Genome Survey Sequencing of Betula platyphylla
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Sequencing
2.2. Data Cleaning
2.3. Genome Size Estimation by k-mer Analysis
2.4. Preliminary Genome Assembly and GC Content Analysis
2.5. Identification and Filtration of Genomic Pollutants
2.6. Scaffold Construction, Gap Closing, and Sequence Polishing
2.7. SSR Identification and Analysis
2.8. Acquisition and Culture of Tissue Culture Seedlings of B. platyphylla
2.9. Sequencing and GC Content Analysis of Tissue Culture Plantlets
3. Results
3.1. Sequencing and Data Cleaning
3.2. Genome Size Estimation by k-mer Analysis
3.3. Preliminary Genome Assembly and GC Content Analysis
3.4. Identification and Filtration of Genomic Pollutants
3.5. Scaffold Construction, Gap Closure, and Sequence Polishing
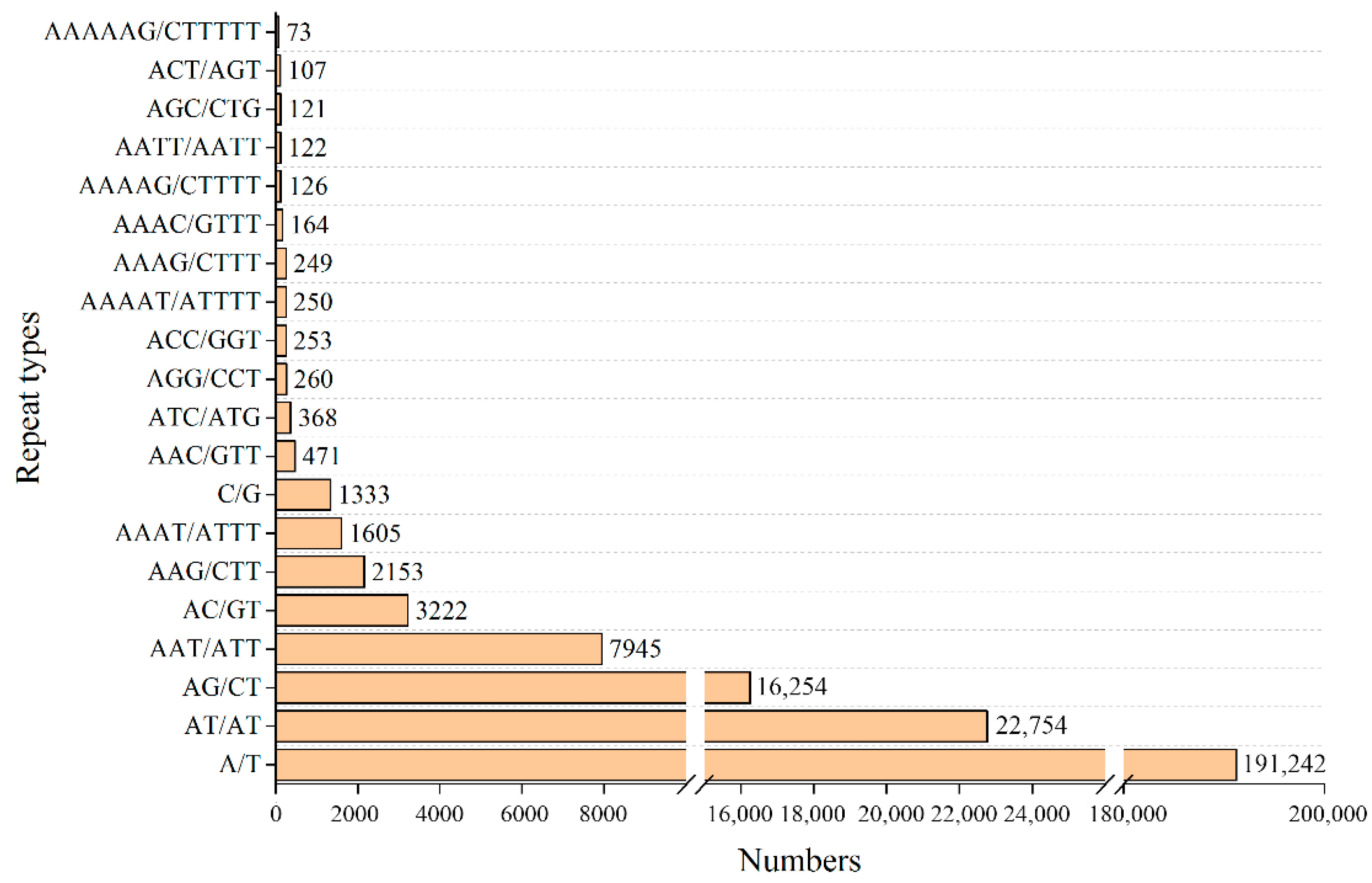
3.6. SSR Identification and Analysis
3.7. Sequencing and GC Content Analysis of Tissue Culture Plantlets
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Zhang, Z.X. Dendrology, 2nd ed.; China Forestry Publishing House: Beijing, China, 2008. [Google Scholar]
- Mijiti, M.; Zhang, Y.M.; Zhang, C.R.; Wang, Y.C. Physiological and molecular responses of Betula platyphylla Suk to salt stress. Trees 2017, 31, 1653–1665. [Google Scholar] [CrossRef]
- MobileReference. The Illustrated Encyclopedia of Trees and Shrubs: An Essential Guide to Trees and Shrubs of the World; MobileReference: Boston, MA, USA, 2008. [Google Scholar]
- Wei, Z.G.; Zhang, K.X.; Yang, C.P.; Liu, G.F.; Liu, G.J.; Lian, L.; Zhang, H.G. Genetic linkage maps of Betula platyphylla Suk. based on ISSR and AFLP markers. Plant Mol. Biol. Report. 2010, 28, 169. [Google Scholar] [CrossRef]
- Krasutsky, P.A. Birch bark research and development. Nat. Prod. Rep. 2006, 23, 919–942. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zhao, H.; Jiang, J.; Liu, G.; Yang, C. Analysis of three types of triterpenoids in tetraploid white birches (Betula platyphylla Suk.) and selection of plus trees. J. For. Res. 2015, 26, 623–633. [Google Scholar] [CrossRef]
- Yogeeswari, P.; Sriram, D. Betulinic acid and its derivatives: A review on their biological properties. Curr. Med. Chem. 2005, 12, 657–666. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.Y.; Qian, L.B.; Zhu, L.G.; Liang, H.T.; Tan, Y.N.; Lu, H.T.; Lu, J.F.; Wang, H.P.; Xia, Q. Betulinic acid ameliorates endothelium-dependent relaxation in L-NAME-induced hypertensive rats by reducing oxidative stress. Eur. J. Pharm. Sci. 2011, 44, 385–391. [Google Scholar] [CrossRef]
