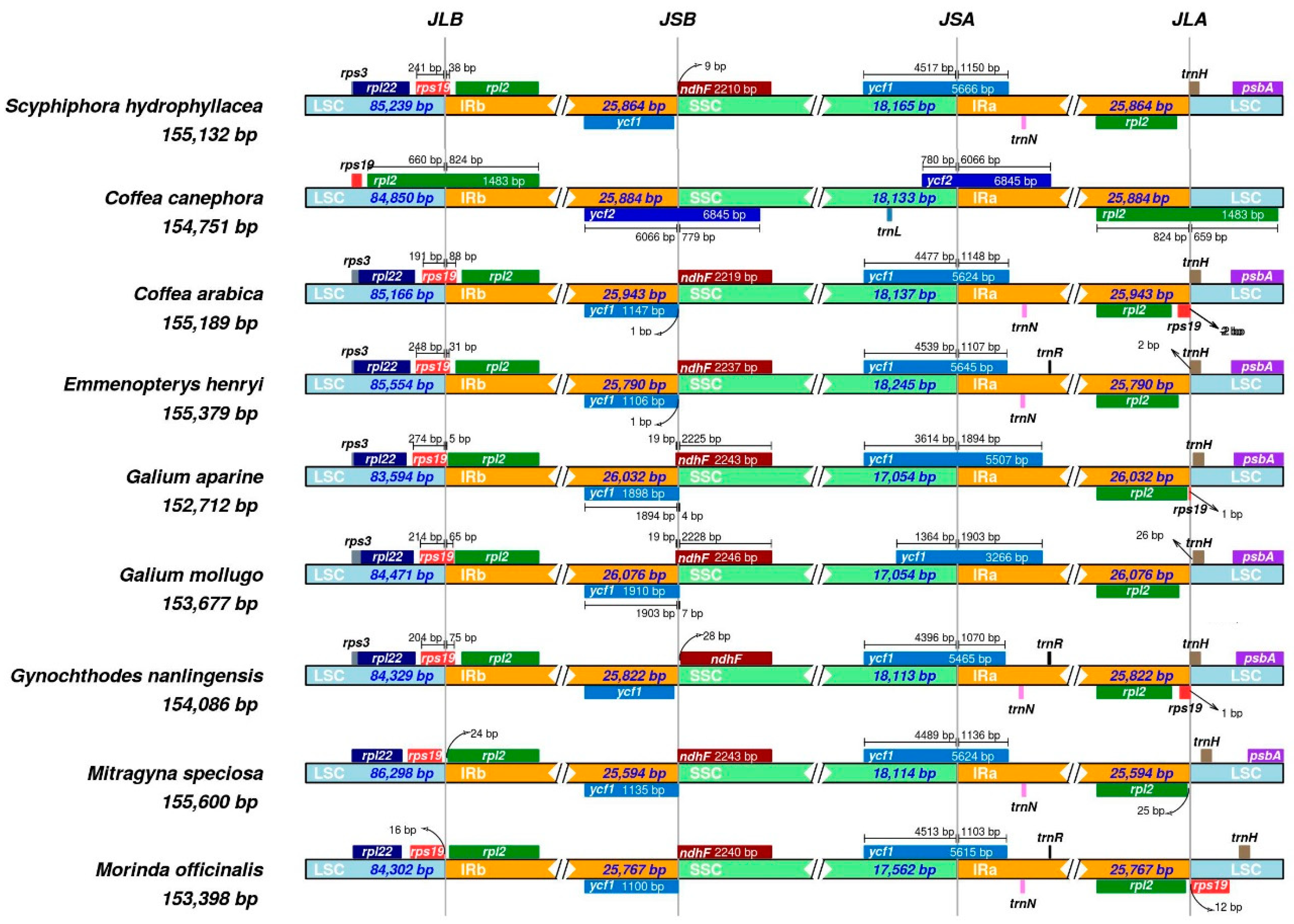
3.4. Comparison of Basic Characteristics of the cp Genome of Nine Rubiaceae Species
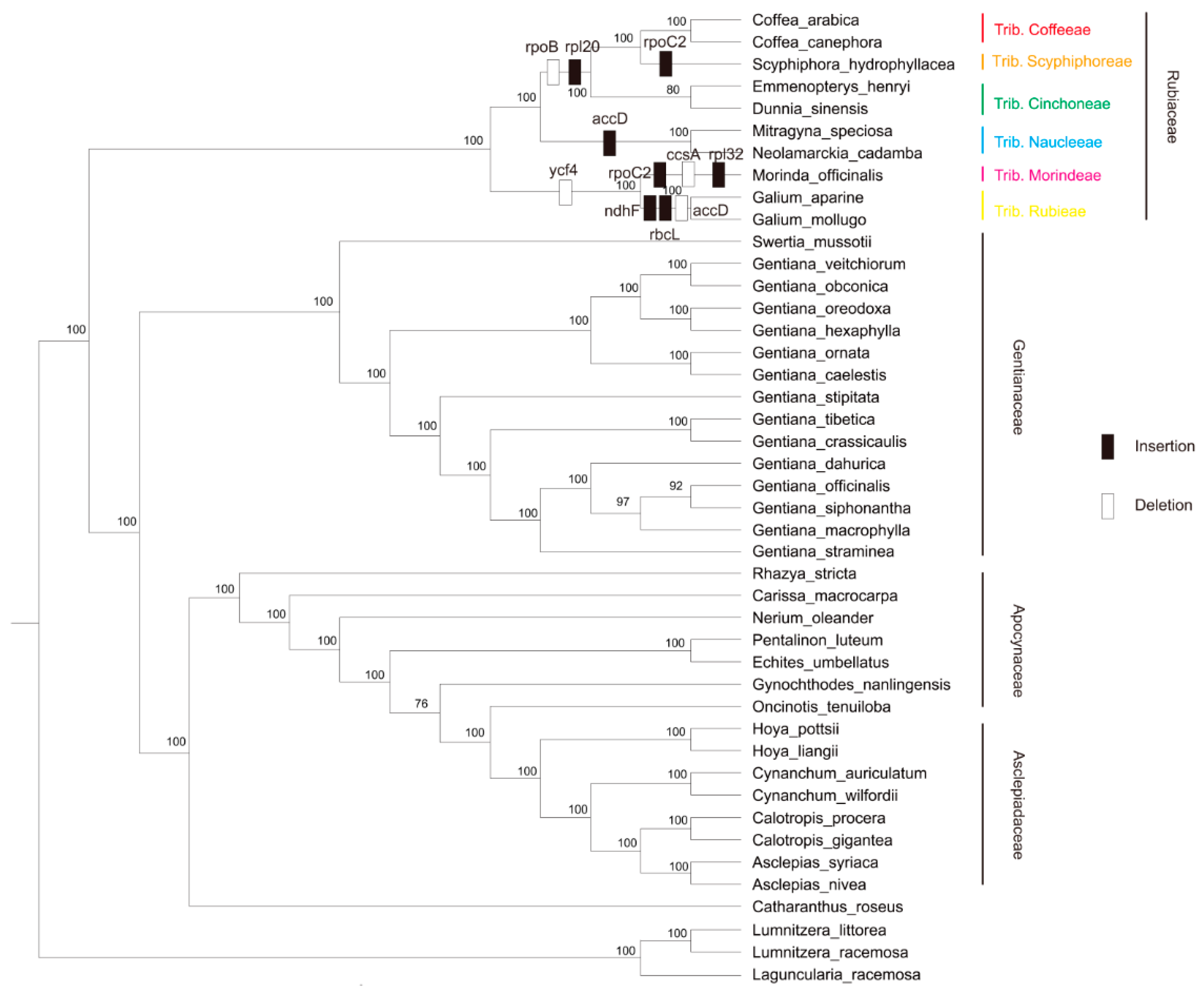
3.5. Phylogenetic Relationships in Gentianales
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Tomlinson, P. The Botany of Mangroves, 1st ed.; Cambridge University Press: New York, NY, USA, 1986; pp. 261–264. [Google Scholar]
- Duke, N.C. Australia’s Mangroves: The Authoritative Guide to Australia’s Mangrove Plants, 1st ed.; University of Queensland Press: Brisbane, QLD, Australia, 2006; pp. 172–173. [Google Scholar]
- Polidoro, B.A.; Carpenter, K.E.; Collins, L.; Duke, N.C.; Ellison, A.M.; Ellison, J.C.; Farnsworth, E.J.; Fernando, E.S.; Kathiresan, K.; Koedam, N.E.; et al. The loss of species: Mangrove extinction risk and geographic areas of global concern. PLoS ONE 2010, 5, e10095. [Google Scholar] [CrossRef] [PubMed]
- Samarakoon, S.R.; Shanmuganathan, C.; Ediriweera, M.K.; Piyathilaka, P.; Tennekoon, K.H.; Thabrew, I.; Galhena, P.; De Silva, E.D. Anti-hepatocarcinogenic and Anti-oxidant Effects of Mangrove Plant Scyphiphora hydrophyllacea. Pharm. Mag. 2017, 13, S76–S83. [Google Scholar] [CrossRef] [PubMed]
- Feng, C.L.; Gong, M.F.; Zeng, Y.B.; Dai, H.F.; Mei, W.L. Scyphiphin C, a new iridoid from Scyphiphora Hydrophyllacea. Molecules 2010, 15, 2473–2477. [Google Scholar] [CrossRef] [PubMed]
- Neuhaus, H.E.; Emes, M.J. Nonphotosynthetic metabolism in plastids. Annu. Rev. Plant Biol. 2000, 51, 111–140. [Google Scholar] [CrossRef]
- Daniell, H.; Lin, C.S.; Yu, M.; Chang, W.J. Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biol. 2016, 17, 134. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, J.; Li, L.; Gao, L.; Xu, J. Structural and Comparative Analysis of the Complete Chloroplast Genome of Pyrus hopeiensis-“Wild Plants with a Tiny Population”-and Three Other Pyrus Species. Int. J. Mol. Sci. 2018, 19, 3262. [Google Scholar] [CrossRef]
- Andreasen, K.; Bremer, B. Combined phylogenetic analysis in the Rubiaceae-Ixoroideae: Morphology, nuclear and chloroplast DNA data. Am. J. Bot. 2000, 87, 1731–1748. [Google Scholar] [CrossRef]
