Diversity and Molecular Barcoding of Stink Bugs (Hemiptera: Pentatomidae) Associated with Macadamia in South Africa
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
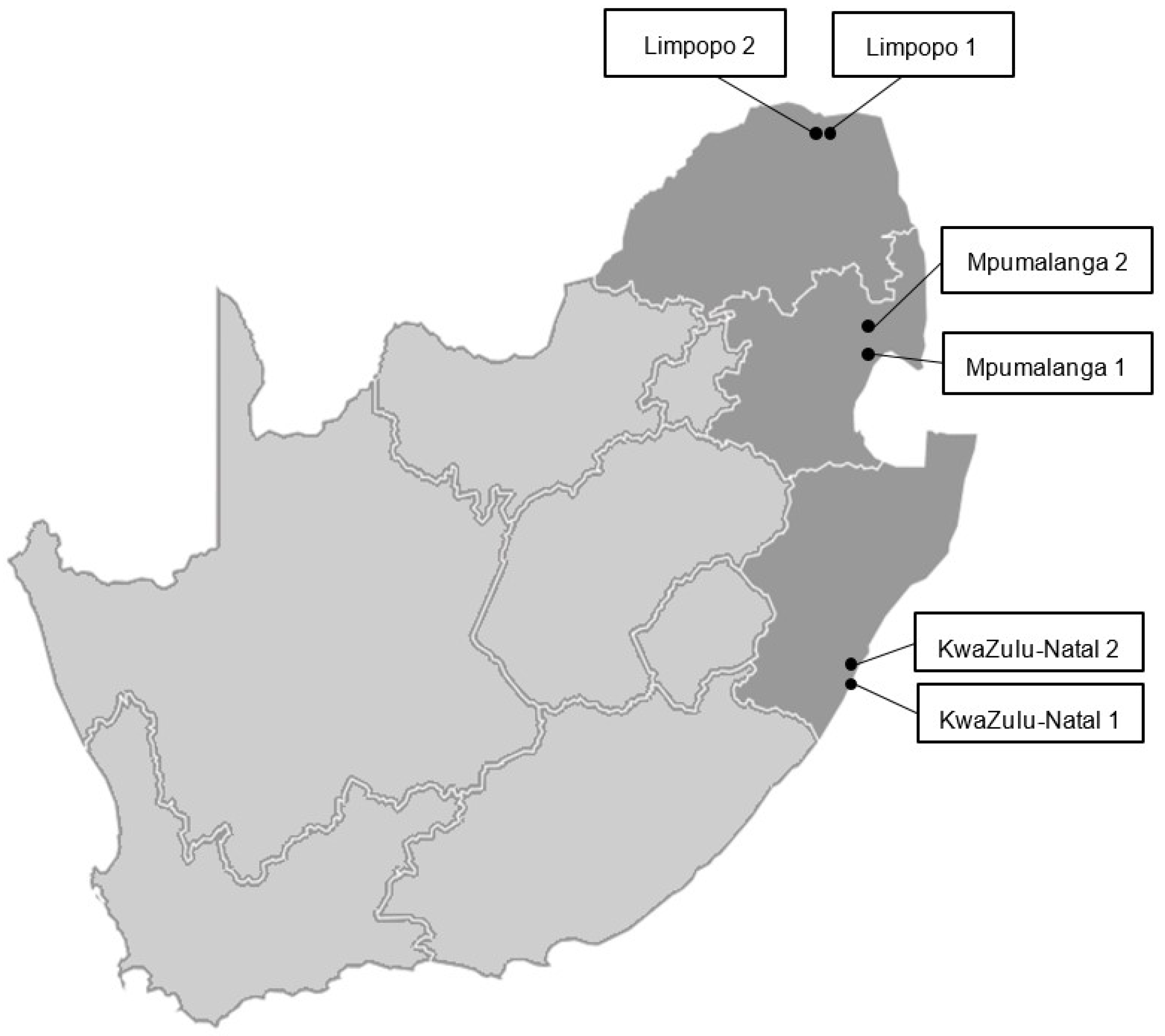
2.1. Insect Collections
2.2. Morphological Identification
2.3. DNA Extraction, Amplification and Sequencing
2.4. Phylogenetic Analyses
2.5. Sequence Divergence Analysis
3. Results
3.1. Morphological Identification
