Abstract
Cigarette smoke (CS), a complex chemical mixture, contains more than 4,800 different compounds, including oxidants, heavy metals, and carcinogens, that individually or in combination initiate or promote pathogenesis in the lung accounting for 82% of chronic obstructive pulmonary disease (COPD) deaths and 87% of lung cancer deaths. Lysyl oxidase (LO), a Cu-dependent enzyme, oxidizes peptidyl lysine residues in collagen, elastin and histone H1, essential for stabilization of the extracellular matrix and cell nucleus. Considerable evidences have shown that LO is a tumor suppressor as exemplified by inhibiting transforming activity of ras, a proto oncogene. CS condensate (CSC), 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) and cadmium (Cd), major components of CS, down-regulate LO expression at such multiple levels as mRNA, protein and catalytic activity in lung cells in vitro and in vivo indicating LO as a critical intra- and extracellular target for CS pathogenesis in the lung. In view of multiple biological functions and regulation characteristics of the LO gene, molecular mechanisms for CS damage to lung LO and its role in emphysema and cancer pathogenesis are discussed in this review.
1. Introduction
Cigarette smoke (CS) is one of the most important risk factors for human disease, accounting for 82% of chronic obstructive pulmonary disease (COPD) deaths and 87% of lung cancer deaths [1–4]. Approximately 23% of the US adults currently smoke because of addiction to nicotine, an important component of CS. More than 440,000 Americans and approximately five million people in the World die prematurely each year as a result of tobacco use and tobacco smoke exposure [5]. Cigarettes kill more Americans than alcohol, car accidents, suicide, AIDS, homicide and illegal drugs combined [6].
Long-term exposure of humans to CS usually results in COPD in some people, often manifested as bronchitis, emphysema or both. Emphysema is a pathological lesion characterized by abnormal enlargement of the respiratory airspaces with destruction of the alveolar walls. Most studies of emphysema pathogenesis have focused on the “elastase-antielastase imbalance” hypothesis [7]. CS-activated macrophages and neutrophils release elastase that proteolyses elastin fibers contributing to lung injury [8,9]. However, this hypothesis cannot explain all aspects of the emphysema pathology. For example, the CS-induced emphysematous lung in guinea pigs exhibited breakdown of collagen without significant changes in elastin [10]. Transgenic mice that over-express the human collagenase gene developed early onset emphysema [11]. Thereby, the “elastase-antielastase imbalance” is not the only mechanism for emphysema pathogenesis.
Strong evidence supports the association of CS with lung cancer. CS is responsible for 772,000 and 265,000 new lung cancer cases in men and women, respectively, each year worldwide [12]. Passive smoke, also known as sidestream smoke (SS), environmental tobacco smoke (ETS) or second-hand smoke, may have more carcinogenic potentials than the mainstream smoke (MS) since it contains larger quantities of organic chemical compounds [13]. Cessation of smoking sharply declines pulmonary carcinogenesis in ex-smokers after approximately five years since quitting in comparison to continuous smokers [12], indicating CS as an important risk factor for lung cancers. Smoke-induced lung malignancies are mainly classified as squamous cell carcinoma, small cell carcinoma, adenocarcinoma, etc. [12]. Note that in addition to the lung, CS can also act as a critical cancer risk factor involved in other organs such as the larynx, bladder, esophagus, pancreas, kidney, oral cavity, etc. [12]. In light of its severe insults to the lung and other organs, CS is of a continuing human health concern. Thus, it is important and necessary to deeply investigate molecular mechanisms and preventing and therapeutic strategies for CS-induced lung diseases.
2. Major Pathogenic Agents of CS
2.1. CS, a Complex Chemical Mixture
CS contains more than 4,800 different compounds, including oxidants, heavy metals, and carcinogens, that individually or in combination initiate or promote pathogenesis in the lung [14,15]. Each puff of smoke produces 1014–1016 free radicals inducing oxidative damage to the lung [16]. The mainstream smoke coming from a burning cigarette is composed of a vapor phase (92%) and a particulate phase (8%). The major constituents of the vapor phase include carbon monoxide, ammonia, nitrogen oxides, hydrogen cyanide, isoprene, butadiene, formaldehyde, acetaldehyde, etc. The particulate phase or CS condensate (CSC) contains at least 3,500 compounds. There are 5 × 109 particles/mL with diameters of 0.1–1 μm in the mainstream CS aerosol in comparison to 105/mL aerosol particles with diameters 0.1–>10 μm in urban air. Thus, all of the particles in CS are within the size range reaching the alveolar space, known to be 0.1–3 μm [14]. Although the chemical composition of both MS and SS are quantitatively similar they still exhibit significant quantitative variety because of the temperature differences at which MS and SS are formed. Inhaled fresh sidestream cigarette smoke is approximately four times more toxic per gram total particulate matter (TPM) than mainstream cigarette smoke [17]. Based on our research focuses, two important classes of toxic/carcinogenic components in CSC such as the nicotine-derived nitrosamines, e.g., 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK), and heavy metals, e.g., cadmium (Cd) [14,15] will be discussed in this review.
2.2. NNK and Cd, Two of Major Pathogenic and Carcinogenic Components of CS
NNK, a tobacco-specific nicotine-nitrosated derivative, has been demonstrated as a very potent carcinogen in rodents, particularly in rats [18]. Its sales-weighted concentration in one current “full-flavored cigarette” is 131 ng [19]. The estimated life time exposure to NNK of a cigarette smoker is about 1.1 mg/kg. close to the lowest NNK dose (1.8 mg/kg) that induced tumors in rats [18,20]. NNK exhibits a remarkable affinity for the lung, resulting in adenoma and adenocarcinoma, independently of the route of administration [18]. Metabolic activation of NNK by cytochrome P-450 (CYP) is required to exert its carcinogenic activity [21]. Although it is established that NNK damages DNA via formation of methyl adducts [22,23] and mutation of p53, a tumor suppressor gene [24], the intracellular events elicited by CS/NNK and their underlying pathological mechanisms leading to cell injury and cancer development remain unclear.
Cd is a toxic metal without any biological availability. In addition to occupational exposure, cigarette smoke constitutes a major source of Cd exposure for humans since tobacco naturally absorbs mobile forms of Cd, e.g., CdCl2, CdSO4, Cd(NO3)2, Cd(CH3COO)2, etc., from soil and accumulates relatively high concentrations of Cd in the leaves [25,26]. The lung, a major CS-target organ, not only absorbs but also accumulates Cd with a biological half-life of 9.4 years [25,27]. Each cigarette contains 1–2 μg Cd, of which 10% is inhaled [25,26]. Pulmonary Cd levels in smokers reached 7.5-fold greater than those in non-smokers [28]. Long-term exposure to Cd resulted in emphysema [29–31]. Fatal emphysema developed in Cd-poisoned patients who had survived acute lung injuries in industrial accidents [32]. Clinical correlation studies have established a causal relationship between smoking and human emphysema [33]. Smokers with severe emphysema have a high Cd levels in their lungs [25,28]. Animal studies indicated that the nature of Cd-induced lung lesions varied with the exposure protocols and species used. Both emphysema and fibrosis with and without airspace enlargement were produced in rodents [7,34]. Cd was listed by the US Environmental Protection Agency as one of 126 priority pollutants [35,36]. Cd is a potent human carcinogen and occupational exposure to Cd is associated with cancers in multiple organs such as the lung, the prostate, the pancreas, the kidney, etc. Because of its characteristics as a lung carcinogen, Cd was classified as a category 1 carcinogen by the International Agency for Research on Cancer (IARC) and the National Toxicology Program of the USA [25,36]. Animal studies provide strong evidence for Cd carcinogenesis. With the rodent model, mast relevant to human exposure to air-borne Cd, various Cd compounds produced adenocarcinomas in the lung of rats after inhalation [37,38]. Due to its negative results in bacterial mutagenic tests, Cd is predominantly considered as a non-genotoxic carcinogen [25], thus, the mechanism(s) of Cd carcinogenesis remain poorly understood.
Recent studies by this lab have shown that CSC down-regulates lysyl oxidase (LO), an intra- and extracellular enzyme with multiple biological functions, at mRNA, protein and catalytic levels [39–41]. This review intends to summarize our current findings indicating LO as a critical intra- and extracellular target for CS pathogenesis in the lung.
3. Lysyl Oxidase
3.1. Multiple Functions of LO in Biology
LO initiates the crosslinking of collagen and elastin by oxidizing specific peptidyl lysine residues in these proteins stabilizing collagen or elastin as fibers in the extracellular matrix (ECM) [41]. Thus, LO plays a central role in ECM morphogenesis and tissue repair. LO can also oxidize lysine residues in various globular proteins with basic isoelectric points (pI > 8), e.g., nuclear proteins histone H1 and H2 [42,43], bFGF [44], etc., suggesting that electrostatic relationships between the anionic enzyme (pI ~ 6.0) and basic protein substrates are critical to substrate potential. In addition to extracellular secretion, LO is also localized within the nucleus [45] where it appears to modulate the packing state of nuclear chromatin [46]. Labeled 32 kDa mature LO readily enters the intracellular compartments and accumulates within the nucleus in vascular smooth muscle cells (VSMC) although mechanistic features of its intracellular transport into and through the cytosol are not known [47]. The 32 kDa purified LO displays chemotactic activities for monocytes and VSMC [48–50]. LO, as an extracellular signal, stimulates stress fiber formation and focal adhesion assembly [50] possibly implicating in mechanisms for tumor metastasis in hypoxia conditions [51]. The LO reaction produces H2O2 which may regulate gene expression and cell behavior [50]. LO is considered as a tumor suppressor gene initiating from the finding that expression of transfected LO cDNA suppressed Ha-ras-induced cell transformation indicating a ras-suppressor effect of LO [52]. Our previous studies have demonstrated bFGF as a substrate of LO. Oxidation of bFGF by LO blocked the proliferation of bFGF-stimulated normal cells and bFGF-autocrine transformed cells with high tumorigenic potential [44]. Based on the widening intra- and extracellular distribution of possible substrates of LO, this enzyme may display multiple functions in biology beyond its role in stabilization of macromolecules in the ECM.
3.2. LO Molecular Structure
As shown in Figures 1A and 1B, LO is synthesized by fibrogenic cells as a 46 kD preproenzyme. Following signal peptide cleavage at the CA site (SPCS in Figure 1B) and N-glycosylation at the NRT site (GS in Figure 1B), the resulting 50 kD proenzyme is secreted and then proteolytically cleaved at the GDD site (PPCS in Figure 1B) to produce the 32 kD functional species in the extracellular space [41]. The sequences within the mature enzyme at which the active site (AS in Figure 1B) is located are highly conserved (95%) in rat and human (Figure 1B). LO specifically binds 1 g-atom of Cu(II) at its active site per mole of enzyme [41]. Five histidine residues located at the sequence of His 283–His 297 in rat LO (CuBS in Figure 1B) constitute the copper-binding motif. Cu binding to proLO occurs in the enzyme secreted pathway such as the Golgi apparatus [53]. In addition to Cu, LO also contains a covalently bound carbonyl cofactor identified as lysine tyrosylquinone (LTQ) deriving from Lys 314 and Tyr 349 (bold letter in Figure 1B) based on the sequence of rat LO [54]. This peptidyl orthoquinone functions as a transient electron sink during amine oxidation. The precise role of Cu in the LO action remains poorly understood. This metal ion may be essential for maintaining the structural integrity of LTQ and/or the protein [41].
