Oral Microbiota—One Habitat or Diverse Niches? A Pilot Study of Sampling and Identification of Oral Bacterial and Fungal Biota in Patients with Type I Diabetes Mellitus Treated with Insulin Pump
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Participants
2.2. Oral Cavity Sampling Methods
2.3. Microbiological Cultures
2.4. Statistical Analysis
2.5. Ethics Statement
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Caselli, E.; Fabbri, C.; D’Accolti, M.; Soffritti, I.; Bassi, C.; Mazzacane, S.; Franchi, M. Defining the oral microbiome by whole-genome sequencing and resistome analysis: The complexity of the healthy picture. BMC Microbiol. 2020, 20, 120. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.; Yin, J.; Zhao, J.; Ma, S.-R.; Wang, H.-R.; Wang, M.; Chen, W.; Wei, W.-Q. Microbial Similarity and Preference for Specific Sites in Healthy Oral Cavity and Esophagus. Front. Microbiol. 2018, 9, 1603. [Google Scholar] [CrossRef] [PubMed]
- Pease, A.J.; Lo, C.; Earnest, A.; Kiriakova, V.; Liew, D.; Zoungas, S. The Efficacy of Technology in Type 1 Diabetes: A Systematic Review, Network Meta-analysis, and Narrative Synthesis. Diabetes Technol. Ther. 2020, 22, 411–421. [Google Scholar] [CrossRef] [PubMed]
- A DiMeglio, L.; Evans-Molina, C.; A Oram, R. Type 1 diabetes. Lancet 2018, 391, 2449–2462. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, X.; Li, H.; Ni, C.; Du, Z.; Yan, F. Human oral microbiota and its modulation for oral health. Biomed. Pharmacother. 2018, 99, 883–893. [Google Scholar] [CrossRef]
- Dicembrini, I.; Serni, L.; Monami, M.; Caliri, M.; Barbato, L.; Cairo, F.; Mannucci, E. Type 1 diabetes and periodontitis: Prevalence and periodontal destruction—A systematic review. Acta Diabetol. 2020, 57, 1405–1412. [Google Scholar] [CrossRef]
- Mauri-Obradors, E.; Estrugo-Devesa, A.; Jane-Salas, E.; Vinas, M.; Lopez-Lopez, J. Oral manifestations of Diabetes Mellitus. A systematic review. Med. Oral Patol. Oral Cir. Bucal. 2017, 22, e586–e594. [Google Scholar] [CrossRef]
- Ferizi, L.; Dragidella, F.; Spahiu, L.; Begzati, A.; Kotori, V. The Influence of Type 1 Diabetes Mellitus on Dental Caries and Salivary Composition. Int. J. Dent. 2018, 2018, 1–7. [Google Scholar] [CrossRef]
- Rodrigues, C.; Rodrigues, M.; Henriques, M. Candida sp. Infections in Patients with Diabetes Mellitus. J. Clin. Med. 2019, 8, 76. [Google Scholar] [CrossRef]
- Li, X.; Liu, Y.; Yang, X.; Li, C.; Song, Z. The Oral Microbiota: Community Composition, Influencing Factors, Pathogenesis, and Interventions. Front. Microbiol. 2022, 13. [Google Scholar] [CrossRef]