- Bejaoui, F.; Salas, J.J.; Nouairi, I.; Smaoui, A.; Abdelly, C.; Martinez-Force, E.; Youssef, N.B. Changes in chloroplast lipid contents and chloroplast ultrastructure in Sulla carnosa and Sulla coronaria leaves under salt stress. J. Plant Physiol. 2016, 198, 32–38. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: Single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Lowe, T.M.; Chan, P.P. tRNAscan-SE On-line: Integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016, 44, W54–W57. [Google Scholar] [CrossRef]
- Wyman, S.K.; Jansen, R.K.; Boore, J.L. Automatic annotation of organellar genomes with DOGMA. Bioinformatics 2004, 20, 3252–3255. [Google Scholar] [CrossRef] [PubMed]
- Greiner, S.; Lehwark, P.; Bock, R. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: Expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019, 47, W59–W64. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.Y.; Yu, Y.; Deng, Y.Q.; Li, J.; Huang, Z.X.; Zhou, S.D. The Chloroplast Genome of Lilium henrici: Genome Structure and Comparative Analysis. Molecules 2018, 23, 1276. [Google Scholar] [CrossRef] [PubMed]
- Mower, J.P. The PREP suite: Predictive RNA editors for plant mitochondrial genes, chloroplast genes and user-defined alignments. Nucleic Acids Res. 2009, 37, W253–W259. [Google Scholar] [CrossRef] [PubMed]
- Frazer, K.A.; Pachter, L.; Poliakov, A.; Rubin, E.M.; Dubchak, I. VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 2004, 32, W273–W279. [Google Scholar] [CrossRef] [PubMed]
- Amiryousefi, A.; Hyvönen, J.; Poczai, P. IRscope: An online program to visualize the junction sites of chloroplast genomes. Bioinformatics 2018, 34, 3030–3031. [Google Scholar] [CrossRef]
- Nakamura, T.; Yamada, K.D.; Tomii, K.; Katoh, K. Parallelization of MAFFT for large-scale multiple sequence alignments. Bioinformatics 2018, 34, 2490–2492. [Google Scholar] [CrossRef]
- Amiryousefi, A.; Hyvonen, J.; Poczai, P. The chloroplast genome sequence of bittersweet (Solanum dulcamara): Plastid genome structure evolution in Solanaceae. PLoS ONE 2018, 13, e0196069. [Google Scholar] [CrossRef]
- Xu, C.; Dong, W.; Li, W.; Lu, Y.; Xie, X.; Jin, X.; Shi, J.; He, K.; Suo, Z. Comparative Analysis of Six Lagerstroemia Complete Chloroplast Genomes. Front. Plant Sci. 2017, 8, 15. [Google Scholar] [CrossRef]