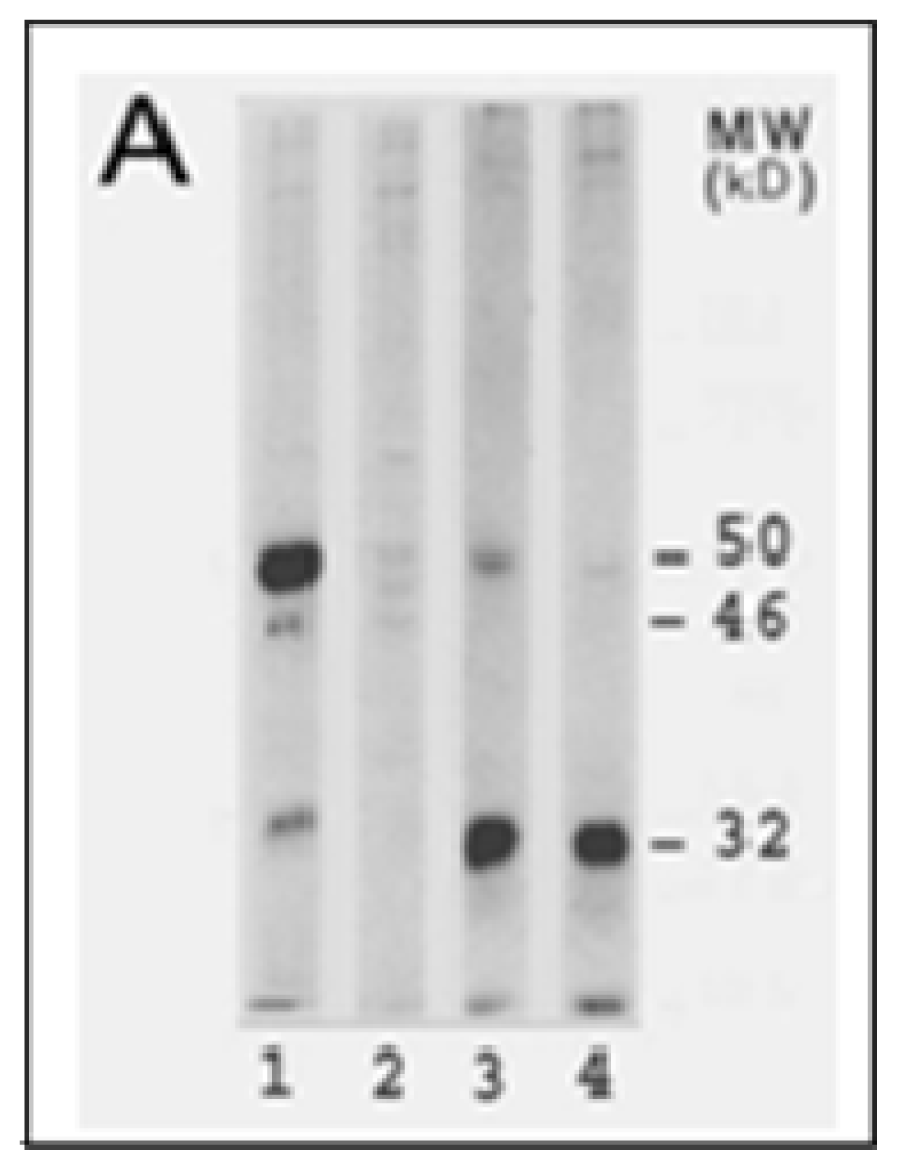

Figure 1.
LO synthesis, processing and molecular composition. A, LO synthesis and processing. LO synthesis and processing were determined by [35S]-methionine labeling and cycloheximide chase (5 h) assays. 1 and 3, 0 time chase; 2 and 4, 5 h chase; 1 and 2, cell extracts; 3 and 4, conditioned media. Note that at 0 time chase the cell extract contains the 46 kDa prepro-LO, the 50 kDa pro-LO, and the membrane-bound 32 kDa mature LO while the conditioned medium contains the secreted 50 kDa pro-LO and the processed 32 kDa mature-LO; at 5 h chase all LO species in the cell extract were processed and secreted into the extracellular space, and further cleaved to the 32 kD mature enzyme. B, amino acid sequences of rat and human LO. SPCS, the signal peptide cleavage site; GS, the N-glycosylation site; PPCS, the propeptide cleavage site; AS, the active site; CuBS, the copper-binding site; asterisks indicate the different amino acid composition of LO between human and rat.
The activity of purified LO can be severely inhibited by metal chelators, e.g., α,α′-dipyridyl [55]. The reaction with peptidyl lysines catalyzed by LO is: RCH2NH2 + H2O + O2 → RCHO + NH3 + H2O2 [41]. Additional LO-like cDNA species encode a variety of LO-like proteins, including LOXL, LOXL2, LOXL3 and LOXL4 [56] which have the homology at the C-terminal domain in comparison to the mature LO. Both LO and LO-like proteins at the C-terminal domain contain ten conserved cysteine residues, copper binding domains as well as tyrosine and lysine residues for forming the LTQ cofactor. Notably, LO also contains a growth factor and cytokine binding domain with the consensus: C-x9–10-C-x-W-x26–32-C-x10–15-C (C is Cys, W is Trp, xn is a defined number of any amino acids) found in a superfamily of receptors for growth factors and cytokines evolved from fibronectin III sequence modules common in cell surface adhesion molecules [56–58].
3.3. LO Gene Core Promoter
In eukaryotic cells, the gene activation is regulated by the interaction of cis-elements with various protein factors at the gene promoter region. The RNA polymerase II (RNA-Poly II) complex binds to the core promoter initiating the gene transcription. Comparative LO gene sequence alignment across species of rat, human and mouse showed that the proximal promoter region is highly conserved whereas the distal promoter region was more divergent among three species suggesting a fundamental role of the proximal promoter region in the regulation of LO transcription. The maximal promoter activities were detected in the 804, 808 and 924 base pair regions, respectively, immediately upstream of ATG in rat, mouse and human LO genes [59]. Thus, the LO transcription control is likely to be regulated by similar elements in these species. The 5′-flanking region of the rat LO gene appears to contain no typical TATA box, the first core promoter element identified in RNA-Poly II-transcribed genes. Multiple transcription start sites cluster in the rat LO promoter region from −78 to −51 relative to ATG, one of which overlaps with the adenosine residue of the INR element (5′-TCATTTTT-3′) mapped from −53 to −46 in the rat LO promoter [60,61]. Furthermore, the sequence from −18 to −14, i.e., 5′-GGACG-3′, a consensus sequence of the DPE that is located approximately 30 bp after the adenosine residue in the INR motif [61]. The INR and the DPE conserved in rat, human and mouse species coordinately function as a single core promoter unit for the RNA-Poly II-directed gene transcription [61]. It should be noted that the DPE in the rat LO gene promoter also worked as an independent core promoter module playing a key role in transactivation of the TATA-less LO gene at multiple transcription starts sites [59].
3.4. Redox-sensitive Cis-elements—the NFI-binding Site, HRE, MRE and ARE in the LO Promoter
Other transcription factor binding sites (cis-acting elements) are generally located upstream of the core promoter and act through binding of different transcriptional activators and/or repressors to alter the level of gene transcription. In addition to putative binding sites for the following transcription factors such as nuclear factor I (NFI), SP1, Oct-1, C/EBPγ, etc., the cloned rat LO promoter also contains the hypoxia response element (HRE), the metal response element (MRE) and antioxidant response element (ARE), etc. [59]. The cloned rat LO promoter −804/−1 yielding the maximal promoter activity contains three putative NFI-binding sites [59]. NFI binds to the consensus sequence TTGGC(N5)GCCAA (N = any nucleotides) on duplex DNA as a dimer. Notably, it can also bind to the individual half site, TTGGC or GCCAA, with a somewhat reduced affinity [62]. The highly conserved N-terminal residues of NFI contain the DNA binding domain whereas the proline-rich C-terminal residues constitute the transcriptional regulation domain [62]. Four cysteine residues are conserved in the DNA binding domain of all rat NFI isoforms sensitive to oxidative damage [63,64]. In addition, the rat LO promoter region −804/−1 also contains four putative hypoxia response elements (HRE, core sequence = RCGTG, R = purine) [65,66] including one on the coding strand and three on the noncoding strand at the region −457/−453, −387/−383, −194/−190 and −37/−33 (relative to ATG) [59], two putative metal response elements (MRE, core sequence = TGCRCNC, R = purine, N = any nucleotide) [67–69] located at −269/−263 and −248/−241, and one antioxidant response element (ARE, core sequence = RTGACNNNGC, R = purine, N = any nucleotides) [70] at the region −581/−572 [59]. The HIF1 is composed of HIF1α and HIF1β subunits [71]. The HIF1β is constitutively expressed whereas HIF1α is maintained at a low level in normoxic cells. Upon hypoxia, HIF1α is upregulated and HIF1 complexes bind to HREs transactivating hypoxia-inducible genes [65]. HIF1α has a unique domain sensitive to oxygen-dependant degradation. In normaxia, specific degradation of HIF1α is triggered through this domain by the proteasome [66]. Hypoxia is an important complication associated with lung diseases and tumors [72]. Reduced expression of HIF1α has been detected in the emphysema lung tissues in severe COPD patients [73], suggesting deregulation of HRE activities occurred in HIF1α targeting genes such as LO under this condition. Since cobalt chloride (CoCl2) stabilizes the HIF1α protein, this metal ion is used to mimic effects of hypoxia in cell study systems [74,75]. The MRE was initially found in multiple copies in metallothionein (MT) genes. MTs are cysteine-rich metal binding proteins essential for cellular metal metabolism and detoxification [68,69]. A protein that binds specifically to MREs is termed as MRE-binding transcription factor-1 (MTF-1) [67]. MTF-1 is a Zn finger transcription factor mediating the expression of MT genes which contain more copies of MREs. Cd and oxidative stress (such as H2O2) activate MTF-1 binding to the MT-I gene promoter by binding to or oxidation of other cellular protein sulfhydryls releasing bound Zn that in turn enhances MTF-1 affinity for DNA, thus activating MRE containing genes [67]. The transcriptional factor interacted with the ARE is the NF-E2-related factor-2 (Nrf2) which drives expressions of a variety xenobiotic metabolizing enzymes such as GST, NQO1, UGT, GCL, HO-1, GSH synthetase, γ-glutamyl transpeptidase, etc., for antioxidant-detoxification [70]. Nrf2 is inactive in the cytoplasm by binding to the cysteine-rich Keap1 protein in the actin filaments. Upon oxidative stress, modification of cysteine residues in the Keap1 protein induces the Nrf2 release and nuclear translocation. After forming the heterodimer with Maf, Nrf2 binds to the ARE for transcriptional activation of the genes. CS exposure has been shown to generate reactive oxygen species (ROS) [16]. Transcription factors for the NFI binding site, HRE, MRE and ARE, are known sensitive to oxidative stress [62,66,69,70]. These redox-sensitive transcription factors will be addressed as potential targets for CS insult. A schematic linear map of the cloned rat LO promoter is shown in Figure 2.