- Verma, D.; Garg, P.K.; Dubey, A.K. Insights into the human oral microbiome. Arch. Microbiol. 2018, 200, 525–540. [Google Scholar] [CrossRef] [PubMed]
- Zaura, E.; Pappalardo, V.Y.; Buijs, M.J.; Volgenant, C.M.C.; Brandt, B.W. Optimizing the quality of clinical studies on oral microbiome: A practical guide for planning, performing, and reporting. Periodontology 2021, 85, 210–236. [Google Scholar] [CrossRef]
- Jensen, E.D.; Selway, C.A.; Allen, G.; Bednarz, J.; Weyrich, L.S.; Gue, S.; Peña, A.S.; Couper, J. Early markers of periodontal disease and altered oral microbiota are associated with glycemic control in children with type 1 diabetes. Pediatr. Diabetes 2021, 22, 474–481. [Google Scholar] [CrossRef] [PubMed]
- van Heck, J.I.; Gacesa, R.; Stienstra, R.; Fu, J.; Zhernakova, A.; Harmsen, H.J.; Weersma, R.K.; Joosten, L.A.; Tack, C.J. The Gut Microbiome Composition Is Altered in Long-standing Type 1 Diabetes and Associates with Glycemic Control and Disease-Related Complications. Diabetes Care 2022, 45, 2084–2094. [Google Scholar] [CrossRef] [PubMed]
- de Groot, P.F.; Belzer, C.; Aydin, Ö.; Levin, E.; Levels, J.H.; Aalvink, S.; Boot, F.; Holleman, F.; van Raalte, D.H.; Scheithauer, T.P.; et al. Distinct fecal and oral microbiota composition in human type 1 diabetes, an observational study. PLoS ONE 2017, 12, e0188475. [Google Scholar] [CrossRef] [PubMed]
- Araszkiewicz, A.; Bandurska-Stankiewicz, E.; Borys, S.; Budzyński, A.; Cyganek, K.; Cypryk, K.; Czech, A.; Czupryniak, L.; Drzewoski, J.; Dzida, G.; et al. 2021 Guidelines on the management of patients with diabetes. A position of Diabetes Poland. Clin. Diabetol. 2021, 10, 1–113. [Google Scholar] [CrossRef]
- Chalmers, J.; King, P.; Spencer, A.; Wright, F.; Carter, K. The Oral Health Assessment Tool—Validity and reliability. Aust. Dent. J. 2005, 50, 191–199. [Google Scholar] [CrossRef]
- Demuyser, T.; De Geyter, D.; Van Dorpe, D.; Vandoorslaer, K.; Wybo, I. Extensive evaluation of fastidious anaerobic bacteria recovery from the Copan eSwab® transport system. J. Microbiol. Methods 2018, 144, 73–78. [Google Scholar] [CrossRef]
- Dental Supplies—Dental Products|KerrDental.com. (n.d.) Available online: https://www.kerrdental.com/ (accessed on 22 January 2023).
- Guentsch, A.; Kramesberger, M.; Sroka, A.; Pfister, W.; Potempa, J.; Eick, S. Comparison of Gingival Crevicular Fluid Sampling Methods in Patients With Severe Chronic Periodontitis. J. Periodontol. 2011, 82, 1051–1060. [Google Scholar] [CrossRef]
- Produkty—GRASO Biotech. (n.d.) Available online: https://grasobiotech.pl/produkty/ (accessed on 22 January 2023).