- Samson, N.; Bausher, M.G.; Lee, S.B.; Jansen, R.K.; Daniell, H. The complete nucleotide sequence of the coffee (Coffea arabica L.) chloroplast genome: Organization and implications for biotechnology and phylogenetic relationships amongst angiosperms. Plant Biotechnol. J. 2007, 5, 339–353. [Google Scholar] [CrossRef]
- Zhang, R.; Li, Q.; Gao, J.; Qu, M.; Ding, P. The complete chloroplast genome sequence of the medicinal plant Morinda officinalis (Rubiaceae), an endemic to China. Mitochondrial DNA Part A 2016, 27, 4324–4325. [Google Scholar] [CrossRef] [PubMed]
- Hildebrand, M.B.; Hallick, R.; Passavant, C.; Bourque, D. Trans-Splicing in Chloroplasts: The rps12 Loci of Nicotiana tabacum. Proc. Natl. Acad. Sci. USA 1988, 85, 372–376. [Google Scholar] [CrossRef] [PubMed]
- Fu, P.C.; Zhang, Y.Z.; Geng, H.M.; Chen, S.L. The complete chloroplast genome sequence of Gentiana lawrencei var. farreri (Gentianaceae) and comparative analysis with its congeneric species. PeerJ 2016, 4, e2540. [Google Scholar] [CrossRef] [PubMed]
- George, B.; Bhatt, B.S.; Awasthi, M.; George, B.; Singh, A.K. Comparative analysis of microsatellites in chloroplast genomes of lower and higher plants. Curr. Genet. 2015, 61, 665–677. [Google Scholar] [CrossRef]
- Saina, J.K.; Li, Z.Z.; Gichira, A.W.; Liao, Y.Y. The Complete Chloroplast Genome Sequence of Tree of Heaven (Ailanthus altissima (Mill.) (Sapindales: Simaroubaceae), an Important Pantropical Tree. Int. J. Mol. Sci. 2018, 19, 929. [Google Scholar] [CrossRef]
- Yang, Y.; Zhu, J.; Feng, L.; Zhou, T.; Bai, G.; Yang, J.; Zhao, G. Plastid Genome Comparative and Phylogenetic Analyses of the Key Genera in Fagaceae: Highlighting the Effect of Codon Composition Bias in Phylogenetic Inference. Front. Plant Sci. 2018, 9, 82. [Google Scholar] [CrossRef]
- Morton, B.R. The role of context-dependent mutations in generating compositional and codon usage bias in grass chloroplast DNA. J. Mol. Evol. 2003, 56, 616–629. [Google Scholar] [CrossRef]
- Lopes, A.D.S.; Pacheco, T.G.; Nimz, T.; Vieira, L.D.N.; Guerra, M.P.; Nodari, R.O.; De Souza, E.M.; Pedrosa, F.D.O.; Rogalski, M. The complete plastome of macaw palm [Acrocomia aculeata (Jacq.) Lodd. ex Mart.] and extensive molecular analyses of the evolution of plastid genes in Arecaceae. Planta 2018, 247, 1011–1030. [Google Scholar]