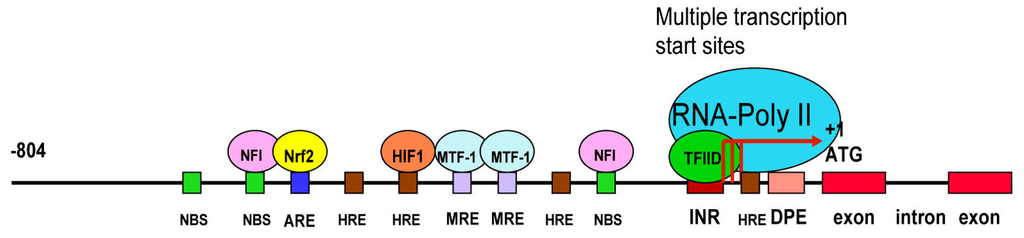
Figure 2.
The schematic linear map of the cloned rat LO promoter. ATG, the translational start site; INR, the initiator element; DPE the down-stream core promoter element; NBS, the NFI binding site; HRE, the hypoxia response element; MRE, the metal response element; ARE, the antioxidant response element.
4. LO as a Target of CS at Multiple Molecular Levels
4.1. Inactivation of LO Catalytic Activity by CSC and Cd
Previous studies by this lab have shown inhibition by CSC and Cd, major components of CS, of LO catalytic activity in treated cells [40,76,77]. LO is a metalloenzyme requiring Cu(II) as a cofactor for enzymatic function [41,55]. Dietary deprivation of Cu induced lathyritic injuries in animals due to suppression of LO activity [78–80]. It is known that thiol-containing MT is a critical modulator for Cu metabolism [81,82]. MT can chelate 11–12 moles of Cu/mole of protein [81] which are in the cuprous state [Cu(I)]. MT has a higher affinity for Cu than Cd since Cd selectively displaced Zn but not Cu in native calf liver MT, which binds four moles of Zn and three moles of Cu per mole of MT [83]. In addition to MT, glutathione (GSH) also functions in cellular Cu metabolism. GSH can form a complex with Cu-MT, providing the biological basis for Cu transfer between these two cellular thiols [82]. Reduction of Cu(II) to Cu(I) by GSH is thought to be a preliminary step in cellular transport of Cu. Cellular concentrations of GSH range from 0.1 to 10 mM, accounting for 90% of total cellular non-protein thiols. A rapid depletion followed by a later rebound increase of intra-cellular GSH was shown to occur in CS-treated lung cells and tissues consistent with marked increases of total GSH concentration in epithelial lining fluid (ELF) in cigarette smokers [84–87]. Our studies have illustrated CSC and Cd enhancement of GSH levels in cultured lung cells and in lung tissues of the animal model, and upregulation of the catalytic unit of γ-glutamylcysteine synthetase (GCS), a rate-limiting enzyme for GSH synthesis, at protein and mRNA levels indicating the activation status of GSH biosynthesis under CSC and Cd stress conditions [40,76,88]. Markedly elevated levels of MT and GSH were associated with considerably decreased catalytic levels of LO in CSC or Cd-treated RFL6 lung fibroblasts [40,76,77]. In light of the function of MT and GSH in Cu metabolism, CSC or Cd-induced elevation of MT and GSH in treated cells may largely trap cellular Cu, thus limiting its biological availability for LO leading to downregulation of LO activity that requires Cu as a cofactor [40,76,77]. Notably, isotope labeling assays have demonstrated that in MT-overexpressing RFL6 cells following long-term Cd exposure, radioactivity of 64Cu bound to the LO fraction amounted to only 9% of the control as compared to 1,400% of the control of 64Cu associated with the MT fraction [77]. Direct correlation of elevated levels of cellular thiols with downregulation of LO in CSC or Cd treated cells came from experiments in which RFL6 cells were exposed to GSH monoethyl ester (GME), a GSH delivery system [40], or transfected with MT cDNA [88]. Clearly, elevation of cellular GSH and MT by these molecular approaches was associated with reduction of LO activity in cell conditioned media. These results provide evidence directly linking perturbation of the homeostasis of cellular thiol and copper to the reduction of LO catalytic activity in model cells in response to CSC or Cd.
4.2. Inhibition of LO Protein Synthesis and Processing by CSC and Cd
CSC and Cd reduced levels of all LO protein species including the 46 kDa preproenzyme, the 50 kDa proenzyme and the 32 kDa mature enzyme in cultured lung fibroblasts [40,76,77]. This may result from upstream mechanisms such as decreases in LO mRNA. Notably, a stronger inhibition of downstream LO species, i.e., the preproLO → the proLO → the mature LO, in cells treated with CSC under the same conditions, suggests that several steps in LO translational and posttranslational processing may be susceptible to CSC [40]. A major difference between Cd-resistant (CdR) RFL6 cells derived from long-term Cd exposure and parental RFL6 cells is the presence of the variant 52 kDa LO precursor in the former. The 52 kDa protein existed in the cell extract fraction but not the conditioned media, reflecting deficiency of secretion [76]. As revealed by double immunofluorescent staining, a number of large LO positive patches colocalized with the ER and nucleus structures in CdR cells, indicating an abnormal processing of LO precursor with its product accumulated in these organelles [76,77]. Since this variant LO precursor is apparently 2 kDa larger than the normal LO precursor, it may represent a molecular species without cleavage of the signal peptide, known to be a total of 21 amino acids in rat LO (Figure 1B). Another possibility to account for the variant LO proenzyme with a higher MW in CdR cells may be a different degree of N-glycosylation. It should be noted that the same 52 kDa protein recognized by the anti-LO antibody was also found in CdR-3T3 cells derived from Swiss 3T3 fibroblasts using the same Cd exposure regimen [89]. These results suggest that the variant proenzyme derived from abnormal LO processing at translational and posttranslational levels may represent an important characteristic of long term Cd exposure.
4.3. Downregulation of LO Transcription and Posttranscription by CSC/NNK and Cd
Both CSC and Cd decreased the steady-state level of LO mRNA in treated cells [39,40,76]. The steady-state concentrations of mRNA as determined by Northern blot are dependent on both the transcriptional rate and the mRNA stability. CSC treatment induced decreased LO transcriptional rates as determined by the nuclear run-on assay and enhanced LO mRNA instability as illustrated by the actinomycin D chase assay [39]. The data further indicated that lower doses of CSC (<40 μg/mL) were sufficient to perturb the LO mRNA stability whereas higher doses of CSC (>80 μg/mL) were required to interfere with its transcriptional initiation. Thus, inhibition of the transcriptional rate and the mRNA stability both collectively contributed to decreased levels of steady-state LO transcripts in cells exposed to CSC.
To further investigate CSC modulation of LO transcription, an approximately 4 kb genomic fragment spanning nucleotides from −3,979 to −1 (Prom −3,979) was inserted in front of the luciferase gene in the reporter gene vector. A series of 5′-deletions of the Prom -−3,979 indicated that the maximal luciferase activity was found in the 804 base pair region immediately upstream of ATG [59]. CSC significantly inhibited LO promoter-directed luciferase expression in treated cells consistent with its effects on the LO transcriptional initiation [39]. Redox-sensitive transcription factors such as NFI, HIF1, MTF-1, etc., are critical targets for CSC or Cd. CSC/NNK or Cd suppressed biological activities of the NFI binding sites, HRE, MRE except the ARE at the LO promoter region in interactions with corresponding transcription factors revealed by reporter gene activation, transcription factor cDNA expression and ChIP assays (Figures 3 and 4, and references [59,90,91]). At least two functional NFI binding sites (−594/−580 and −147/−133) are active in the 804 base pair promoter region of the LO gene. CSC inhibited NFI-binding site-directed luciferase activities in transfected cells in a dose-dependent manner and abolished the reporter gene activation by NFI A and B, two isoforms expressed in rat lung fibroblasts. The ChIP assays further confirmed CSC interferences with endogenous NFI incorporation with the LO gene in the RFL6 cell model [59,90]. Electrophoretic mobility shift, oligonucleotide competition and in vitro translation of HIF 1α assays indicated that only one HRE mapped at −387/−383 relative to the ATG was functionally active among 4 consensuses within the LO promoter region −804/−1 upstream of ATG. Cd enhanced cellular levels of reactive oxygen species (ROS) [92], thus inhibited HIF1α expression and binding to the LO gene in treated cells in the presence of CoCl2 (Co) that mimics hypoxia conditions in cell culture system revealed by ChIP and qPCR (Figure 3 and reference [91]). Cd significantly suppressed HIF1α mRNA expression in Cd-pulsed RFL6 cells and in Cd resistant cells stimulated by Co [91]. The cloned rat LO promoter (−804/−1 relative to ATG) also contains at least two MREs located at −269/−263 and −248/−241 and one ARE mapped at −581/−572. Unexpected, treatment of cells with Cd, a heavy metal, weakened but not enhanced MTF-1 binding to the MREs in the LO promoter thus inhibited LO mRNA expression (Figure 4A). Note that upregulation of MT as a major phenotypic change was accompanied by downregulation of LO in CSC or Cd treated cells [40,76] and both MT and LO promoters contain MREs. Different phenotype changes in LO and MT genes in response to Cd may be explained by that many MRE-related transcription factors would be recruited to bind to MREs in the MT promoters for gene activation thus competing with their binding to the LO promoter. As reported, MT-I and MT-II genes each contain six MREs in their promoter regions, respectively, which display the different affinities for MTF-1 [67]. Apparently, LO MREs may have lower affinity than MT MREs for MTF-1 binding. In contrast to the LO MREs in response to Cd, the LO ARE is activated in Cd treated cells as evidenced by enhancement of Nrf2 binding to the LO ARE as examined by the ChIP assay (Figure 4B). Presumably, elevated ARE activities may play a critical role in maintaining the low basal level of the LO gene transcription under Cd stress conditions. NNK, a tobacco-specific carcinogen, is believed to contribute to the cancer burden in cigarette smokers [18]. Like Cd, NNK also inhibited activities of the LO MREs, but enhanced activities of the LO ARE (Figures 4A and 4B; reference [93]). NNK and Cd both have been shown to enhance methylation of CpG at the core promoter region of the LO gene (Figure 4E).