- Ríos, J.L.; Manez, S. New pharmacological opportunities for betulinic acid. Planta Med. 2018, 84, 8–19. [Google Scholar] [CrossRef] [PubMed]
- Liang, D.Y.; Zhang, X.X.; Wang, C.; Wang, X.W.; Li, K.L.; Liu, G.F.; Zhao, X.Y.; Qu, G.Z. Evaluation of Betula platyphylla Families Based on Growth and Wood Property Traits. For. Sci. 2018, 64, 663–670. [Google Scholar] [CrossRef]
- Zhao, X.Y.; Bian, X.Y.; Liu, M.R.; Li, Z.X.; Li, Y.; Zheng, M.; Teng, W.H.; Jiang, J.; Liu, G.F. Analysis of genetic effects on a complete diallel cross test of Betula platyphylla. Euphytica 2014, 200, 221–229. [Google Scholar] [CrossRef]
- Porebski, S.; Bailey, L.G.; Baum, B.R. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol. Biol. Report. 1997, 15, 8–15. [Google Scholar] [CrossRef]
- Patel, R.K.; Jain, M. NGS QC Toolkit: A Toolkit for Quality Control of Next Generation Sequencing Data. PLoS ONE 2012, 7, e30619. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.H.; Shi, Y.J.; Yuan, J.Y.; Hu, X.S.; Zhang, H.; Li, N.; Li, Z.Y.; Chen, Y.X.; Mu, D.S.; Fan, W. Estimation of genomic characteristics by analyzing k-mer frequency in de novo genome projects. arXiv 2013, arXiv:1308.2012. [Google Scholar]
- Hernandez, D.; François, P.; Farinelli, L.; Osterås, M.; Schrenzel, J. De novo bacterial genome sequencing: Millions of very short reads assembled on a desktop computer. Genome Res. 2008, 18, 802–809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357. [Google Scholar] [CrossRef] [PubMed]
- Boetzer, M.; Henkel, C.V.; Jansen, H.J.; Butler, D.; Pirovano, W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 2011, 27, 578–579. [Google Scholar] [CrossRef] [PubMed]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.D.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef] [PubMed]
- Robert, M.W.; Mathieu, S.; Felipe, A.S.; Mose, M.; Panagiotis, I.; Guennadi, K.; Evgenia, V.K.; Evgeny, M.Z. BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol. Biol. Evol. 2017, 35, 543–548. [Google Scholar]
- Salojarvi, J.; Smolander, O.P.; Nieminen, K.; Rajaraman, S.; Safronov, O.; Safdari, P.; Lamminmaki, A.; Immanen, J.; Lan, T.Y.; Tanskanen, J.; et al. Genome sequencing and population genomic analyses provide insights into the adaptive landscape of silver birch. Nat. Genet. 2017, 49, 904–912. [Google Scholar] [CrossRef]
- Wang, N.; Thomson, M.; Bodles, W.J.; Crawford, R.M.; Hunt, H.V.; Featherstone, A.W.; Pellicer, J.; Buggs, R.J. Genome sequence of dwarf birch (Betula nana) and cross-species RAD markers. Mol. Ecol. 2013, 22, 3098–3111. [Google Scholar] [CrossRef] [PubMed]