- Guisinger, M.M.; Kuehl, J.V.; Boore, J.L.; Jansen, R.K. Extreme reconfiguration of plastid genomes in the angiosperm family Geraniaceae: Rearrangements, repeats, and codon usage. Mol. Biol. Evol. 2011, 28, 583–600. [Google Scholar] [CrossRef]
- Duan, R.Y.; Huang, M.Y.; Yang, L.M.; Liu, Z.W. Characterization of the complete chloroplast genome of Emmenopterys henryi (Gentianales: Rubiaceae), an endangered relict tree species endemic to China. Conserv. Genet. Resour. 2017, 9, 1–3. [Google Scholar] [CrossRef]
- Tsudzuki, T.; Wakasugi, T.; Sugiura, M. Comparative analysis of RNA editing sites in higher plant chloroplasts. J. Mol. Evol. 2001, 53, 327–332. [Google Scholar] [CrossRef]
- Li, P.; Lu, R.S.; Xu, W.Q.; Ohi-Toma, T.; Cai, M.Q.; Qiu, Y.X.; Cameron, K.M.; Fu, C.X. Comparative Genomics and Phylogenomics of East Asian Tulips (Amana, Liliaceae). Front. Plant Sci. 2017, 8, 451. [Google Scholar] [CrossRef] [PubMed]
- Dong, W.; Xu, C.; Cheng, T.; Zhou, S. Complete chloroplast genome of Sedum sarmentosum and chloroplast genome evolution in Saxifragales. PLoS ONE 2013, 8, e77965. [Google Scholar] [CrossRef] [PubMed]
- Tosh, J.; Dessein, S.; Buerki, S.; Groeninckx, I.; Mouly, A.; Bremer, B.; Smets, E.F.; De, B.P. Evolutionary history of the Afro-Madagascan Ixora species (Rubiaceae): Species diversification and distribution of key morphological traits inferred from dated molecular phylogenetic trees. Ann. Bot. 2013, 112, 1723–1742. [Google Scholar] [CrossRef]
- Guyot, R.; Lefebvre-Pautigny, F.; Tranchant-Dubreuil, C.; Rigoreau, M.; Hamon, P.; Leroy, T.; Hamon, S.; Poncet, V.; Crouzillat, D.; Kochko, A.D. Ancestral synteny shared between distantly-related plant species from the asterid (Coffea canephora and Solanum Sp.) and rosid (Vitis vinifera) clades. BMC Genom. 2012, 13, 103. [Google Scholar] [CrossRef]
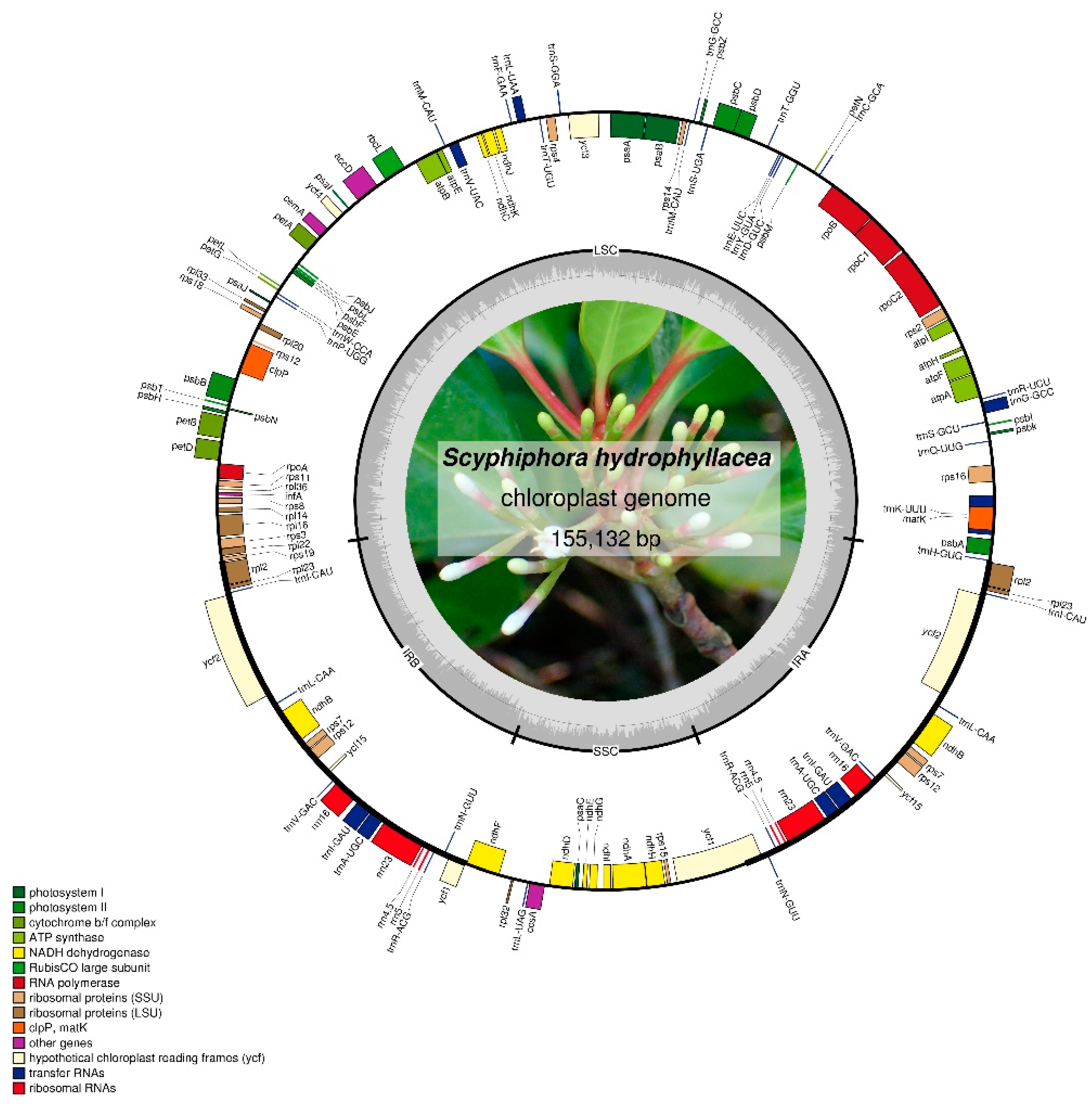
| Functions Category | Group of Genes | Gene Name |
|---|---|---|
| Self-replication | Small subunit of ribosome | rps2, rps3, rps4, rps7a, rps8, rps11,rps12acde, rps14, rps15,rps16, rps18, rps19 |
| large subunit of ribosome | rpl2ab, rpl14, rpl16, rpl20,rpl22, rpl23a, rpl32,rpl33, rpl36 | |