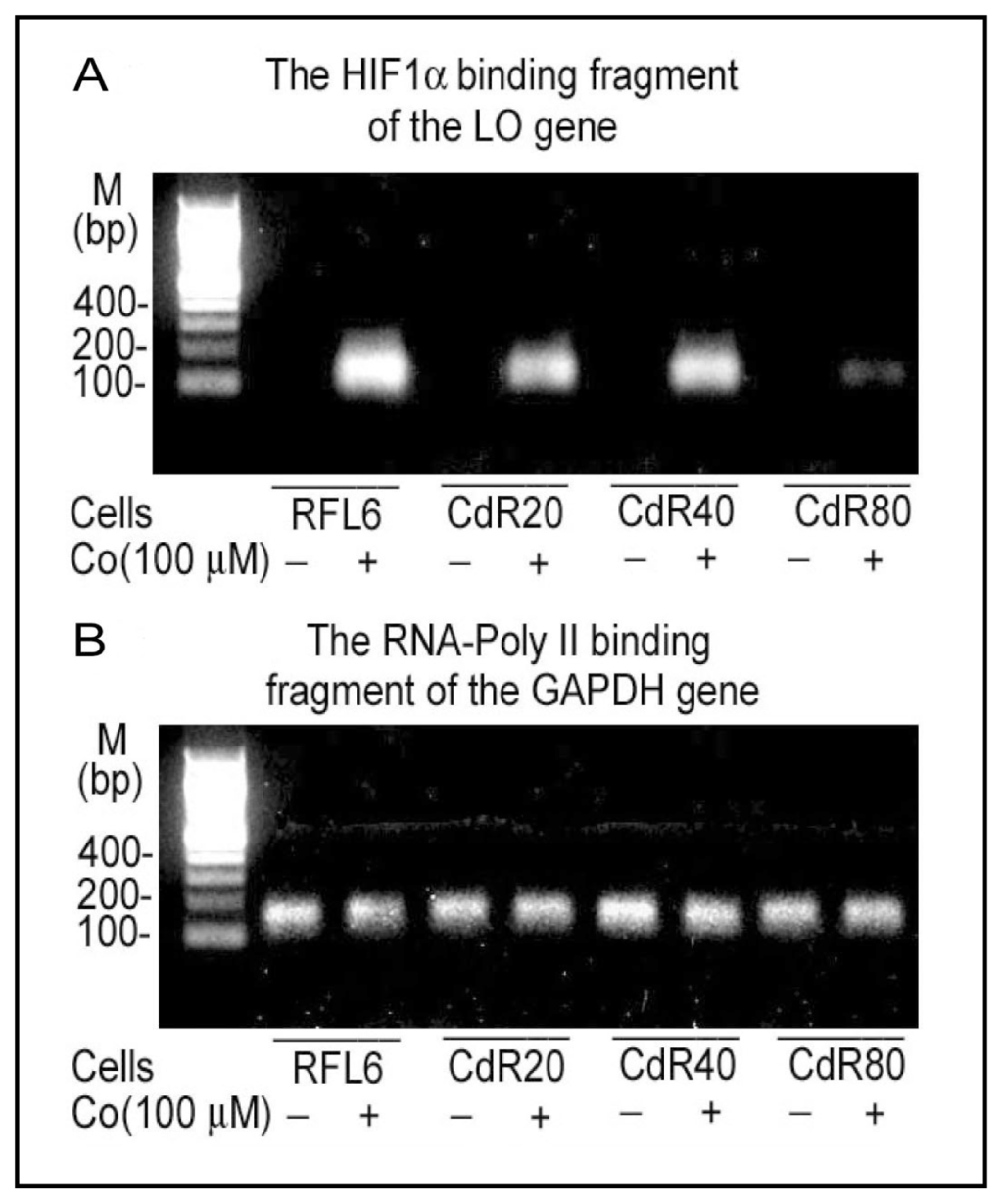
Figure 3.
Cd effects on HIF1α binding to the LO promoter HRE. RFL6 cells and Cd-resistant (CdR) cells derived from RFL6 cells by long-term Cd exposure were treated without or with Co to mimic the hypoxia condition in the cell culture system. Cells resistant to 20, 40, and 80 μM Cd were referred to as CdR20, CdR40 and CdR80, respectively. HIF1α binding to the LO promoter HRE (−387/−383) in control and CdR cells were assessed by the ChIP assay (A) using RNA-Poly II binding to the GAPDH gene as an internal control (B) as described [59]. Primer pairs used in PCR are 5′-ctccctgtgcaacgtgtct-3′ [forward (F)] and 5′-tgcagttacacaagccgttc-3′ [Reverse (R)] for amplification of the HRE containing LO gene fragment (152 bp), and 5′-ttgcttggcttcttctttgg-3′ (F) and 5′-gagacgaggctggtactcca-3′ (R) for amplification of the GAPDH gene fragment (160 bp).
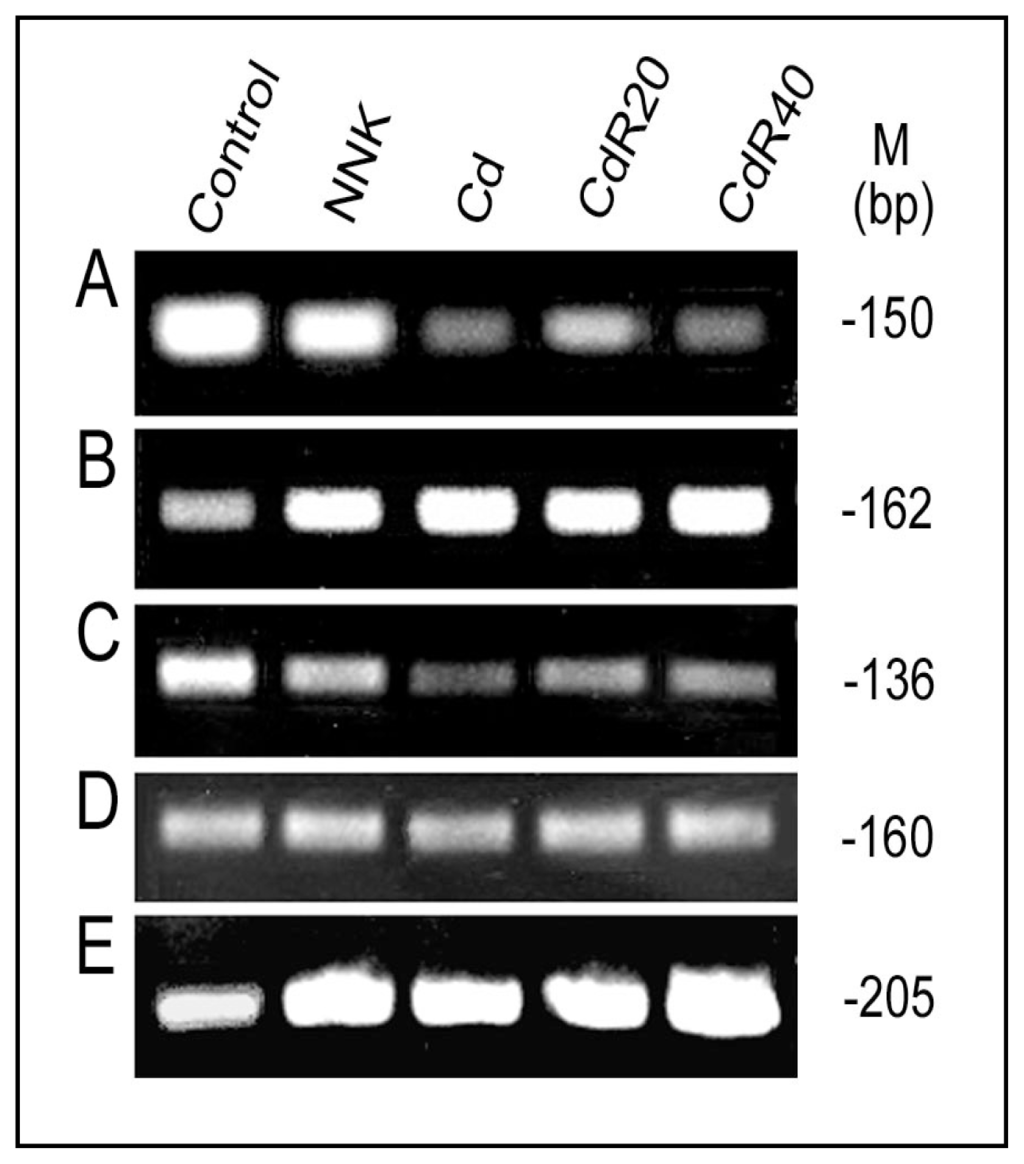
Figure 4.
NNK and Cd effects on MTF-1, Nrf2 and acetylated histone H3 binding to and methylation of the LO promoter. MTF-1 binding to the LO MRE (A), Nrf2 binding to the LO ARE (B) and acetylated histone H3 binding to the LO core promoter region (C) in RFL6 control, NNK (300 μM for 48 h) and Cd (5 μM for 24 h) treated cells, and in CdR cells were assessed by the ChIP assays as described [59] using RNA-Poly II binding to the GAPDH gene as an internal control (D). Methylation of the LO core promoter region (E) was assessed by using the promoter methylation PCR kit (Panomics, Fremont, CA., USA). Primer pairs used in PCR are 5′-cttcagacactgtgcgctct-3′ (F) and 5′-gcagggactggtgccaag-3′ (R) for amplification of the MTF-1-bound LO MRE fragment (150 bp); 5′-tttggccctcatcgctct-3′ (F) and 5′-gacttaatctgggccgaaca-3′ (R) for amplification of the Nrf2-bound LO ARE fragment (162 bp); 5′-gaagaggtctccctccttcg-3′ (F) and 5′-actgcagctgtcccagaaag-3′ (R) for amplification of the acetylated histone H3-bound LO core promoter (INR-DPE) region (136 bp); 5′-ttgcttggcttcttctttgg-3′ (F) and 5′-gagacgaggctggtactcca-3′ (R) for amplification of the RNA Poly II-bound GAPDH gene fragment (160 bp); 5′-ttcagacactgtgcgctctc-3′ (F) and 5′-aggagggagacctcttcgag-3′ (R) for amplification of the methylated LO core promoter (INR-DPE) region (205 bp) containing 15 CpG islands.
Cytosine methylation in the CpG islands interferes with the interaction of DNA with proteins thus resulting in stable transcriptional repression [94,95]. Aberrant hypermethylation of genomic DNAs and increased levels of DNA methyltransferase (DNA MeTase) activity have been detected in NNK and Cd treated cells [22,23,96]. Furthermore, both NNK and Cd also suppressed histone H3 acetylation at the LO core promoter region (Figure 4C). The appropriate acetylation and deacetylation of H3 K56 is critical for chromatin assembly and genome stability [97]. Thus, inhibition of histone H3 acetylation is an important marker for inactivation of the LO gene by NNK and Cd. It should be noted that in addition to its action on histone H3, LO also oxidizes histone H1, a substrate of LO [42,43]. Thus, LO may be a key factor for the modification of nuclear structure proteins relevant to changes in chromatin configuration. Cells long-term treated with CSC or β-aminoproprionitrile (BAPN), that inhibits LO activity, exhibited an abnormal DNA organization manifested by appearance of nuclear blebs (Figure 5), the indices for chromosomal instability [98,99].