- Xia, E.X.; Yao, Q.Y.; Zhang, H.B.; Jiang, J.J.; Zhang, L.P.; Gao, L.Z. CandiSSR: An Efficient Pipeline used for Identifying Candidate Polymorphic SSRs Based on Multiple Assembled Sequences. Front. Plant Sci. 2016, 6, 1171. [Google Scholar] [CrossRef]
- Cournac, A.; Koszul, R.; Mozziconacci, J. The 3D folding of metazoan genomes correlates with the association of similar repetitive elements. Nucleic Acids Res. 2015, 44, 245–255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shapiro, J.A.; Von Sternberg, R. Why repetitive DNA is essential to genome function. Biol. Rev. 2005, 80, 227–250. [Google Scholar] [CrossRef]
- Martinez-Garcia, P.J.; Crepeau, M.W.; Puiu, D.; Gonzalez-Ibeas, D.; Whalen, J.; Stevens, K.A.; Paul, R.; Butterfield, T.S.; Britton, M.T.; Reagan, R.L.; et al. The walnut (Juglans regia) genome sequence reveals diversity in genes coding for the biosynthesis of non-structural polyphenols. Plant J. 2016, 87, 507–532. [Google Scholar] [CrossRef]
- Hibrand Saint-Oyant, L.; Ruttink, T.; Hamama, L.; Kirov, I.; Lakhwani, D.; Zhou, N.N.; Bourke, P.M.; Daccord, N.; Leus, L.; Schulz, D.; et al. A high-quality genome sequence of Rosa chinensis to elucidate ornamental traits. Nat. Plants 2018, 4, 473. [Google Scholar] [CrossRef]
- Raymond, O.; Gouzy, J.; Just, J.; Badouin, H.; Verdenaud, M.; Lemainque, A.; Vergne, P.; Moja, S.; Choisne, N.; Pont, C.; et al. The Rosa genome provides new insights into the domestication of modern roses. Nat. Genet. 2018, 50, 772. [Google Scholar] [CrossRef] [PubMed]
- Donmez, N.; Brudno, M. Hapsembler: An assembler for highly polymorphic genomes. In Proceedings of the International Conference on Research in Computational Molecular Biology, Vancouver, BC, Canada, 28–31 March 2011; pp. 38–52. [Google Scholar]
- Aguiar, D.; Istrail, S. HapCompass: A fast cycle basis algorithm for accurate haplotype assembly of sequence data. J. Comput. Biol. 2012, 19, 577–590. [Google Scholar] [CrossRef] [PubMed]
- Kajitani, R.; Toshimoto, K.; Noguchi, H.; Toyoda, A.; Ogura, Y.; Okuno, M.; Yabana, M.; Harada, M.; Nagayasu, E.; Maruyama, H.; et al. Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Genome Res. 2014, 24, 1384–1395. [Google Scholar] [CrossRef] [Green Version]