- Biomaxima. (n.d.) Available online: https://biomaxima.com/ (accessed on 22 January 2023).
- Thermo Fisher Scientific—PL. (n.d.) Available online: https://www.thermofisher.com/pl/en/home.html (accessed on 22 January 2023).
- Jang, K.-S.; Kim, Y.H. Rapid and robust MALDI-TOF MS techniques for microbial identification: A brief overview of their diverse applications. J. Microbiol. 2018, 56, 209–216. [Google Scholar] [CrossRef]
- Mosler, B.; Twardawa, H.; Trzcionka, A.; Korkosz, R.; Rahnama, M.; Tanasiewicz, M. Oral Cavity Status of Type 1 Diabetic Patients Who Underwent an Oral Hygiene Tuition. Healthcare 2022, 10, 606. [Google Scholar] [CrossRef] [PubMed]
- Pachoński, M.; Koczor-Rozmus, A.; Mocny-Pachońska, K.; Łanowy, P.; Mertas, A.; Jarosz-Chobot, P. Oral microbiota in children with type 1 diabetes mellitus. Pediatr. Endocrinol. Diabetes Metab. 2021, 27. [Google Scholar] [CrossRef]
- Verhulst, M.J.L.; Loos, B.G.; Gerdes, V.E.A.; Teeuw, W.J. Evaluating All Potential Oral Complications of Diabetes Mellitus. Front. Endocrinol. 2019, 10, 56. [Google Scholar] [CrossRef]
- Latti, B.R.; Kalburge, J.V.; Birajdar, S.B.; Latti, R.G. Evaluation of relationship between dental caries, diabetes mellitus and oral microbiota in diabetics. J. Oral Maxillofac. Pathol. 2018, 22, 282. [Google Scholar] [CrossRef] [PubMed]
- Matsha, T.; Prince, Y.; Davids, S.; Chikte, U.; Erasmus, R.; Kengne, A.; Davison, G. Oral Microbiome Signatures in Diabetes Mellitus and Periodontal Disease. J. Dent. Res. 2020, 99, 658–665. [Google Scholar] [CrossRef] [PubMed]
- Surowiec, P.; Matejko, B.; Kopka, M.; Filemonowicz-Skoczek, A.; Klupa, T.; Cyganek, K.; Romanowska-Dixon, B.; Malecki, M.T. Low prevalence of diabetic retinopathy in patients with long-term type 1 diabetes and current good glycemic control - one-center retrospective assessment. Endocrine 2021, 75, 427–436. [Google Scholar] [CrossRef]
- Deo, P.N.; Deshmukh, R. Oral microbiome: Unveiling the fundamentals. J. Oral Maxillofac. Pathol. 2019, 23, 122–128. [Google Scholar] [CrossRef]
- Zaura, E.; Keijser, B.J.F.; Huse, S.M.; Crielaard, W. Defining the healthy "core microbiome" of oral microbial communities. BMC Microbiol. 2009, 9, 259. [Google Scholar] [CrossRef]
- Moskovitz, M.; Nassar, M.; Moriel, N.; Cher, A.; Faibis, S.; Ram, D.; Zangen, D.; Yassour, M.; Steinberg, D. Characterization of the Oral Microbiome Among Children with Type 1 Diabetes Compared with Healthy Children. Front. Microbiol. 2021, 12. [Google Scholar] [CrossRef]