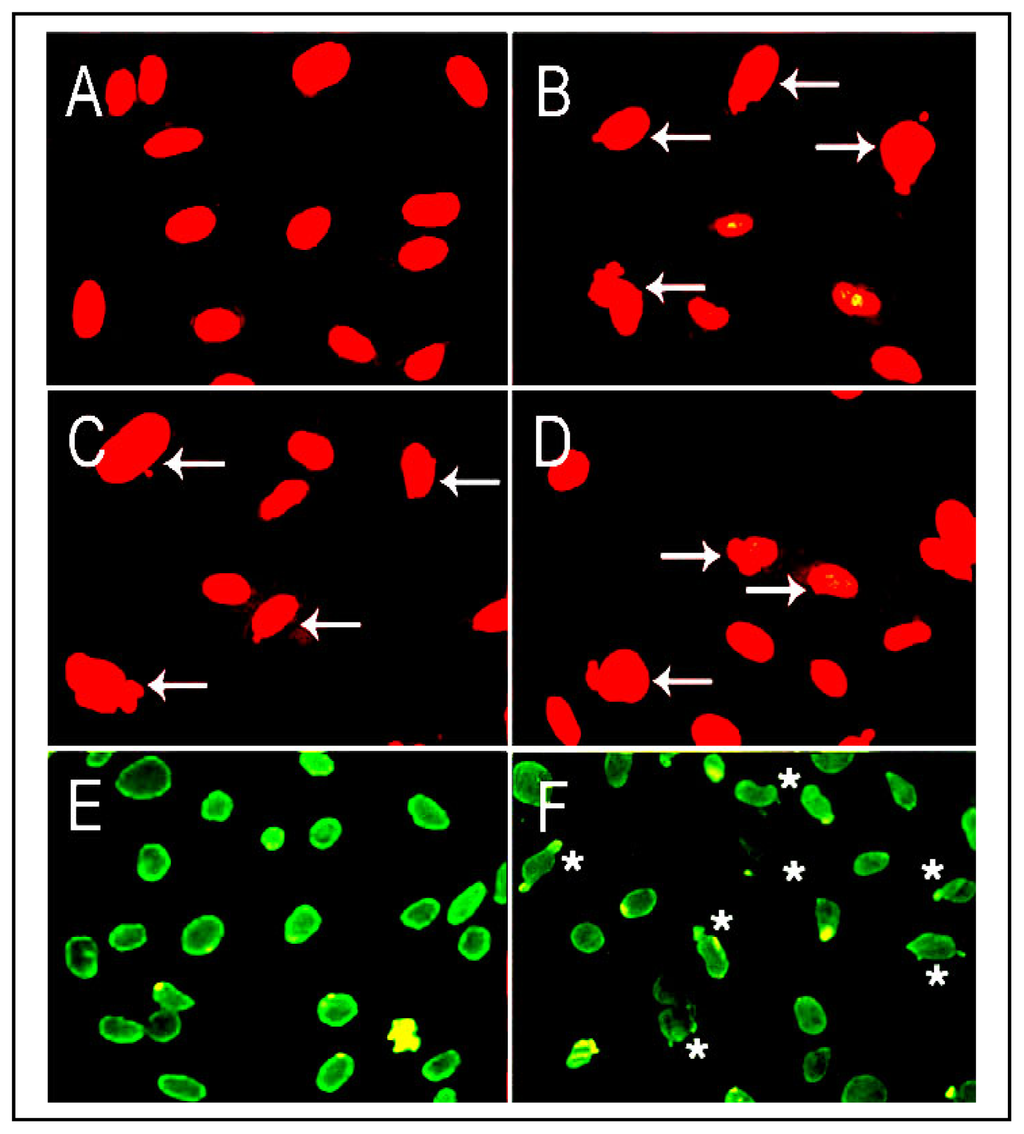
Figure 5.
Alterations in nuclear organization in cells treated with CSC and BAPN, an inhibitor of LO. RFL6 cells were long-term incubated with graded concentrations of CSC from 80 to 160 μg/mL. Survival cells isolated with different degrees of CSC-resistance (CSCR) such as to 80, 120 and 160 μg/mL were referred to as CSCR80, CSCR120 and CSCR160. Arrows show altered nuclear morphology revealed by fluorescent DNA staining with propidium iodide. A, control; B, CSCR80; C, CSCR120; and D, CSCR160. Alterations in nuclear organization in RFL6 cells treated with BAPN, an LO inhibitor, at 100 μM for 7 days were revealed by histone H1 staining using the primary antibody against histone H1 and the secondary antibody conjugated with FITC. E, control; F, cells treated with BAPN. Asterisks show altered nuclear organization in BAPN-treated cells. Notably, both CSC and BAPN induced nuclear blebs.
Thus, CSC, NNK and Cd, the major components of CS, perturbed LO at transcriptional, translational and catalytic levels representing a critical mechanism for CS toxicity to the lung (Figure 6).
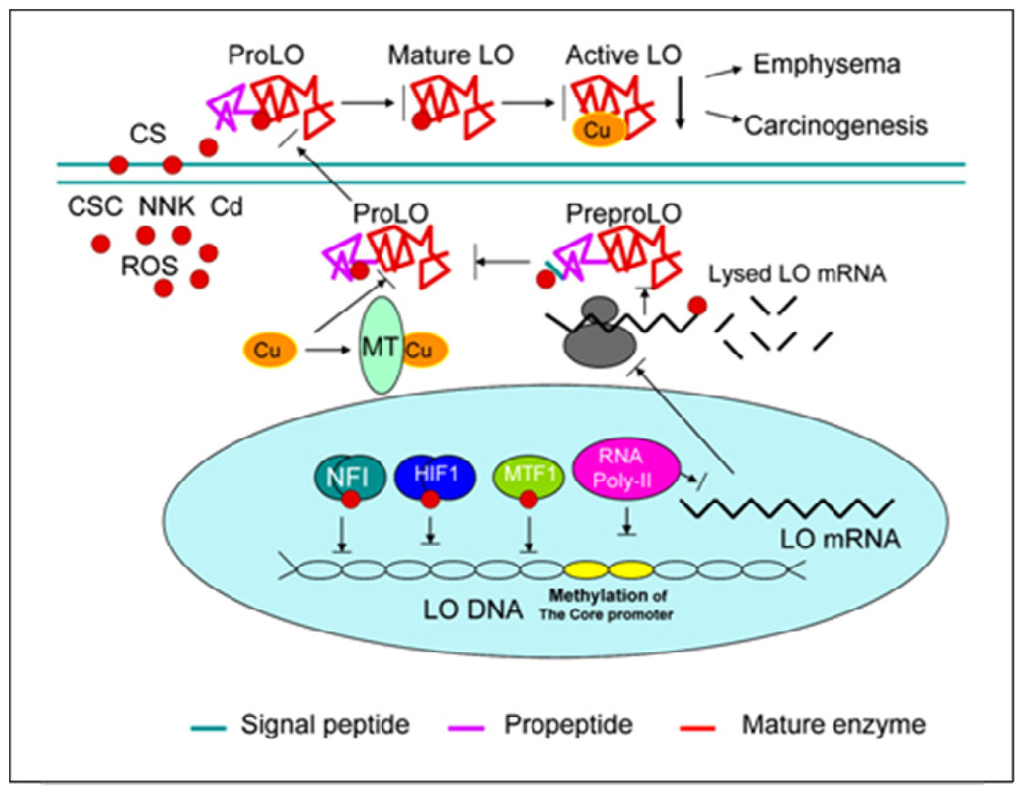
Figure 6.
CS perturbs LO at transcriptional, translational and catalytic levels. At the promoter level, CS interferes with the interaction of redox-sensitive transcription factors such as NFI, HIF1, MTF-1 with corresponding cis-elements and induces methylation of CpG at the core promoter region inhibiting the initiation of LO gene transcription. At the mRNA levels, CS inhibition of both the synthesis and the stability of LO transcripts leads to decreased levels of steady-state LO mRNA. At the protein level, CS suppresses synthesis and processing of the preproLO such as signal peptide cleavage, and Cu binding to the proLO due to elevation of cellular thiols, resulting in reduction of the ProLO secretion to the ECM. At the catalytic levels mature apoLO without Cu cofactor binding exhibits the defect in enzyme activity. Thus, CS inhibition of the LO gene expression at multiple levels collectively contributes to decreased levels of the mature LO in the lung ECM favoring emphysema pathogenesis and carcinogenesis in the lung.
5. Downregulation of LO and Pathogenesis of the Lung
5.1. LO Deficiency and Emphysema
Lung ECM, a dynamic structure, is composed of a number of functionally diverse elements which are integrated mainly by interstitial cells, e.g., fibroblasts [100,101]. The overall pattern of the lung ECM results from an intricate balance between the synthesis and degradation of its major structural components, e.g., collagen and elastin. Thus, the pathogenesis of emphysema, a lung ECM disease, is initiated by multiple mechanisms as revealed by animal model studies [7]. CS induced emphysema in humans [102]. It has been established that CS-enhanced degradation of the lung ECM proteins resulted from either the excess of protease production or the deficiency in α1-antitrypsin, an inhibitor of proteases, contributes to the onset of emphysema [7]. This review article aims at evaluation of the lung ECM protein synthesis and processing mechanism for emphysema pathology by emphasizing LO as a CS target. The critical role of LO in emphysema development was reflected in the disruption of the lung structure in chicks and rats following diet-induced deficiency of Cu, a cofactor for LO [78,79]. The lung lesions in these animals resembled panlobular emphysema in humans [80]. A mild form of emphysema was also produced in pigs raised on Cu deficient, Zn supplemented diet [103]. Zn, a known MT inducer, was used in this study based on its reported inhibitory effect on LO activity [104].
Patients with Menkes disease, an X-linked recessive disorder of Cu transport associated with the deficiency in LO, developed severe diffuse emphysema leading to respiratory failure and early death [105]. BAPN, an irreversible inhibitor of LO, can limit the fibrogenic response in certain surgical procedures and decrease collagen/elastin deposition in models of lung fibrosis. Feeding of BAPN markedly enhanced elastase-induced emphysema in hamsters [106]. Moreover, with simultaneous feeding of hamsters with BAPN, intratracheally instilled Cd resulted in lesions of emphysema but not fibrosis in the lung [107]. Additional evidence correlating LO deficiency with emphysema derives from studies of genetic variants of animals, e.g., blotchy mice with abnormal Cu transport, exhibited a deficiency of LO, inducing emphysema [108]. Furthermore, knockout mice deficient in LO, thus failed in crosslinking of collagen and elastin, resulted in major development problems in the lung manifested by emphysema [109]. Inactivation of LO and LO-like protein 1 individually caused enlarged airspaces of the lung [110,111]. It has long been known that cigarette smoke blocks cross-linking of elastin in vitro [112]. Our recent studies using CSC/NNK and Cd treated cells and Cd intratracheally instilled rats as in vitro and in vivo models have demonstrated downregulation of LO at multiple levels accompanied by inhibition of collagen and elastin expression in cell models and emphysema pathology in the lung of the animal model [40,76,88]. Modulation of the ECM gene expression by LO is supported by the observation that addition of BAPN, an inhibitor of LO, reduced mRNA levels of elastin in VSMC [113] and that overexpression of LO in COS-7 cells enhanced the promoter activity of the collagen type III gene [114].