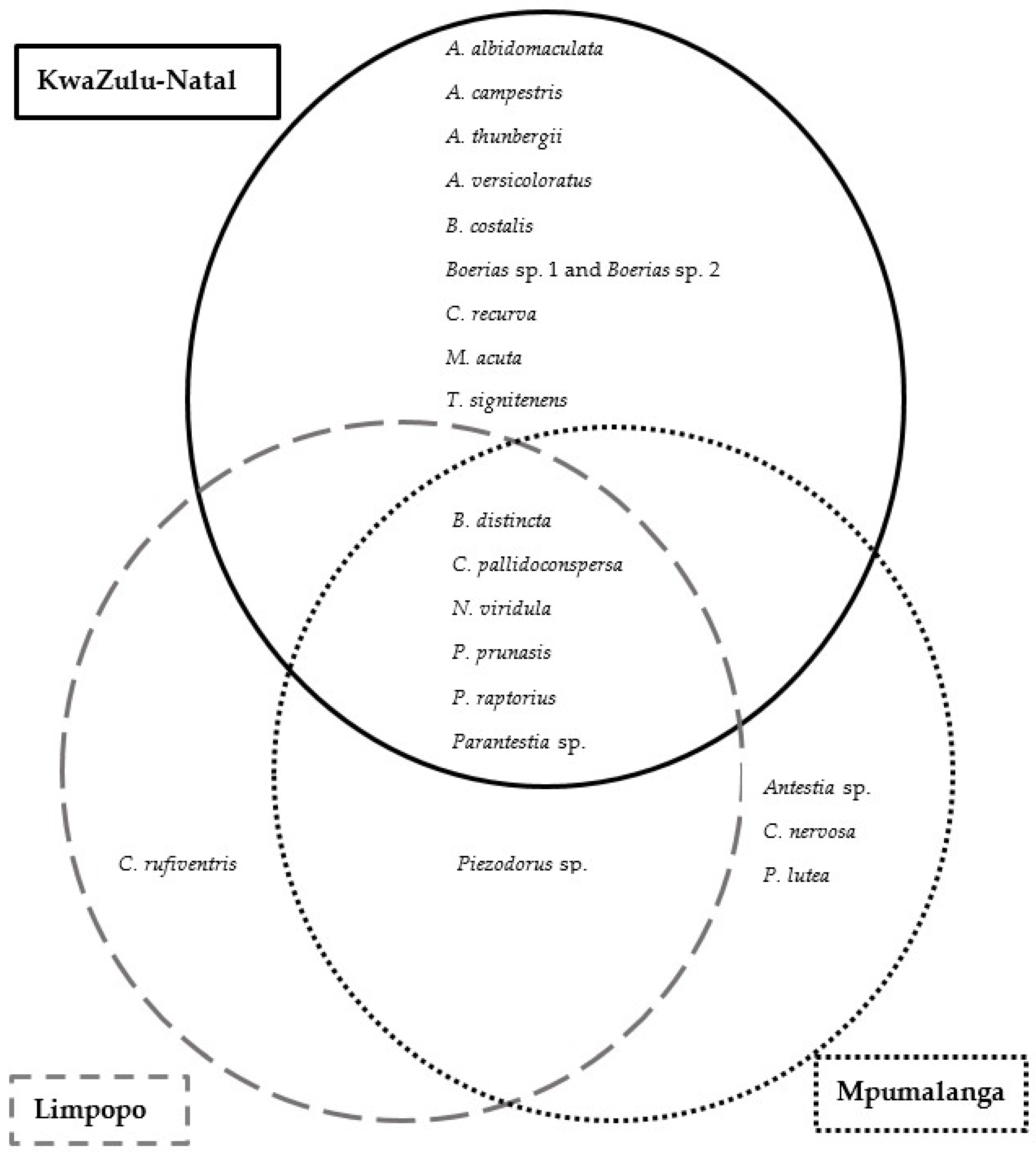
3.2. Species Composition
3.2.1. Species Composition across Growing Regions
3.2.2. Species Composition across Growing Seasons in Limpopo
3.3. Phylogenetic Analysis
3.4. Sequence Divergence Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- SAMAC SAMAC-Industry Statistics. Available online: www.samac.org.za/industry-statistics (accessed on 14 April 2021).