- Kunath, B.J.; Hickl, O.; Queirós, P.; Martin-Gallausiaux, C.; Lebrun, L.A.; Halder, R.; Laczny, C.C.; Schmidt, T.S.B.; Hayward, M.R.; Becher, D.; et al. Alterations of oral microbiota and impact on the gut microbiome in type 1 diabetes mellitus revealed by integrated multi-omic analyses. Microbiome 2022, 10, 243. [Google Scholar] [CrossRef]
- Carneiro, V.L.; Fraiz, F.C.; Ferreira, F.D.M.; Pintarelli, T.P.; Oliveira, A.C.B.; Boguszewski, M.C.D.S. The influence of glycemic control on the oral health of children and adolescents with diabetes mellitus type 1. Arq. Bras. Endocrinol. Metabol. 2015, 59, 535–540. [Google Scholar] [CrossRef]
- Yuan, X.; Wu, J.; Chen, R.; Chen, Z.; Su, Z.; Ni, J.; Zhang, M.; Sun, C.; Zhang, F.; Liu, Y.; et al. Characterization of the oral microbiome of children with type 1 diabetes in the acute and chronic phases. J. Oral Microbiol. 2022, 14. [Google Scholar] [CrossRef]
- Mrozinska, S.; Kapusta, P.; Gosiewski, T.; Sroka-Oleksiak, A.; Ludwig-Słomczyńska, A.H.; Matejko, B.; Kiec-Wilk, B.; Bulanda, M.; Malecki, M.T.; Wolkow, P.P.; et al. The Gut Microbiota Profile According to Glycemic Control in Type 1 Diabetes Patients Treated with Personal Insulin Pumps. Microorganisms 2021, 9, 155. [Google Scholar] [CrossRef] [PubMed]
- Ghannoum, M.A.; Jurevic, R.J.; Mukherjee, P.K.; Cui, F.; Sikaroodi, M.; Naqvi, A.; Gillevet, P.M. Characterization of the Oral Fungal Microbiome (Mycobiome) in Healthy Individuals. PLoS Pathog. 2010, 6, e1000713. [Google Scholar] [CrossRef]
- Dehghan, P.; Mohammadi, F.; Javaheri, M.R.; Nekoeian, S. Identification of Candida species in the oral cavity of diabetic patients. Curr. Med Mycol. 2016, 2, 1–7. [Google Scholar] [CrossRef]
- Unniachan, A.S.; Jayakumari, N.K.; Sethuraman, S. Association between Candida species and periodontal disease: A systematic review. Curr. Med Mycol. 2020, 6, 63–68. [Google Scholar] [CrossRef]
- Eliskases-Lechner, F.; Guéguen, M.; Panoff, J. Yeasts and Molds|Geotrichum candidum. Encycl. Dairy Sci. Second. Ed. 2011, 11, 765–771. [Google Scholar] [CrossRef]
- Yamashita, Y.; Takeshita, T. The oral microbiome and human health. J. Oral Sci. 2017, 59, 201–206. [Google Scholar] [CrossRef]
- Marsh, P. In Sickness and in Health—What Does the Oral Microbiome Mean to Us? An Ecological Perspective. Adv. Dent. Res. 2018, 29, 60–65. [Google Scholar] [CrossRef]
- Zhou, Y.; Gao, H.; Mihindukulasuriya, K.A.; La Rosa, P.S.; Wylie, K.M.; Vishnivetskaya, T.; Podar, M.; Warner, B.; Tarr, P.I.; Nelson, D.E.; et al. Biogeography of the ecosystems of the healthy human body. Genome Biol. 2013, 14, R1. [Google Scholar] [CrossRef]