- Safonova, Y.; Bankevich, A.; Pevzner, P.A. dipSPAdes: Assembler for Highly Polymorphic Diploid Genomes. J. Comput. Biol. 2015, 22, 528–545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chin, C.S.; Peluso, P.; Sedlazeck, F.J.; Nattestad, M.; Concepcion, G.T.; Clum, A.; Dunn, C.; O’Malley, R.; Figueroa-Balderas, R.; Morales-Cruz, A.; et al. Phased diploid genome assembly with single-molecule real-time sequencing. Nat. Methods 2016, 13, 1050. [Google Scholar] [CrossRef]
- Koren, S.; Rhie, A.; Walenz, B.P.; Dilthey, A.T.; Bickhart, D.M.; Kingan, S.B.; Hiendleder, S.; Williams, J.L.; Smith, T.; Phillippy, A.M. De novo assembly of haplotype-resolved genomes with trio binning. Nat. Biotechnol. 2018, 36, 1174. [Google Scholar] [CrossRef]
- Roach, M.J.; Schmidt, S.A.; Borneman, A.R. Purge Haplotigs: Allelic contig reassignment for third-gen diploid genome assemblies. BMC Bioinform. 2018, 19, 460. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.F.; Kang, M.J.; Xu, A.L. HaploMerger2: Rebuilding both haploid sub-assemblies from high-heterozygosity diploid genome assembly. Bioinformatics 2017, 33, 2577–2579. [Google Scholar] [CrossRef] [PubMed]
- Miskoff, J.A.; Chaudhri, M. Mycobacterium Chimaera: A Rare Presentation. Cureus 2018, 10, e2750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quambusch, M.; Pirttila, A.M.; Tejesvi, M.V.; Winkelmann, T.; Bartsch, M. Endophytic bacteria in plant tissue culture: Differences between easy- and difficult-to-propagate Prunus avium genotypes. Tree Physiol. 2014, 34, 524–533. [Google Scholar] [CrossRef] [PubMed]
- Koskimaki, J.J.; Hankala, E.; Suorsa, M.; Nylund, S.; Pirttila, A.M. Mycobacteria are hidden endophytes in the shoots of rock plant [Pogonatherum paniceum (Lam.) Hack.](Poaceae). Environ. Microbiol. Rep. 2010, 2, 619–624. [Google Scholar] [CrossRef] [PubMed]
- Taber, R.A.; Thielen, M.A.; Falkinham, J.O., III; Smith, R.H. Mycobacterium scrofulaceum: A bacterial contaminant in plant tissue culture. Plant Sci. 1991, 78, 231–236. [Google Scholar] [CrossRef]
- Goh, C.H.; Vallejos, D.F.V.; Nicotra, A.B.; Mathesius, U. The impact of beneficial plant-associated microbes on plant phenotypic plasticity. J. Chem. Ecol. 2013, 39, 826–839. [Google Scholar] [CrossRef]
- Ulrich, K.; Stauber, T.; Ewald, D. Paenibacillus—A predominant endophytic bacterium colonising tissue cultures of woody plants. Plant Cell Tissue Organ Cult. 2008, 93, 347–351. [Google Scholar] [CrossRef]
- Laukkanen, H.; Soini, H.; Kontunen-Soppela, S.; Hohtola, A.; Viljanen, M. A mycobacterium isolated from tissue cultures of mature Pinus sylvestris interferes with growth of Scots pine seedlings. Tree Physiol. 2000, 20, 915–920. [Google Scholar] [CrossRef]