5.2. LO Deficiency and Carcinogenesis in the Lung and Other Organs
Suppression of a set of tumor suppressor genes is a prerequisite to prosecute CS oncogenic programs in the lung. LO is a tumor suppressor gene as evidenced by its inhibitory effects on ras transformation activities [52]. Consistent with the anti-tumorigenic function of LO, normal rat kidney fibroblasts became transformed by expression of LO antisense displaying anchorage-independent growth and elevation of p21-ras expression [115]. Low levels of LO mRNA were detected in a variety of spontaneous human cancers [116] such as lung bronchogenic carcinoma [117], gastric cancers [118], head and neck squamous cell carcinoma [119], etc. Reduced LO mRNA expression was detected in various lung cancer cells, one of which (VRMC-LCD) exhibited complete methylation of the LO promoter and loss of LO expression which was reversed upon treatment with 5-aza-dC [120]. LO mRNA expressions in human bronchogenic carcinomas displayed a 3.4-fold graded reduction as tumors progressed from Stage I to IV, with a similar pattern of downregulation in LO proteins [117]. Moreover, microarray analysis demonstrated decreased LO mRNA expression in primary lung adenocarcinoma tissues and lung cancer cell lines compared to normal lung tissues and lung cells [120]. LO transcripts were progressively reduced in malignant prostate tumors either at primary or at metastatic lesions [121]. In breast tumors abundant amount of LO was observed in benign breast lesions surrounding in situ ductal carcinomas and in reactive fibrosis at the invasion front of infiltrating tumors. The level of LO was markedly decreased in late stromal reactions and nodetectable in the loose scirrhous stroma of invading ductal carcinomas [122,123], but increased in hypoxia-induced metastasis of breast cancers [51]. LO was absent in basal and squamous cell carcinoma and its knockout induced an invading phenotype in a skin equivalent model [124]. The LO gene was inactivated by methylation and loss of heterozygosity in human gastric cancers [118]. Reduction of LO expression as a result of somatic mutation of the LO gene was associated with human colorectal tumors [125]. Our recent studies have demonstrated that purified mature LO suppressed the proliferation of transformed cells by inactivation of growth factors such as bFGF [44]. Repression of BCL2, a proto-oncogene, by proLO and LO propeptide inhibited transformed phenotype of lung cancer cells (H1299) [120]. Moreover, LO modification of histone H1, H2 and H3, critical nuclear structural proteins, has been identified in vitro or in cells (Figure 4C, references [42,43,126]) and inhibition of LO enhanced chromatin instability (Figure 5). Therefore, modulating the expression of oncogenes such as ras and BCL2, regulating the activity of growth factors such as bFGF, and stabilizing nuclear proteins such as histone H1, H2 and H3 and thus maintaining chromatin and nuclear stabilities may be key mechanisms for LO tumor suppression. In view of novel functions of LO, downregulation of LO by CSC/NNK/Cd may severely impair its tumor suppressor properties triggering and facilitating carcinogenic programs in the lung.
6. Conclusions
In this paper, we have reviewed the literature for CS toxicity and our research findings, presented evidence for LO as a critical target for the insult of CSC, NNK and Cd, major components of CS, and emphasized the role of downregulation of LO, an enzyme with multiple biological functions, in the lung pathogenesis such emphysema and cancers. Further tracing CS effects on the interactions of LO with its substrates or binding ligands and exploring their underlying mechanisms are expected to provide the basis for prevention and treatment of CS-elicited lung diseases.
Acknowledgements
National Institutes of Health (R01-ES 11340) and the External Research Program of Philip Morris USA Inc. and Philip Morris International. There are no conflicts of interest.
References
- American Lung Association in Washington. Lung Disease, 2011. Available online: http://www.alaw.org/lungdisease (accessed on 17 January 2011).
- E-MedicineHealth. Lung Cancer, 2011. Available online: http://www.emedicinehealth.com/lung_cancer/article_em.htm (accessed on 17 January 2011).
- Mackay, J; Eriksen, M. The Tobacco Atlas; WHO; Geneva, Switzerland, 2002. [Google Scholar]
- Hecht, SS. Cigarette smoke and lung cancer: chemical mechanisms and approaches to prevention. Lancet Oncol 2002, 3, 461–469. [Google Scholar]
- Leischow, SJ; Djordjevic, MV. Smoking reduction and tobacco-related cancers: The more things change, the more they stay the same. J. Natl. Cancer Inst 2004, 96, 86–87. [Google Scholar]
- American Cancer Society. Tobacco-related Cancers Fact Sheet, 2011. Available online: http://www.cancer.org/Cancer/CancerCauses/TobaccoCancer/tobacco-related-cancer-fact-sheet (accessed on 17 January 2011).
- Snider, GL; Lucey, EC; Stone, PJ. Animal models of emphysema. Am. Rev. Respir. Dis 1986, 133, 149–169. [Google Scholar]
- Hautamaki, RD; Kobayashi, DK; Senior, RM; Shapiro, SD. Requirement for macrophage elastase for cigarette smoke-induced emphysema in mice. Science 1997, 277, 2002–2004. [Google Scholar]
- Shapiro, SD; Goldstein, NM; Houghton, AM; Kobayashi, DK; Kelley, D; Belaaouaj, A. Neutrophil elastase contributes to cigarette smoke-induced emphysema in mice. Am. J. Pathol 2003, 163, 2329–2335. [Google Scholar]
- Wright, JL; Churg, A. Smoke-induced emphysema in guinea pigs is associated with morphometric evidence of collagen breakdown and repair. Am. J. Physiol 1995, 268, L17–20. [Google Scholar]
- D’Armiento, JL; Dalal, SS; Okada, Y; Berg, RA; Chada, K. Collagenase expression in the lungs of transgenic mice causes pulmonary emphysema. Cell 1992, 71, 955–961. [Google Scholar]
- Kuper, H; Boffetta, P; Adami, HO. Tobacco use and cancer causation: association by tumour type. J. Intern. Med 2002, 252, 206–224. [Google Scholar]
- Mulcahy, S. The toxicology of cigarette smoke and environmental tobacco smoke. 2011. Available online: http://www.skynet.ie/~stephen/reports/bc4927.html (accessed on 17 January 2011).
- Hoffmann, D; Wynder, EL. Marquardt, H, Schaefer, SG, McClellan, R, Welsch, F, Eds.; Active and passive smoking. In Toxicology; Academic Press: New York, NY, USA, 1999; pp. 879–898. [Google Scholar]
- Hoffmann, D; Hoffmann, I; El-Baypumy, K. The less harmful cigarette: a controversial issue. A tribute to Ernst L. Wynder. Chem. Res. Toxicol 2001, 14, 767–790. [Google Scholar]
- Church, T; Pryor, WA. Free-radical chemistry of cigarette smoke and its toxilogical implications. Environ. Health Perspect 1985, 64, 111–126. [Google Scholar]
- Schick, S; Glantz, S. Philip Morris toxicological experiments with fresh sidestream smoke: More toxic than mainstream smoke. Tob. Control 2005, 14, 396–404. [Google Scholar]
- Hecht, SS. Biochemistry, biology and carcinogenicity of tobacco-specific N-nitrosamines. Chem. Res. Toxicol 1998, 11, 560–603. [Google Scholar]
- Chepiga, T; Morton, M; Murphy, P; Avalos, J; Bombick, B; Doolittle, D; Borgerding, M; Swauger, J. A comparison of the mainstream smoke chemistry and mutagenicity of a representative sample of the US cigarette market with two Kentucky reference cigarettes (K1R4F and K1R5F). Food Chem. Toxicol 2000, 38, 949–962. [Google Scholar]
- Belinsky, SA; Foley, JF; White, CM; Anderson, MW; Maronpot, RR. Dose-response relationship between O6-methylguanine formation in Clara cells and induction of pulmonary neoplasia in rat by 4-(methyl nitrosamino)-1-(3-pyridyl)-1-butanone. Cancer Res 1990, 50, 3772–3780. [Google Scholar]
- Jalas, JR; Ding, X; Murphy, SE. Comparative metabolism of the tobacco-specific nitrosamines 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone and 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol by rat cytochrome P450 2A3 and human cytochrome P450 2A13. Drug Metab. Dispos 2003, 31, 1199–1202. [Google Scholar]
- Lin, RK; Hsieh, YS; Lin, P; Hsu, HS; Chen, CY; Tang, YA; Lee, CF; Wang, YC. The tobacco-specific carcinogen NNK induces DNA methyltransferase 1 accumulation and tumor suppressor gene hypermethylation in mice and lung cancer patients. J. Clin. Invest 2010, 120, 521–532. [Google Scholar]
- Castonguay, A; Foiles, PG; Trushin, N; Hecht, SS. Study of DNA methylation by tobacco-specific N-nitrosamines.
- Pfeifer, GP; Denissenko, MF; Olivier, M; Tretyakova, N; Hecht, SS; Hainaut, P. Tobacco smoke carcinogens, DNA damage and p53 mutations in smoking-associated cancers. Oncogene 2002, 21, 7435–7451. [Google Scholar]
- Beryllin, Cadmium. Mercury and Exposures in the Glass Manufacturing Industry. Monographs on the Evaluation of Carcinogenic Risk to Humans; IARC: Lyon, France, 1993; 58, pp. 119–236.
- Tobacco Smoke and Involuntary Smoking. In Monographs on the Evaluation of Carcinogenic Risk of Chemicals to Humans; IARC: Lyon, France, 2004; Volume 83.
- Post, C; Johansson, B; Allenmark, S. Organ distribution and protein binding of cadmium in autopsy material from heavy smokers. Environ. Res 1984, 34, 29–37. [Google Scholar]
- Paakko, P; Kokkonen, P; Anttila, S; Kalliomaki, P-L. Cadmium and chromium as markers of smoking in human lung tissue. Environ. Res 1989, 49, 197–207. [Google Scholar]
- Coultas, DB; Samet, JM. Hensley, MJ, Saunders, NA, Eds.; Cigarette smoking. In Clinical Epidemiology of Chronic Obstractive Pulmonary Disease; Marcel Dekker: New York, NY, USA, 1989; pp. 109–138. [Google Scholar]
- Davision, AG; Newman-Taylor, AJ; Darbyshire, J; Chettle, DR; Guthrie, CJG; O’Malley, D; Mason, HJ; Fayers, PM; Venables, KM; Pickering, CAC; Franklin, D; Scott, MC; Holden, H; Wright, AL. Cadmium fume inhalation and emphysema. Lancet 1988, 1, 663–667. [Google Scholar]
- Kazantzis, G; Blacks, RG. Cook, ME, Hiscock, SA, Morrow, H, Volpe, RA, Eds.; A mortality study of cadmium exposed workers. Proceedings of the Seventh International Cadmium Conference, New Orleans, LA, USA, 6–8 April 1992, Cadmium Association/Cadmium Council: New Orleans, LA, USA/Reston, VA, USA, 1992; pp. 150–157. [Google Scholar]
- Lane, RE; Campbell, ACP. Fatal emphysema in two men making a copper cadmium alloy. Brit. J. Indust. Med 1954, 11, 118–122. [Google Scholar]
- Kimbel, P. Petty, TL, Ed.; Proteolytic damage and emphysema pathogenesis. In Chronic Obstructive Pulmonary Disease; Marcel Dekker: New York, NY, USA, 1985; pp. 105–127. [Google Scholar]
- Snider, GL; Hayes, JA; Korthy, AL; Lewis, GP. Centrilobular emphysema experimentally induced by cadmium chloride aerosol. Am. Rev. Respir. Dis 1973, 108, 40–47. [Google Scholar]
- Waisberg, M; Joseph, P; Hale, B; Beyersmann, D. Molecular and cellular mechanisms of cadmium carcinogenesis. Toxicology 2003, 192, 95–117. [Google Scholar]
- Ninth Report on Carcinogens. In National Toxicology Program; Research Triangle Park: NC, USA, 2000.