- McPherson, J.E.; Scott Bundy, C.; Wheeler, A. Introduction: Overview of the Superfamily Pentatomoidea. In Invasive Stink Bugs and Related Species (Pentatomoidea): Biology, Higher Systematics, Semiochemistry, and Management; McPherson, J.E., Ed.; Taylor & Francis: New York, NY, USA, 2018; pp. 3–24. [Google Scholar]
- McPherson, J.E.; McPherson, R. General Introduction to Stink Bugs. In Stink Bugs of Economic Importance in America North of Mexico; CRC Press: Boca Raton, FL, USA, 2000; ISBN 9780429115028. [Google Scholar]
- Jones, V.P.; Caprio, L.C. Southern Green Stink Bug (Hemiptera: Pentatomidae) Feeding on Hawaiian Macadamia Nuts: The Relative Importance of Damage Occurring in the Canopy and on the Ground. J. Econ. Entomol. 1994, 87, 431–435. [Google Scholar] [CrossRef]
- Panizzi, A.R.; McPherson, J.E.; James, D.G.; Jahavery, M.; McPherson, R. Stink Bugs (Pentatomidae). In Heteroptera of Economic Importance; Schaefer, C.W., Panizzi, A.R., Eds.; CRC Press: Boca Raton, FL, USA, 2000; pp. 421–474. ISBN 9780849306952. [Google Scholar]
- Tavella, L.; Arzone, A.; Miaja, M.L.; Sonnati, C. Influence of Bug (Heteroptera, Coreidae and Pentatomidae) Feeding Activity on Hazelnut in Northwestern Italy. Acta Hortic. 2001, 556, 461–467. [Google Scholar] [CrossRef]
- Daane, K.; Yokota, G.; Krugner, R.; Steffan, S.; da Silva, P.; Beede, R.; Bentley, W.; Weinberger, G. Large Bugs Damage Pistachio Nuts Most Severely during Midseason. Calif. Agric. 2005, 59, 95–102. [Google Scholar] [CrossRef] [Green Version]
- Rijal, J.; Gyawaly, S. Characterizing Brown Marmorated Stink Bug Injury in Almond, a New Host Crop in California. Insects 2018, 9, 126. [Google Scholar] [CrossRef] [Green Version]
- Schoeman, P.S. Relative Seasonal Occurrence of Economically Significant Heteropterans (Pentatomidae and Coreidae) on Macadamias in South Africa: Implications for Management. Afr. Entomol. 2018, 26, 543–549. [Google Scholar] [CrossRef]
- Schoeman, P.S. Phytophagous Stink Bugs (Hemiptera: Pentatomidae; Coreidae) Associated with Macadamia in South Africa. Open J. Anim. Sci. 2013, 3, 179. [Google Scholar] [CrossRef] [Green Version]
- van den Berg, M.A.; Steyn, W.P.; Greenland, J. Hemiptera Occurring on Macadamia in the Mpumalanga Lowveld of South Africa. Afr. Plant Prot. 1999, 5, 89–92. [Google Scholar]
- Taylor, P.J.; Grass, I.; Alberts, A.J.; Joubert, E.; Tscharntke, T. Economic Value of Bat Predation Services—A Review and New Estimates from Macadamia Orchards. Ecosyst. Serv. 2018, 30, 372–381. [Google Scholar] [CrossRef]
- Dent, D.; Binks, R.H. Insect Pest Management, 3rd ed.; CABI Publishing: Wallingford, UK, 2000. [Google Scholar]
- McPherson, J.E. Management. In Invasive Stink Bugs and Related Species (Pentatomoidea): Biology, Higher Systematics, Semiochemistry, and Management; McPherson, J.E., Ed.; Taylor & Francis: New York, NY, USA, 2018; pp. 729–774. ISBN 9781315371221. [Google Scholar]
- Jinbo, U.; Kato, T.; Ito, M. Current Progress in DNA Barcoding and Future Implications for Entomology. Entomol. Sci. 2011, 14, 107–124. [Google Scholar] [CrossRef]
- Emam, A.K.; Ibrahim, H.E.; Helmi, A.; Sharaf, A. Identification of Some Egyptian Leafhopper Species (Hemiptera: Cicadellidae) Using DNA Barcoding. Biologia 2020, 75, 1–10. [Google Scholar] [CrossRef]
- Ball, S.L.; Armstrong, K.F. DNA Barcodes for Insect Pest Identification: A Test Case with Tussock Moths (Lepidoptera: Lymantriidae). Can. J. For. Res. 2006, 36, 337–350. [Google Scholar] [CrossRef]