- Taheri, S.; Lee, A.T.; Yusop, M.R.; Hanafi, M.M.; Sahebi, M.; Azizi, P.; Shamshiri, R.R. Mining and Development of Novel SSR Markers Using Next Generation Sequencing (NGS) Data in Plants. Molecules 2018, 23, 399. [Google Scholar] [CrossRef]
- Zhou, X.J.; Dong, Y.; Zhao, J.J.; Huang, L.; Ren, X.P.; Chen, Y.N.; Huang, S.M.; Liao, B.S.; Lei, Y.; Yan, L.Y.; et al. Genomic survey sequencing for development and validation of single-locus SSR markers in peanut (Arachis hypogaea L.). BMC Genom. 2016, 17, 420. [Google Scholar] [CrossRef] [PubMed]
- An, J.Y.; Yin, M.Q.; Zhang, Q.; Gong, D.T.; Jia, X.W.; Guan, Y.J.; Hu, J. Genome Survey Sequencing of Luffa Cylindrica L. and Microsatellite High Resolution Melting (SSR-HRM) Analysis for Genetic Relationship of Luffa Genotypes. Int. J. Mol. Sci. 2017, 18, 1942. [Google Scholar]
- Li, G.Q.; Song, L.X.; Jin, C.Q.; Li, M.; Gong, S.P.; Wang, Y.F. Genome survey and SSR analysis of Apocynum venetum. Biosci. Rep. 2019, 39, BSR20190146. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.; Guo, J.J.; Yin, M.Y.; Wang, H.; Dong, W.P.; Zeng, J.; Zhou, S.L. Next Generation Sequencing-Based Molecular Marker Development: A Case Study in Betula Alnoides. Molecules 2018, 23, 2963. [Google Scholar] [CrossRef] [PubMed]




| Medium | Basic Medium | Plant Hormone (mg/L) | Sucrose (g/L) | Agar (g/L) | pH | Others (g/L) |
|---|---|---|---|---|---|---|
| Dedifferentiation medium | WPM | NAA 0.02 6-BA 1.0 | 30 | 6.0 | 5.8~6.0 | -- |
| Redifferentiation medium | WPM | NAA 0.02 | 30 | 6.0 | 5.8~6.0 | -- |
| 6-BA 1.0 GA3 0.5 | ||||||
| Rooting medium | 1/2 MS | IBA 0.4 | 20 | 6.5 | 5.8~6.0 | Activated carbon 2.0 |
| NAA 0.02 |
| Library | 200 bp | 500 bp | 800 bp | 300 bp * |
|---|---|---|---|---|
| Raw reads length (bp) | 100 | 100 | 100 | 150 |
| Raw data (Gb) | 14.5 | 12.0 | 5.0 | 23.5 |
| Raw Q20 (%) | 96.9 | 94.4 | 86.5 | 94.8 |
| Clean reads length | 90 | 90 | 90 | 135 |
| Clean data (Gb) | 12.3 | 9.6 | 3.2 | 18.4 |
| Clean Q20 (%) | 99.1 | 98.6 | 97.4 | 97.7 |
| Species Name | Hit Number | Species Name | Hit Number |
|---|---|---|---|
| Mycobacterium chimaera | 229 | Cutibacterium avidum | 1 |
| Mycobacterium intracellulare | 187 | Mycobacterium gilvum | 1 |
| Mycobacterium marseillense | 186 | Mycobacterium vanbaalenii | 1 |
| Mycobacterium yongonense | 106 | Malus x | 1 |
| Mycobacterium indicus | 76 | Mycobacterium dioxanotrophicus | 1 |
| Mycobacterium avium | 26 | Kocuria palustris | 1 |