| rRNA genes | rrn4.5a, rrn5a, rrn16a, rrn23a | |
| DNA-dependent RNA polymerase | rpoA, rpoB, rpoC1b, rpoC2 | |
| rRNA Genes | trnY-GUA, trnW-CCA, trnV-UAC, trnV-GACa, trnT-UGU, trnT-GGU, trnS-UGA, trnS-GGA, trnS-GCU, trnR-UCU, trnR-ACGa,trnQ-UUG, trnP-UGG, trnN-GUUa,trnM-CAU, trnL-UAG, trnL-UAA, trnL-CAAa, trnK-UUU, trnI-GAUa, trnI-CAUa,trnH-GUG, trnG-GCC, trnG-UCC, trnfM-CAU, trnF-GAA, trnE-UUC, trnD-GUC, trnC-GCA, trnA-UGCa | |
| Genes for Photosynthesis | Subunits of ATP synthase | atpA, atpB, atpE, atpFb, atpH, atpI |
| Subunits of NADH-dehydrogenase | ndhAb, ndhBab, ndhC, ndhD, ndhE, ndhF, ndhG, ndhHb, ndhI, ndhJ, ndhK | |
| Subunits of cytochrome b/f comples | petA, petB, petD, petG, petL, petN | |
| Subunits of photosystem I | psaA, psaB, psaC, psaI, psaJ | |
| Subunits of photosystem II | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ | |
| Subunits of rubisco | rbcL | |
| Other Genes | Subunits of Acetyl-CoA-carboxylase | accD |
| Envelop membrane protein | cemA | |
| c-type cytochrome synthesis gene | ccsA | |
| Protease | clpPc | |
| Translational initiation factor | infA | |
| Maturase | matK | |
| Elongation factor | ||
| Genes of Unknown Function | Conserved open reading frames | ycf1a, ycf2a, ycf3c, ycf4, ycf15a |
| Gene | Nucleotide Position | Amino Acid Position | Codon Conversion | Effect | Score |
|---|---|---|---|---|---|
| accD | 280 | 94 | CCC => TCC | P => S | 1 |
| 845 | 282 | TCG => TTG | S => L | 0.8 | |
| 887 | 296 | GCC => GTC | A => V | 0.8 | |
| atpA | 773 | 258 | TCA => TTA | S => L | 1 |
| 791 | 264 | CCC => CTC | P => L | 1 | |
| 914 | 305 | TCA => TTA | S => L | 1 | |
| matK | 292 | 98 | CTT => TTT | L => F | 0.86 |
| 454 | 152 | CAT => TAT | H => Y | 1 | |
| 643 | 215 | CAT => TAT | H => Y | 1 | |
| ndhA | 341 | 114 | TCA => TTA | S => L | 1 |
| 566 | 189 | TCA => TTA | S => L | 1 | |
| 1073 | 358 | TCC => TTC | S => F | 1 | |
| ndhB | 149 | 50 | TCA => TTA | S => L | 1 |
| 467 | 156 | CCA => CTA | P => L | 1 | |
| 586 | 196 | CAT => TAT | H => Y | 1 | |
| 611 | 204 | TCA => TTA | S => L | 0.8 | |
| 737 | 246 | CCA => CTA | P => L | 1 | |
| 746 | 249 | TCT => TTT | S => F | 1 | |
| 830 | 277 | TCA => TTA | S => L | 1 | |
| 836 | 279 | TCA => TTA | S => L | 1 | |