- Hebert, P.D.N.; Ratnasingham, S.; De Waard, J.R. Barcoding Animal Life: Cytochrome c Oxidase Subunit 1 Divergences among Closely Related Species. Proc. R. Soc. Lond. Ser. B Biol. Sci. 2003, 270, S96–S99. [Google Scholar] [CrossRef] [Green Version]
- Hebert, P.D.N.; Gregory, T.R. The Promise of DNA Barcoding for Taxonomy. Syst. Biol. 2005, 54, 852–859. [Google Scholar] [CrossRef]
- Grazia, J.; Schwertner, C.F. Review of Parachinavia Roche (Hemiptera, Pentatomidae, Pentatominae). In Advances In Heteroptera Research. Festschrift in Honour of 80th Anniversary of Michail Josifov; Pensoft Publishers: Sofia, Bulgaria, 2008; pp. 159–169. [Google Scholar]
- Ferrari, A.; Schwertner, C.F.; Grazia, J. Review, Cladistic Analysis and Biogeography of Nezara Amyot & Serville (Hemiptera: Pentatomidae). Zootaxa 2010, 2424, 1–41. [Google Scholar]
- Staddon, B.W.; Ahmad, I. Species Problems and Species Groups in the Genus Piezodorus Fieber (Hemiptera: Pentatomidae). J. Nat. Hist. 1995, 29, 787–802. [Google Scholar] [CrossRef]
- Salini, S.; Viraktamath, C.A. Genera of Pentatomidae (Hemiptera: Pentatomoidea) from South India-an Illustrated Key to Genera and Checklist of Species. Zootaxa 2015, 3924, 1–76. [Google Scholar] [CrossRef] [Green Version]
- Rolston, L.H. A Revision of the Genus Acrosternum Fieber, Subgenus Chinavia Orian, in the Western Hemisphere (Hemiptera: Pentatomidae). J. N. Y. Entomol. Soc. 1983, 91, 97–176. [Google Scholar]
- Kment, P.; Jindra, Z. A Revision of Tripanda and Tenerva (Hemiptera: Heteroptera: Pentatomidae: Pentatominae). Zootaxa 2009, 1978, 1–47. [Google Scholar] [CrossRef]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA Primers for Amplification of Mitochondrial Cytochrome c Oxidase Subunit I from Diverse Metazoan Invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar]
- Hall, T.; Biosciences, I.; Carlsbad, C. BioEdit: An Important Software for Molecular Biology. GERF Bull. Biosci. 2011, 2, 60–61. [Google Scholar]
- Park, D.-S.; Foottit, R.; Maw, E.; Hebert, P.D.N. Barcoding Bugs: DNA-Based Identification of the True Bugs (Insecta: Hemiptera: Heteroptera). PLoS ONE 2011, 6, e18749. [Google Scholar] [CrossRef]
- Gariepy, T.D.; Haye, T.; Fraser, H.; Zhang, J. Occurrence, Genetic Diversity, and Potential Pathways of Entry of Halyomorpha Halys in Newly Invaded Areas of Canada and Switzerland. J. Pest Sci. 2014, 87, 17–28. [Google Scholar] [CrossRef]
- Gariepy, T.D.; Bruin, A.; Konopka, J.; Scott-Dupree, C.; Fraser, H.; Bon, M.-C.; Talamas, E. A Modified DNA Barcode Approach to Define Trophic Interactions between Native and Exotic Pentatomids and Their Parasitoids. Mol. Ecol. 2019, 28, 456–470. [Google Scholar] [CrossRef]
- Raupach, M.J.; Hendrich, L.; Küchler, S.M.; Deister, F.; Morinière, J.; Gossner, M.M. Building-up of a DNA Barcode Library for True Bugs (Insecta: Hemiptera: Heteroptera) of Germany Reveals Taxonomic Uncertainties and Surprises. PLoS ONE 2014, 9, e106940. [Google Scholar] [CrossRef]
- Tembe, S.; Shouche, Y.; Ghate, H.V. DNA Barcoding of Pentatomomorpha Bugs (Hemiptera: Heteroptera) from Western Ghats of India. Meta Gene 2014, 2, 737–745. [Google Scholar] [CrossRef]
- Zhao, W.; Zhao, Q.; Zhang, H. The Determination of Doubted Species in Eurydema Based on DNA Barcoding (Heteroptera: Pentatomidae). J. Shanxi Agric. Univ. 2015, 35, 169–174. [Google Scholar]
- Gwiazdowski, R.A.; Foottit, R.G.; Maw, H.E.L.; Hebert, P.D.N. The Hemiptera (Insecta) of Canada: Constructing a Reference Library of DNA Barcodes. PLoS ONE 2015, 10, e0125635. [Google Scholar] [CrossRef] [Green Version]