| Mycobacterium colombiense | 12 | Acidiphilium multivorum | 1 |
| Mycobacterium abscessus | 4 | Gluconobacter oxydans | 1 |
| Juglans regia | 4 | Mycobacterium ulcerans | 1 |
| Mycobacterium marinum | 4 | Mycobacterium sinense | 1 |
| Roseomonas gilardii | 2 | Theobroma cacao | 1 |
| Mycobacterium thermoresistibile | 2 | Mycobacterium lepraemurium | 1 |
| Xanthobacter autotrophicus | 1 | Prunus avium | 1 |
| Cajanus cajan | 1 | Paracoccus yeei | 1 |
| Mycobacterium liflandii | 1 | Glycine max | 1 |
| Mycobacterium rhodesiae | 1 | Prunus persica | 1 |
| Acidiphilium cryptum | 1 | Herrania umbratica | 1 |
| Rhodococcus jostii | 1 | Vitis vinifera | 1 |
| Item | Scaffold | Contig |
|---|---|---|
| No. of sequences | 79,580 | 97,097 |
| Total length (bp) | 250,090,936 | 249,878,124 |
| N50 length (bp) | 4312 | 3081 |
| N90 length (bp) | 1336 | 1244 |
| Max length (bp) | 85,240 | 85,240 |
| GC content (%) | 34.8 | 34.8 |
| Species Name | Hit Number | Align Length (bp) | Species Name | Hit Number | Align Length (bp) |
|---|---|---|---|---|---|
| Juglans regia | 8726 | 5,851,287 | Ziziphus jujuba | 89 | 51,320 |
| Vitis vinifera | 638 | 658,331 | Prunus persica | 89 | 70,721 |
| Theobroma cacao | 332 | 194,640 | Betula nana | 84 | 57,369 |
| Betula pendula | 263 | 667,040 | Malus x | 66 | 27,903 |
| Corylus avellana | 217 | 211,681 | Hevea brasiliensis | 65 | 36,133 |
| Betula platyphylla | 197 | 64,844 | Fragaria vesca | 55 | 26,960 |
| Betula luminifera | 191 | 98,488 | Prunus mume | 55 | 33,181 |
| Quercus robur | 162 | 110,642 | Pyrus x | 54 | 23,152 |
| Prunus avium | 141 | 77,758 | Glycine max | 54 | 29,659 |
| Populus trichocarpa | 135 | 83,113 | Citrus clementina | 43 | 29,869 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, S.; Chen, S.; Liu, C.; Liu, Y.; Zhao, X.; Yang, C.; Qu, G.-Z. Genome Survey Sequencing of Betula platyphylla. Forests 2019, 10, 826. https://doi.org/10.3390/f10100826
Wang S, Chen S, Liu C, Liu Y, Zhao X, Yang C, Qu G-Z. Genome Survey Sequencing of Betula platyphylla. Forests. 2019; 10(10):826. https://doi.org/10.3390/f10100826
Chicago/Turabian StyleWang, Sui, Su Chen, Caixia Liu, Yi Liu, Xiyang Zhao, Chuanping Yang, and Guan-Zheng Qu. 2019. "Genome Survey Sequencing of Betula platyphylla" Forests 10, no. 10: 826. https://doi.org/10.3390/f10100826
APA StyleWang, S., Chen, S., Liu, C., Liu, Y., Zhao, X., Yang, C., & Qu, G.-Z. (2019). Genome Survey Sequencing of Betula platyphylla. Forests, 10(10), 826. https://doi.org/10.3390/f10100826