| 1481 | 494 | CCA => CTA | P => L | 1 | |
| ndhD | 29 | 10 | ACG => ATG | T => M | 1 |
| 626 | 209 | TCA => TTA | S => L | 1 | |
| 701 | 234 | TCG => TTG | S => L | 1 | |
| 905 | 302 | TCA => TTA | S => L | 1 | |
| 1103 | 368 | GCT => GTT | A => V | 1 | |
| 1325 | 442 | TCA => TTA | S => L | 0.8 | |
| 1337 | 446 | TCA => TTA | S => L | 0.8 | |
| ndhF | 290 | 97 | TCA => TTA | S => L | 1 |
| ndhG | 314 | 105 | ACA => ATA | T => I | 0.8 |
| petB | 418 | 140 | CGG => TGG | R => W | 1 |
| 611 | 204 | CCA => CTA | P => L | 1 | |
| psaI | 80 | 27 | TCT => TTT | S => F | 0.86 |
| psbE | 214 | 72 | CCT => TCT | P => S | 1 |
| rpl20 | 320 | 107 | TCA => TTA | S => L | 0.86 |
| rpoB | 473 | 158 | TCA => TTA | S => L | 0.86 |
| 551 | 184 | TCA => TTA | S => L | 1 | |
| 566 | 189 | TCG => TTG | S => L | 1 | |
| 2414 | 805 | TCA => TTA | S => L | 0.86 | |
| rpoC1 | 41 | 14 | TCA => TTA | S => L | 1 |
| rpoC2 | 2296 | 766 | CGG => TGG | R => W | 1 |
| 3746 | 1249 | TCA => TTA | S => L | 0.86 | |
| rps14 | 80 | 27 | TCA => TTA | S => L | 1 |
| 149 | 50 | CCA => CTA | P => L | 1 | |
| rps2 | 248 | 83 | TCA => TTA | S => L | 1 |
| rps8 | 143 | 48 | GCG => GTG | A => V | 1 |
| Scyphiphora hydrophyllacea | Coffea arabica | Coffea canephora | Emmenopterys henryi | Galium aparine | Galium mollugo | Gynochthodes nanlingensis | Mitragyna officinalis | Mitragyna speciosa | |
|---|---|---|---|---|---|---|---|---|---|
| Length (bp) | 153,132 | 155,189 | 154,751 | 155,379 | 152,712 | 153,677 | 154,086 | 153,398 | 155,600 |
| GC content (%) | 37.60 | 37.43 | 37.47 | 37.64 | 37.28 | 37.18 | 38.52 | 38.05 | 37.52 |
| AT content (%) | 62.40 | 62.57 | 62.53 | 62.36 | 62.72 | 62.82 | 61.48 | 61.95 | 62.48 |
| LSC length (bp) | 85,239 | 85,166 | 84,850 | 85,554 | 83,594 | 84,471 | 84,329 | 84,302 | 86,298 |
| SSC length (bp) | 18,165 | 18,137 | 18,133 | 18,245 | 17,054 | 17,054 | 18,113 | 17,562 | 18,114 |
| IR length (bp) | 25,864 | 25,943 | 25,884 | 25,790 | 26,032 | 26,076 | 25,822 | 25,767 | 25,594 |
| Gene number | 132 | 133 | 133 | 132 | 132 | 131 | 133 | 133 | 131 |
| Pseudogene number | 1 | 2 | 2 | 1 | 3 | 1 | 2 | 1 | 1 |
| Gene number in IR regions | 19 | 16 | 17 | 17 | 16 | 16 | 18 | 18 | 16 |
| Protein-coding gene number | 88 | 85 | 86 | 86 | 84 | 85 | 90 | 91 | 85 |