- Glaser, U; Hochrainer, D; Otto, FJ; Oldiges, H. Carcinogenicity and toxicity of four cadmium compounds inhaled by rats. Toxicol. Environ. Chem 1990, 27, 153–162. [Google Scholar]
- Takenaka, S; Oldiges, H; Konig, H; Hochrainer, D; Oberdorster, G. Carcinogenicity of cadmium chloride aerosols in W. rats. J. Natl. Cancer Inst 1983, 70, 367–373. [Google Scholar]
- Gao, S; Chen, K; Zhao, Y; Rich, CB; Chen, L; Li, SJ; Toselli, P; Stone, P; Li, W. Transcriptional and posttranscriptional inhibition of lysyl oxidase expression by cigarette smoke condensate in cultured rat fetal lung fibroblasts. Toxicol. Sci 2005, 87, 197–203. [Google Scholar]
- Chen, L-J; Zhao, Y; Gao, S; Chou, I-N; Toselli, P; Stone, P; Li, W. Downregulation of lysyl oxidase and upregulation of cellular thiols in rat fetal lung fibroblasts treated with cigarette smoke condensate. Toxicol. Sci 2005, 83, 372–379. [Google Scholar]
- Kagan, HM; Li, W. Lysyl oxidase: Properties, specificity and biological roles inside of the cell. J. Cell. Biochem 2003, 88, 660–672. [Google Scholar]
- Giampuzzi, M; Oleggini, R; Di Donato, A. Demonstration of in vitro interaction between tumor suppressor lysyl oxidase and histone H1 and H2: Definition of the regions involved. Biochim. Biophys. Acta 2003, 1647, 245–251. [Google Scholar]
- Kagan, HM; Williams, MA; Calaman, SD; Berkowitz, EM. Histone H1 is a substrate for lysyl oxidase and contains endogenous sodium borotritide-reducible residues. Biochem. Biophys. Res. Commun 1983, 115, 186–192. [Google Scholar]
- Li, W; Nugent, MA; Zhao, Y; Chau, AN; Li, SJ; Chou, IN; Liu, G; Kagan, HM. Lysyl oxidase oxidizes basic fibroblast growth factor and inactivates its mitogenic potential. J. Cell Biochem 2003, 88, 152–164. [Google Scholar]
- Li, W; Nellaiappan, K; Strassmaier, T; Graham, L; Thomas, KM; Kagan, HM. Localization and activity of lysyl oxidase within nuclei of fibrogenic cells. Proc. Natl. Acad. Sci. USA 1997, 94, 12817–12822. [Google Scholar]
- Mello, MLS; Contente, S; Vidal, BC; Planding, W; Schenck, U. Modulation of ras transformation affecting chromatin supraorganization as assessed by image analysis. Exp. Cell Res 1995, 220, 374–382. [Google Scholar]
- Nellaiappan, K; Risitano, A; Liu, G; Nicklas, G; Kagan, HM. Fully processed lysyl oxidase catalyst translocates from the extracellular space into nuclei of aortic smooth muscle cells. J. Cell. Biochem 2000, 79, 576–582. [Google Scholar]
- Lazarus, HM; Cruikshank, WW; Narasimhan, N; Kagan, HM; Center, DM. Induction of human monocyte motility by lysyl oxidase. Matrix Biol 1995, 14, 727–731. [Google Scholar]
- Lucero, HA; Ravid, K; Grimsby, JL; Rich, CB; DiCamillo, SJ; Mäki, JM; Myllyharju, J; Kagan, HM. Lysyl oxidase oxidizes cell membrane proteins and enhances the chemotactic response of vascular smooth muscle cells. J. Biol. Chem 2008, 283, 24103–24117. [Google Scholar]
- Li, W; Liu, G; Chou, I-N; Kagan, HM. Hydrogen peroxide-mediated, lysyl oxidase-dependent chemotaxis of vascular smooth muscle cells. J. Cell. Biochem 2000, 78, 550–557. [Google Scholar]
- Erler, JT; Bennewith, KL; Nicolau, M; Dornhofer, N; Kong, C; Le, QT; Chi, JT; Jeffrey, SS; Giaccia, AJ. Lysyl oxidase is essential for hypoxia-induced metastasis. Nature 2006, 440, 1222–1226. [Google Scholar]
- Kenyon, K; Contente, S; Trackman, PC; Tang, J; Kagan, HM; Friedman, RM. Lysyl oxidase and rrg messenger RNA. Science 1991, 253, 802. [Google Scholar]
- Kosonen, T; Uriu-Hare, JY; Clegg, MS; Keen, CL; Rucker, RB. Incorporation of copper into lysyl oxidase. Biochem. J 1997, 327, 283–289. [Google Scholar]
- Wan, SX; Mure, M; Medzihradszky, KF; Burlingame, AL; Brown, DE; Dooley, DM; Smith, AJ; Kagan, HM; Klinman, JP. A crosslinked cofactor in lysyl oxidase: Redox function for amino acid side chains. Science 1996, 273, 1078–1084. [Google Scholar]
- Gacheru, SN; Trackman, PC; Shah, MA; O’Gara, CY; Spacciapoli, P; Greenaway, FT; Kagan, HM. Structural and catalytic properties of copper in lysyl oxidase. J. Biol. Chem 1990, 265, 19022–19027. [Google Scholar]
- Csiszar, K. Lysyl oxidases: A novel multifunctional amine oxidase family. Prog. Nucleic Acid Res. Mol. Biol 2001, 70, 1–32. [Google Scholar]
- Bazan, JF. A novel family of growth factor receptors: A common binding domain in the growth hormone, prolactin, erythropoietin and IL-6 receptors and the p75 IL-2 receptor β-chain. Biochem. Biophys. Res. Commun 1989, 164, 788–795. [Google Scholar]
- Bazan, JF. Structural design and molecular evolution of a cytokine receptor superfamily. Proc. Natl. Acad. Sci. USA 1990, 87, 6934–6938. [Google Scholar]
- Gao, S; Zhao, Y; Kong, L; Toselli, P; Chou, I-N; Stone, P; Li, W. Cloning and characterization of the rat lysyl oxidase gene promoter: Identification of core promoter elements and functional nuclear factor I binding sites. J. Biol. Chem 2007, 282, 25322–25337. [Google Scholar]
- Corden, J; Wasylyk, B; Buchwalder, A; Sassone-Corsi, P; Kedinger, C; Chambon, P. Promoter sequences of eukaryotic protein-coding genes. Science 1980, 209, 1406–1414. [Google Scholar]
- Smale, ST; Kadonaga, JT. The RNA polymerase II core promoter. Annu. Rev. Biochem 2003, 72, 449–479. [Google Scholar]
- Gronostajski, RM. Roles of the NFI/CTF gene family in transcription and development. Gene 2000, 249, 31–45. [Google Scholar]
- Bandyopadhyay, S; Gronostajski, RM. Identification of a conserved oxidation-sensitive cysteine residue in the NFI family of DNA binding proteins. J. Biol. Chem 1994, 269, 29949–29955. [Google Scholar]
- Bandyopadhyay, S; Starke, DW; Mieyal, JJ; Gronostajski, RM. Thioltranferase (Glutaredoxin) reactivates the DNA-binding activity of oxidation-inactivated Nuclear Factor I. J. Biol. Chem 1998, 273, 392–397. [Google Scholar]
- Semenza, GL. Expression of hypoxia-inducible factor 1: Mechanism and consequences. Biochem. Pharmacol 2000, 59, 47–53. [Google Scholar]
- Déry, MA; Michaud, MD; Richard, DE. Hypoxia-inducible factor 1: Regulation by hypoxic and non-hypoxic activators. Int. J. Biochem. Cell Biol 2005, 37, 535–540. [Google Scholar]
- Giedroc, DP; Chen, X; Apuy, JL. Metal response element (MRE) binding transcription factor-1 (MTF-1): Structure, function, and regulation. Antioxid. Redox Signal 2001, 3, 577–596. [Google Scholar]
- Stuart, GW; Searle, PF; Chen, HY; Brinster, R; Palmiter, RD. A 12-base pair DNA motif that is repeated several times in metallothione gene promoters confers metal regulation to a heterologous. Proc. Natl. Acad. Sci. USA 1984, 81, 7318–7322. [Google Scholar]
- Lichtlen, P; Wang, Y; Belser, T; Georgiev, O; Certa, U; Sack, R; Schaffner, W. Target gene search for the metal-responsive transcription factor MTF-1. Nuc. Aci Res 2001, 29, 1514–1523. [Google Scholar]
- Copple, IM; Goldring, CE; Kitteringham, NR; Park, BK. The Nrf2-Keap1 defence pathway: Role in protection against drug-induced toxicity. Toxicology 2008, 246, 24–33. [Google Scholar]
- Wang, GL; Jiang, B-H; Rue, EA; Semenza, GL. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc. Natl. Acad. Sci. USA 1995, 92, 5510–5514. [Google Scholar]
- Weissmann, N. Hypoxia-driven mechanisms in lung biology and disease: a new review series of the ERS Lung Science Conference. Eur. Respir. J 2008, 3, 697–698. [Google Scholar]
- Yasuo, M; Mizuno, S; Kraskauskas, D; Bogaard, HJ; Natarajan, R; Cool, CD; Zamora, M; Voelkel, NF. Hypoxia inducible factor-1 {alpha} in human emphysema lung tissue. Eur. Respir. J 2010. [Google Scholar] [CrossRef]
- Wang, GL; Semenza, GL. Desferrioxamine induces erythropoietin gene expression and hypoxia-inducible factor 1 DNA-binding activity: Implications for models of hypoxia signal transduction. Blood 1993, 82, 3610–3615. [Google Scholar]
- Epstein, AC; Gleadle, JM; McNeill, LA; Hewitson, KS; O’Rourke, J; Mole, DR; Mukherji, M; Metzen, E; Wilson, MI; Dhanda, A; Tian, YM; Masson, N; Hamilton, DL; Jaakkola, P; Barstead, R; Hodgkin, J; Maxwell, PH; Pugh, CW; Schofield, CJ; Ratcliffe, PJ. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell 2001, 107, 43–54. [Google Scholar]
- Zhao, Y; Gao, S; Chou, I-N; Toselli, P; Stone, P; Li, W. Inhibition of the expression of lysyl oxidase and its substrates in cadmium-resistant rat fetal lung fibroblasts. Toxicol. Sci 2006, 90, 478–489. [Google Scholar]
- Chou, DK; Zhao, Y; Gao, S; Chou, I-N; Toselli, P; Stone, P; Li, W. Perturbation of copper (Cu) homeostasis and expression of Cu-binding proteins in cadmium-resistant lung fibroblasts. Toxicol Sci 2007, 99, 267–276. [Google Scholar]
- Dubic, MA; Keen, CL; Rucker, RB. Elastin metabolism during prenatal lung development in the copper-deficient rat. Exp. Lung Res 1985, 8, 227–241. [Google Scholar]
- Harris, ED. Biochemical defect in chick lung resulting from copper deficiency. J. Nutr 1986, 116, 252–258. [Google Scholar]
- Soskel, NT; Watanabe, S; Sandberg, LB. Mechanisms of lung injury in the copper-deficient hamster model of emphysema. Chest 1984, 85, 70s–72s. [Google Scholar]
- Richards, MP. Recent developments in trace element metabolism and function: Role of metallothionein in copper and zinc metabolism. J. Nutr 1989, 119, 1062–1070. [Google Scholar]
- Brouwer, M; Hoexum-Brouwer, T; Cashon, RE. A putative glutathione-binding site in CdZn-metallothionein identified by epuilibrium binding and molecular modelling studies. Biochem. J 1993, 294, 219–225. [Google Scholar]
- Briggs, RW; Armitage, IM. Evidence for site-selective metal binding in calf liver metallothionein. J. Biol. Chem 1982, 257, 1259–1262. [Google Scholar]
- Morris, PE; Bernard, GR. Significance of glutathione in lung disease and implications for therapy. Am. J. Med. Sci 1994, 307, 119–127. [Google Scholar]
- Li, XY; Donaldson, K; Rahman, I; MacNee, W. An investigation of the role of glutathione in the increased permeability induced by cigarette smoke in vivo and in vitro. Am. Rev. Respir. Crit. Care Med 1994, 149, 1518–1525. [Google Scholar]
- Rahman, I; Li, XY; Donaldson, K; Harrison, HJ; MacNee, W. Glutathione homeostasis in alveolar epithelial cells in vitro and lung in vivo under oxidative stress. Am. J. Physiol 1995, 269, L285–L292. [Google Scholar]
- Rahman, I. Regulation of glutathione in inflammation and chronic lung diseases. Mutat. Res 2005, 579, 58–80 . [Google Scholar]Int. J. Environ. Res. Public Health 2011, 8, 182.