- Tillman, P.G.; Greenstone, M.H.; Hu, J.S. Predation of Stink Bugs (Hemiptera: Pentatomidae) by a Complex of Predators in Cotton and Adjoining Soybean Habitats in Georgia, USA. Fla. Entomol. 2015, 98, 1114–1126. [Google Scholar] [CrossRef] [Green Version]
- Dhami, M.K.; Dsouza, M.; Waite, D.W.; Anderson, D.; Li, D. Real-Time PCR Assay for the Identification of the Brown Marmorated Stink Bug (Halyomorpha halys). Front. Mol. Biosci. 2016, 3, 5. [Google Scholar] [CrossRef] [Green Version]
- Barman, A.K.; Joyce, A.L.; Torres, R.; Higbee, B.S. Assessing Genetic Diversity in Four Stink Bug Species, Chinavia hilaris, Chlorochroa uhleri, Chlorochroa sayi, and Thyanta pallidovirens (Hemiptera: Pentatomidae), Using DNA Barcodes. J. Econ. Entomol. 2017, 110, 2590–2598. [Google Scholar] [CrossRef] [Green Version]
- Valentin, R.E.; Nielsen, A.L.; Wiman, N.G.; Lee, D.-H.; Fonseca, D.M. Global Invasion Network of the Brown Marmorated Stink Bug, Halyomorpha halys. Sci. Rep. 2017, 7, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gowande, G.G.; Tembe, S.; Ghate, H.V. Revisiting DNA Barcoding of True Bugs of the Infraorder Pentatomomorpha (Hemiptera: Heteroptera) from India. Mitochondrial DNA Part A 2018, 29, 1215–1223. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT Online Service: Multiple Sequence Alignment, Interactive Sequence Choice and Visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stamatakis, A. RAxML Version 8: A Tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Librado, P.; Rozas, J. DnaSP v5: A Software for Comprehensive Analysis of DNA Polymorphism Data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef] [Green Version]
- Nei, M. Molecular Evolutionary Genetics; Columbia University Press: New York, NY, USA, 1987. [Google Scholar]
- Bruwer, I.J. The Influence of Various Hemipteran Species on Macadamia and Some Factors Which Can Limit Nut Damage. Doctoral Thesis, Stellenbosch University, Stellenbosch, South Africa, 1992. Unpublished. [Google Scholar]
- Schoeman, P.S. Aspects Affecting Distribution and Dispersal of the Indigenous Heteroptera Complex (Heteroptera: Pentatomidae & Coreidae) in South African Macadamia Orchards. Afr. Entomol. 2014, 22, 191–197. [Google Scholar]
- Bosco, L.; Moraglio, S.T.; Tavella, L. Halyomorpha halys, a Serious Threat for Hazelnut in Newly Invaded Areas. J. Pest Sci. 2018, 91, 661–670. [Google Scholar] [CrossRef]
- Haddad, C.R.; Louw, S. Phenology and Potential Biological Control of the Stink Bug Atelocera raptoria Germar (Hemiptera: Pentatomidae) in Pistachio Orchards. Afr. Plant Prot. 2006, 12, 23–27. [Google Scholar]
- Joubert, P.H.; Neethling, C. Kernel Damage to Pecan Nuts in the Nelspruit Area of South Africa. J. S. Afr. Soc. Hortic. Sci. 1994, 4, 42–44. [Google Scholar]
- Schoeman, P.S.; Morey, L. Hemiptera (Pentatomidae & Coreidae) Occurring on Litchi chinensis Sonn. in the Nelspruit Region of the Mpumalanga Province of South Africa. Afr. Plant Prot. 2016, 19, 4–7. [Google Scholar]
- Follett, P.A.; Wright, M.G.; Golden, M. Nezara viridula (Hemiptera: Pentatomidae) Feeding Patterns in Macadamia Nut in Hawaii: Nut Maturity and Cultivar Effects. Environ. Entomol. 2009, 38, 1168–1173. [Google Scholar] [CrossRef] [Green Version]
- Esquivel, J.F. Stink Bug Rostrum Length vs. Stylet Penetration Potential. Entomol. Exp. Et Applicata 2019, 167, 323–329. [Google Scholar] [CrossRef] [Green Version]