| Protein-coding gene (%) | 66.67 | 63.91 | 64.66 | 65.15 | 63.64 | 64.89 | 67.67 | 68.42 | 64.89 |
| rRNA gene number | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 8 |
| rRNA (%) | 6.06 | 6.02 | 6.02 | 6.06 | 6.06 | 6.11 | 6.02 | 6.02 | 6.11 |
| tRNA gene number | 36 | 38 | 37 | 37 | 37 | 37 | 33 | 33 | 37 |
| tRNA (%) | 27.27 | 28.57 | 27.82 | 28.03 | 28.03 | 28.24 | 24.81 | 24.81 | 28.24 |
| Gene | A.A Position | Scyphiphora hydrophyllacea | Coffea arabica | Coffea canephora | Emmenopterys henryi | Galium aparine | Galium mollugo | Gynochthodes nanlingensis | Morinda officinalis | Mitragyna speciosa |
|---|---|---|---|---|---|---|---|---|---|---|
| ndhA | 114 | TCA (S) => TTA (L) | TCA (S) => TTA (L) | TCA (S) => TTA (L) | TCA (S) => TTA (L) | TCA (S) => TTA (L) | TCA (S) => TTA (L) | TCA (S) => TTA (L) | TCA (S) => TTA (L) | TCA (S) => TTA (L) |
| ndhD | 10 | ACG (T) =>ATG (M) | ACG (T) => ATG (M) | ACG (T) => ATG (M) | ACG (T) => ATG (M) | ACG (T) => ATG (M) | ACG (T) => ATG (M) | ACG (T) => ATG (M) | ACG (T) => ATG (M) | ACG (T) => ATG (M) |
| 302 | TCA (S) => TTA (L) | TCA (S) => TTA (L) | TCA (S) => TTA (L) | TCA (S) => TTA (L) | TCA (S) => TTA (L) | TCA (S) => TTA (L) | TCA (S) => TTA (L) | TCA (S) => TTA (L) | TCA (S) => TTA (L) | |
| 442 | TCA (S) => TTA (L) | TCA (S) => TTA (L) | TCA (S) => TTA (L) | TCA (S) => TTA (L) | TCA (S) => TTA (L) | TCA (S) => TTA (L) | TCA (S) => TTA (L) | TCA (S) => TTA (L) | TCA (S) => TTA (L) | |
| 446 | TCA (S) => TTA (L) | TCA (S) => TTA (L) | TCA (S) => TTA (L) | TCA (S) => TTA (L) | TCA (S) => TTA (L) | TCA (S) => TTA (L) | TCA (S) => TTA (L) | TCA (S) => TTA (L) | TCA (S) => TTA (L) | |
| ndhF | 97 | TCA (S) => TTA (L) | TCA (S) => TTA (L) | TCA (S) => TTA (L) | TCA (S) => TTA (L) | TCA (S) => TTA (L) | TCA (S) => TTA (L) | TCA (S) => TTA (L) | TCA (S) => TTA (L) | TCA (S) => TTA (L) |
| ndhG | 105 | ACA (T) => ATA (I) | ACA (T) => ATA (I) | ACA (T) => ATA (I) | ACA (T) => ATA (I) | ACA (T) => ATA (I) | ACA (T) => ATA (I) | ACA (T) => ATA (I) | ACA (T) => ATA (I) | ACA (T) => ATA (I) |
| psaI | 27 | TCT (S) => TTT (F) | TCT (S) => TTT (F) | TCT (S) => TTT (F) | TCT (S) => TTT (F) | TCT (S) => TTT (F) | TCT (S) => TTT (F) | TCT (S) => TTT (F) | TCT (S) => TTT (F) | TCT (S) => TTT (F) |