- Zhao, Y; Chen, L; Gao, S; Toselli, P; Stone, P; Li, W. The critical role of the cellular thiol homeostasis in cadmium perturbation of the lung extracellular matrix. Toxicology 2010, 267, 60–69. [Google Scholar]
- Li, W; Chou, I-N; Boak, A; Kagan, HM. Down-regulation of lysyl oxidase in cadmium-resistant fibroblasts. Am. J. Respir. Cell Mol. Biol 1995, 13, 418–425. [Google Scholar]
- Li, W; Zhao, Y; Toselli, P; Stone, P. Cigarette smoke condensate downregulation of lysyl oxidase transcription mediated by inactivation of nuclear factor I. Toxicologist 2008, 102, 227, (abstract). [Google Scholar]
- Li, W; Zhao, Y; Gao, S; Toselli, P. Transcriptional regulation of the lysyl oxidase gene by the hypoxia-inducible factor 1. FASEB J 2010, 24. Meeting Abstract Supplement, No. 833.16, (abstract). [Google Scholar]
- Galan, A; Garcia-Bermejo, L; Troyano, A; Vilaboa, NE; Fernandez, C; de Blas, E; Aller, P. The role of intracellular oxidation in death induction (apoptosis and necrosis) in human promonocytic cells treated with stress inducers (cadmium, heat, X-ray). Eur. J. Cell Biol 2001, 80, 312–320. [Google Scholar]
- Zhao, Y; Gao, S; Zhou, J; Toselli, P; Li, W. NNK, a tobacco-specific carcinogen, inhibits the expression of lysyl oxidase, a tumor suppressor gene. Toxicologist 2011, (abstract in press). [Google Scholar]
- Bestor, TH. The DNA methyltransferase of mammals. Hum. Mol. Genet 2000, 9, 2395–2402. [Google Scholar]
- Ng, HH; Bird, A. DNA methylation and chromatin modification. Curr. Opin. Genet. Dev 1999, 9, 158–163. [Google Scholar]
- Takiguchi, M; Achanzar, WE; Qu, W; Li, G; Waalkes, MP. Effects of cadmium on DNA-(cytosine-5) methyltransferase activity and DNA methylation status during cadmium-induced cellular transformation. Exp. Cell Res 2003, 286, 355–365. [Google Scholar]
- Downs, JA. Histone H3 K56 acetylation, chromatin assembly, and the DNA damage checkpoint. DNA Repair 2008, 7, 2020–2024. [Google Scholar]
- Xu, G-L; Bestor, TH; Bourchis, D; Hsieh, CL; Tommerrup, N; Bugge, M; Hulten, M; Qu, X; Russo, JJ; Viegas-Pequignot, E. Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature 1999, 402, 187–191. [Google Scholar]
- Gisselsson, D; Bjork, J; Hoglund, M; Mertens, F; Cin, PD; Akerman, M; Mandahl, N. Abnormal nuclear shape in solid tumors reflects mitotic instability. Am. J. Pathol 2001, 158, 199–206. [Google Scholar]
- Davidson, JM. Biochemistry and turnover of lung interstitium. Eur. Respir. J 1990, 3, 1048–1063. [Google Scholar]
- Campa, JS; Harrison, NK; Laurent, G. Jolles, G, Karlsson, J-A, Taylor, J, Eds.; Regulation of matrix production in the airways. In T-Lymphocyte and Inflammatory Cell Research in Asthma; Academic Press: London, UK, 1993; pp. 221–235. [Google Scholar]
- Snider, GL. Emphysema: the first two centuries-and beyond. Am. Rev. Respir. Dis 1992, 146, 1615–1622. [Google Scholar]
- Soskel, NT; Watanabe, S; Hammond, E; Sandberg, LB; Renzetti, AD, Jr; Crapo, JD. A copper-deficient, zinc-supplemented diet produces emphysema in pig. Am. Rev. Respir. Dis 1982, 126, 316–325. [Google Scholar]
- Chvapil, M; Misiorowski, R. In vivo inhibition of lysyl oxidase by high dose of zinc. Proc. Soc. Exp. Biol. Med 1980, 164, 137–141. [Google Scholar]
- Grange, DK; Kaler, SG; Albers, GM; Petterchak, JA; Thorpe, CM; DeMello, DE. Severe bilateral panlobular emphysema and pulmonary arterial hypoplasia: unusual manifestations of Menkes disease. Am. J. Med. Genet 2005, 139A, 151–155. [Google Scholar]
- Kuhn, C; Starcher, BC. The effects of lathyrogens on the evolution of elastase-induced emphysema. Am. Rev. Respir. Dis 1980, 122, 453–460. [Google Scholar]
- Niewoehner, DE; Hoidal, JR. Lung fibrosis and emphysema: Divergent responses to a common injury. Science 1982, 217, 359–360. [Google Scholar]
- Fisk, DE; Kuhn, C. Emphysema-like changes in the lungs of blotchy mouse. Am. Rev. Respir. Dis 1976, 113, 787–797. [Google Scholar]
- Warburton, D; Shi, W. Lo, and the niche is knit: Lysyl oxidase activity and maintenance of lung, aorta, and skin integrity. Am. J. Pathol 2005, 167, 921–922. [Google Scholar]
- Liu, X; Zhao, Y; Gao, J; Pawlyk, B; Starcher, B; Spencer, JA; Yanagisawa, H; Zuo, J; Li, T. Elastic fiber homeostasis requires lysyl oxidase-like 1 protein. Nat. Genet 2004, 36, 178–182. [Google Scholar]
- Maki, JM; Sormunen, R; Lippo, S; Kaarteenaho-Wiik, R; Soininen, R; Myllyharju, J. Lysyl oxidase is essential for normal development and function of the respiratory system and for the integrity of elastic and collagen fibers in various tissues. Am. J. Pathol 2005, 167, 927–936. [Google Scholar]
- Laurent, P; Janoff, A; Kagan, HM. Cigarette smoke blocks cross-linking of elastin. in vitro. Am. Rev. Respir. Dis 1983, 127, 189–192. [Google Scholar]
- Jackson, LE; Faris, B; Martin, BM; Jones, HV; Rich, CB; Foster, JA; Franzblau, C. The effect of β-aminopropionitrile on elastin gene expression in smooth muscle cell cultures. Biochem. Biophys. Res. Commun 1991, 179, 939–944. [Google Scholar]
- Giampuzzi, M; Botti, G; Duca, MD; Arata, L; Ghiggeri, G; Gusmano, R; Ravazzolo, R; Donato, AD. Lysyl oxidase activates the transcription activity of human collagene III promoter. J. Biol. Chem 2000, 275, 36341–36349. [Google Scholar]
- Giampuzzi, M; Botti, G; Gilli, M; Gusmano, R; Borel, A; Sommer, P; Di Donato, A. Down-regulation of lysyl oxidase-induced tumorigenic transformation in NRK-49F cells characterized by constitutive activation of ras-proto-oncogene. J. Biol. Chem 1995, 276, 29226–29232. [Google Scholar]
- Hamalainen, ER; Kemppainen, R; Kuivaniemi, H; Tromp, G; Vaheri, A; Pihlajaniemi, T; Kivirikko, KI. Quantitative polymerase chain reaction of lysyl oxidase mRNA in malignantly transformed human cell lines demonstrates that their low lysyl oxidase activity is due to low quantities of its mRNA and low levels of transcription of the respective gene. J. Biol. Chem 1995, 270, 21590–21593. [Google Scholar]
- Woznick, AR; Braddock, AL; Dulai, M; Seymour, ML; Callahan, RE; Welsh, RJ; Chmielewski, GW; Zelenock, GB; Hanley, SCJ. Lysyl oxidase expression in bronchogenic carcinoma. Am. J. Surg 2005, 189, 297–301. [Google Scholar]
- Kaneda, A; Wakazono, K; Tsukamoto, T; Watanabe, N; Yagi, Y; Tatematsu, M; Kaminishi, M; Sugimura, T; Ushijima, T. Lysyl oxidase is a tumor suppressor gene inactivated by methylation and loss of heterozygosity in human gastric cancers. Cancer Res 2004, 64, 6410–6415. [Google Scholar]
- Rost, T; Pyritz, V; Rathcke, IO; Gorogh, T; Dunne, AA; Werner, JA. Reduction of LOX- and LOXL2-mRNA expression in head and neck squamous cell carcinomas. Anticancer Res 2003, 23, 1565–1573. [Google Scholar]
- Wu, M; Min, C; Wang, X; Yu, Z; Kirsch, KH; Trackman, PC; Sonenshein, GE. Repression of BCL2 by the tumor suppressor activity of the lysyl oxidase propeptide inhibits transformed phenotype of lung and pancreatic cancer cells. Cancer Res 2007, 67, 6278–6285. [Google Scholar]
- Ren, C; Yang, G; Timme, TL; Wheeler, TM; Thompson, TC. Reduced lysyl oxidase messenger RNA levels in experimental and human prostate cancer. Cancer Res 1998, 58, 1285–1290. [Google Scholar]
- Decitre, M; Gleyzal, C; Raccurt, M; Peyrol, S; Aubert-Foucher, E; Csiszar, K; Sommer, P. Lysyl oxidase-like protein localizes to sites of de novo fibrinogenesis and in the early stromal reaction of ductal breast carcinomas. Lab. Invest 1998, 78, 143–151. [Google Scholar]
- Peyrol, S; Raccurt, M; Gerard, F; Gleyzal, C; Grimaud, JA; Sommer, P. Lysyl oxidase gene expression in the stromal reaction to in situ and invasive ductal breast carcinoma. Am. J. Pathol 1997, 150, 497–507. [Google Scholar]
- Bouez, C; Reynaud, C; Noblesse, E; Thépot, A; Gleyzal, C; Kanitakis, J; Perrier, E; Damour, O; Sommer, P. The lysyl oxidase (LOX) is absent in basal and squamous cell carcinomas and its knockdown induces an invading phenotype in a skin equivalent model. Clin. Cancer Res 2006, 12, 1463–1469. [Google Scholar]
- Csiszar, K; Fong, FST; Ujfalusi, A; Krawetz, SA; Salvati, EP; Mackenzie, JW; Boyd, CD. Somatic mutations of the lysyl oxidase gene on chromosome 5q23.1 in colorectal tumors. Int. J. Cancer 2002, 97, 636–642. [Google Scholar]
- Li, W; Li, SJ; Zhao, Y; Kagan, HM. Oxidation of histone H1 within cell nuclei by lysyl oxidase: Implications for changes in chromosomal stability. The First Meeting Bulletin of the American Society for Matrix Biology 2002, 1, 119, (abstract). [Google Scholar]
© 2011 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).