- Esquivel, J.F. Estimating Potential Stylet Penetration of Southern Green Stink Bug-a Mathematical Modeling Approach. Entomol. Exp. Et Appl. 2011, 140, 163–170. [Google Scholar] [CrossRef]
- Schoeman, P.S. Damage Potential of Indigenous Heteroptera Species Occurring on Macadamia Nuts (Macadamia integrifolia Maiden & Betche & Macadamia tetraphylla L. Johnson) in South Africa during the Early and Late Season. Int. J. Trop. Insect Sci. 2020, 40, 217–219. [Google Scholar]
- Bruwer, I.J.; Giliomee, J.H.; Pringle, K.L. The Relationship between Proboscis Length and the Ability of Certain Heteroptera to Damage Macadamia Kernels. Afr. Entomol. 2021, 29, 112–124. [Google Scholar] [CrossRef]
- Habou, Z.A.; Adam, T.; Haubruge, E.; Mergeai, G.; Verheggen, F.J. Insects Associated with Jatropha curcas Linn. (Euphorbiaceae) in West Niger. J. Insect Sci. 2014, 14, 255. [Google Scholar] [CrossRef] [Green Version]
- Greathead, D.J. A Taxonomic Study of the Species of Antestiopsis (Hemipteea, Pentatomidae) Associated with Coffea arabica in Africa. Bull. Entomol. Res. 1966, 56, 515–554. [Google Scholar] [CrossRef]
- van Rooyen, A. An Ecological Analysis of Stink Bug and Lepidopteran Borer Complexes Associated with Pecan and Citrus Orchards in the Vaalharts Region. Masters Dissertation, University of the Free State, Bloemfontein, South Africa, 2017. Unpublished. [Google Scholar]
- Taylor, J.S. The Fruit-Piercing Moth, Achaea lienardi Boisduval (Lepidoptera: Noctuidae), in the Eastern Cape Province. J. Entomol. Soc. S. Afr. 1965, 28, 50–56. [Google Scholar]
- Kiritani, K. Studies on the Adult Polymorphism in the Southern Green Stink Bug, Nezara viridula (Hemiptera: Pentatomidae). Popul. Ecol. 1970, 12, 19–34. [Google Scholar] [CrossRef]
- Hebert, P.D.N.; Penton, E.H.; Burns, J.M.; Janzen, D.H.; Hallwachs, W. Ten Species in One: DNA Barcoding Reveals Cryptic Species in the Neotropical Skipper Butterfly Astraptes Fulgerator. Proc. Natl. Acad. Sci. USA 2004, 101, 14812–14817. [Google Scholar] [CrossRef] [Green Version]




| Stink Bug Species | Nucleotide Diversity (π) | % |
|---|---|---|
| Bathycoelia distincta | 0.00428 | 0.43 |
| Boerias sp. 1 | 0.04711 | 4.71 |
| Boerias sp. 2 | 0 | 0 |
| Chinavia pallidoconspersa | 0.00107 | 0.11 |
| Nezara viridula | 0.00257 | 0.26 |
| Parachinavia prunasis | 0.00107 | 0.11 |
| Parantestia sp. | 0.00257 | 0.26 |
| Piezodorus sp. | 0.00857 | 0.86 |
| Pseudatelus raptorius | 0.04283 | 4.28 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sonnekus, B.; Slippers, B.; Hurley, B.P.; Joubert, E.; Stiller, M.; Fourie, G. Diversity and Molecular Barcoding of Stink Bugs (Hemiptera: Pentatomidae) Associated with Macadamia in South Africa. Insects 2022, 13, 601. https://doi.org/10.3390/insects13070601
Sonnekus B, Slippers B, Hurley BP, Joubert E, Stiller M, Fourie G. Diversity and Molecular Barcoding of Stink Bugs (Hemiptera: Pentatomidae) Associated with Macadamia in South Africa. Insects. 2022; 13(7):601. https://doi.org/10.3390/insects13070601
Chicago/Turabian StyleSonnekus, Byron, Bernard Slippers, Brett P. Hurley, Elizabeth Joubert, Michael Stiller, and Gerda Fourie. 2022. "Diversity and Molecular Barcoding of Stink Bugs (Hemiptera: Pentatomidae) Associated with Macadamia in South Africa" Insects 13, no. 7: 601. https://doi.org/10.3390/insects13070601
APA StyleSonnekus, B., Slippers, B., Hurley, B. P., Joubert, E., Stiller, M., & Fourie, G. (2022). Diversity and Molecular Barcoding of Stink Bugs (Hemiptera: Pentatomidae) Associated with Macadamia in South Africa. Insects, 13(7), 601. https://doi.org/10.3390/insects13070601