| rpoB | 158 | TCA (S) => TTA (L) | TCA (S) => TTA (L) | TCA (S) => TTA (L) | TCA (S) => TTA (L) | TCA (S) => TTA (L) | TCA (S) => TTA (L) | TCA (S) => TTA (L) | TCA (S) => TTA (L) | TCA (S) => TTA (L) |
| 184 | TCA (S) => TTA (L) | TCA (S) => TTA (L) | TCA (S) => TTA (L) | TCA (S) => TTA (L) | TCA (S) => TTA (L) | TCA (S) => TTA (L) | TCG (S) => TTG (L) | TCA (S) => TTA (L) | TCA (S) => TTA (L) | |
| rpoC2 | 766 | CGG (R) => TGG (W) | CGG (R) => TGG (W) | CGG (R) => TGG (W) | CGG (R) => TGG (W) | CGG (R) => TGG (W) | CGG (R) => TGG (W) | CGG (R) => TGG (W) | CGG (R) => TGG (W) | CGG (R) => TGG (W) |
| 1249 | TCA (S) => TTA (L) | TCA (S) => TTA (L) | TCA (S) => TTA (L) | TCA (S) => TTA (L) | TCA (S) => TTA (L) | TCG (S) => TTG (L) | TCA (S) => TTA (L) | TCA (S) => TTA (L) | TCA (S) => TTA (L) | |
| rps14 | 27 | TCA (S) => TTA (L) | TCA (S) => TTA (L) | TCA (S) => TTA (L) | TCA (S) => TTA (L) | TCA (S) => TTA (L) | TCA (S) => TTA (L) | TCA (S) => TTA (L) | TCA (S) => TTA (L) | TCA (S) => TTA (L) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Zhang, J.-W.; Yang, Y.; Li, X.-N. Structural and Comparative Analysis of the Complete Chloroplast Genome of a Mangrove Plant: Scyphiphora hydrophyllacea Gaertn. f. and Related Rubiaceae Species. Forests 2019, 10, 1000. https://doi.org/10.3390/f10111000
Zhang Y, Zhang J-W, Yang Y, Li X-N. Structural and Comparative Analysis of the Complete Chloroplast Genome of a Mangrove Plant: Scyphiphora hydrophyllacea Gaertn. f. and Related Rubiaceae Species. Forests. 2019; 10(11):1000. https://doi.org/10.3390/f10111000
Chicago/Turabian StyleZhang, Ying, Jing-Wen Zhang, Yong Yang, and Xin-Nian Li. 2019. "Structural and Comparative Analysis of the Complete Chloroplast Genome of a Mangrove Plant: Scyphiphora hydrophyllacea Gaertn. f. and Related Rubiaceae Species" Forests 10, no. 11: 1000. https://doi.org/10.3390/f10111000
APA StyleZhang, Y., Zhang, J.-W., Yang, Y., & Li, X.-N. (2019). Structural and Comparative Analysis of the Complete Chloroplast Genome of a Mangrove Plant: Scyphiphora hydrophyllacea Gaertn. f. and Related Rubiaceae Species. Forests, 10(11), 1000. https://doi.org/10.3390/f10111000