- Hall, M.W.; Singh, N.; Ng, K.F.; Lam, D.K.; Goldberg, M.B.; Tenenbaum, H.C.; Neufeld, J.D.; Beiko, R.G.; Senadheera, D.B. Inter-personal diversity and temporal dynamics of dental, tongue, and salivary microbiota in the healthy oral cavity. Npj Biofilms Microbiomes 2017, 3, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Richards, V.P.; Alvarez, A.J.; Luce, A.R.; Bedenbaugh, M.; Mitchell, M.L.; Burne, R.A.; Nascimento, M.M. Microbiomes of Site-Specific Dental Plaques from Children with Different Caries Status. Infect. Immun. 2017, 85, e00106-17. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Shi, M.; Zhen, M.; Wang, C.; Hu, W.; Nie, Y.; Wu, X. Comparison of Subgingival and Buccal Mucosa Microbiome in Chronic and Aggressive Periodontitis: A Pilot Study. Front. Cell. Infect. Microbiol. 2019, 9, 53. [Google Scholar] [CrossRef] [PubMed]
- Foxman, B.; Luo, T.; Srinivasan, U.; Ramadugu, K.; Wen, A.; Goldberg, D.; Shedden, K.; Crout, R.; McNeil, D.W.; Weyant, R.; et al. The effects of family, dentition, and dental caries on the salivary microbiome. Ann. Epidemiol. 2016, 26, 348–354. [Google Scholar] [CrossRef]
- Faust, K.; Sathirapongsasuti, J.F.; Izard, J.; Segata, N.; Gevers, D.; Raes, J.; Huttenhower, C. Microbial Co-occurrence Relationships in the Human Microbiome. PLoS Comput. Biol. 2012, 8, e1002606. [Google Scholar] [CrossRef]
- Yu, G.; Phillips, S.; Gail, M.H.; Goedert, J.J.; Humphrys, M.; Ravel, J.; Ren, Y.; Caporaso, N.E. Evaluation of Buccal Cell Samples for Studies of Oral Microbiota. Cancer Epidemiol. Biomark. Prev. 2017, 26, 249–253. [Google Scholar] [CrossRef]

| Clinical Feature | Available Data, N (%) | Mean (SD), Median (Q1–Q3) or Number (%) | p-Value |
|---|---|---|---|
| Age | 22 (100%) | 27.05 (5.95) 26.5 (22–29.25) | - |
| Gender [male] | 22 (100%) | 13 (59.1%) | - |
| Weight [kg] | 15 (68.2%) | 71.78 (17.75) 66.0 (57–90.0) | - |
| HbA1c [%] $ | 22(100%) | 6.97 (0.95) 6.85 (6.3–7.35 | |
| Fasting glycemia [mg/dL] | 21 (95.5%) | 116.24 (38.29) 112.0 (93.0–131.5) | - |
| OHAT [score] | 22 (100%) | 0/16 (19, 86.4%) 1/16 (3, 13.6%) | - |
| Microbial Counts | |||
| Number of genera | 22 (100%) | 0.459 | |
| A | 2 (1.75–4) | ||
| B | 2 (2–3) | ||
| C | 3 (2–4) | ||
| Da | 2 (1–3) | ||
| Db | 3 (2–3) | ||
| E | 2 (2–3) | ||
| Number of species | 22 (100%) | 0.459 | |
| A | 4 (3–5) | ||
| B | 5 (4–6) | ||
| C | 5 (3.75–6.25) | ||
| Da | 3 (2.75–6) | ||
| Db | 5 (3–6) | ||
| E | 4 (3–5) | ||
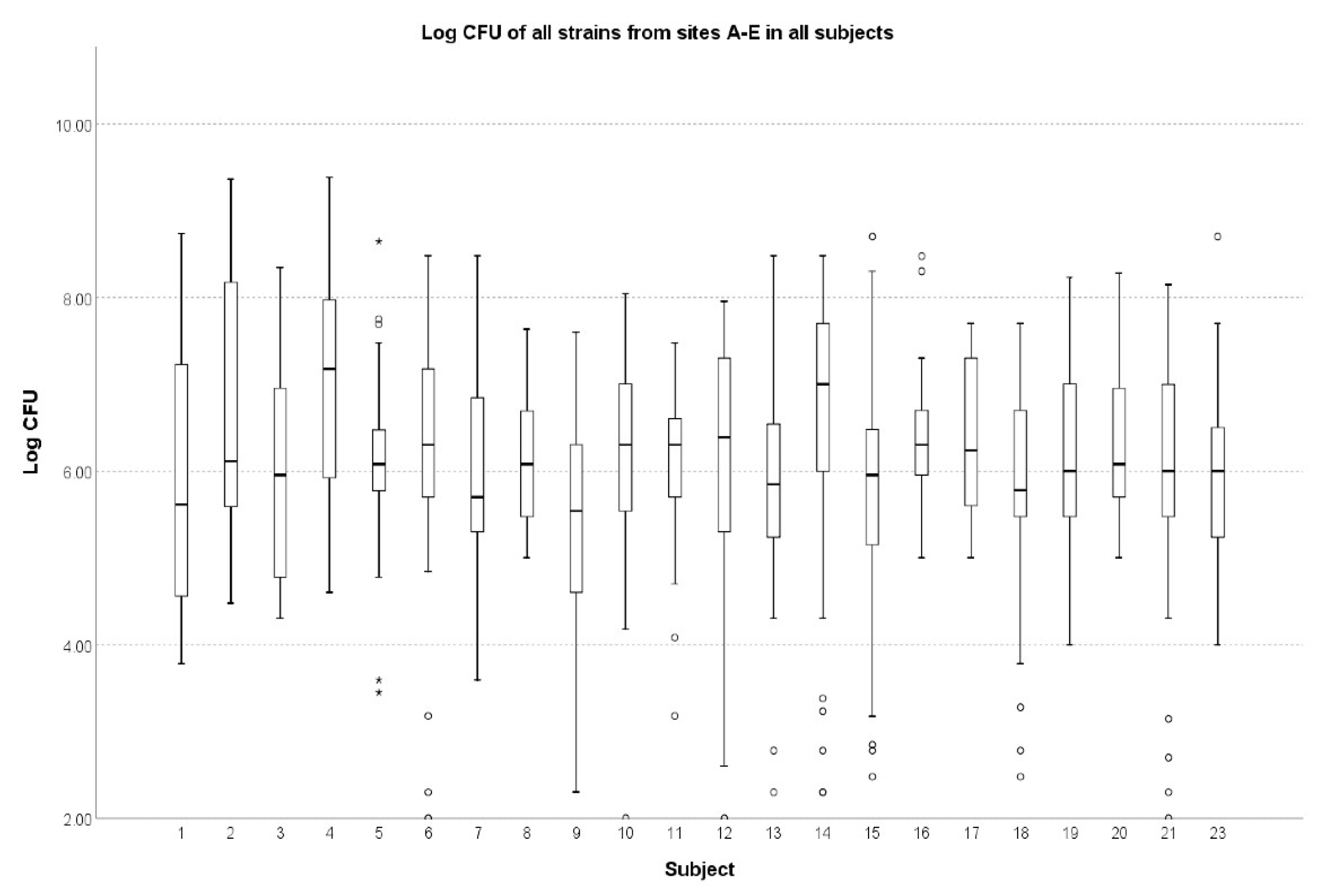
| Mean CFU [CFU/mL] | 22 (100%) | 0.018 # | |
| All sites A–E | 3.88 × 107 ± 1.88 × 108 1.20 × 106 (3.00× 105 –1.00 × 107) | ||
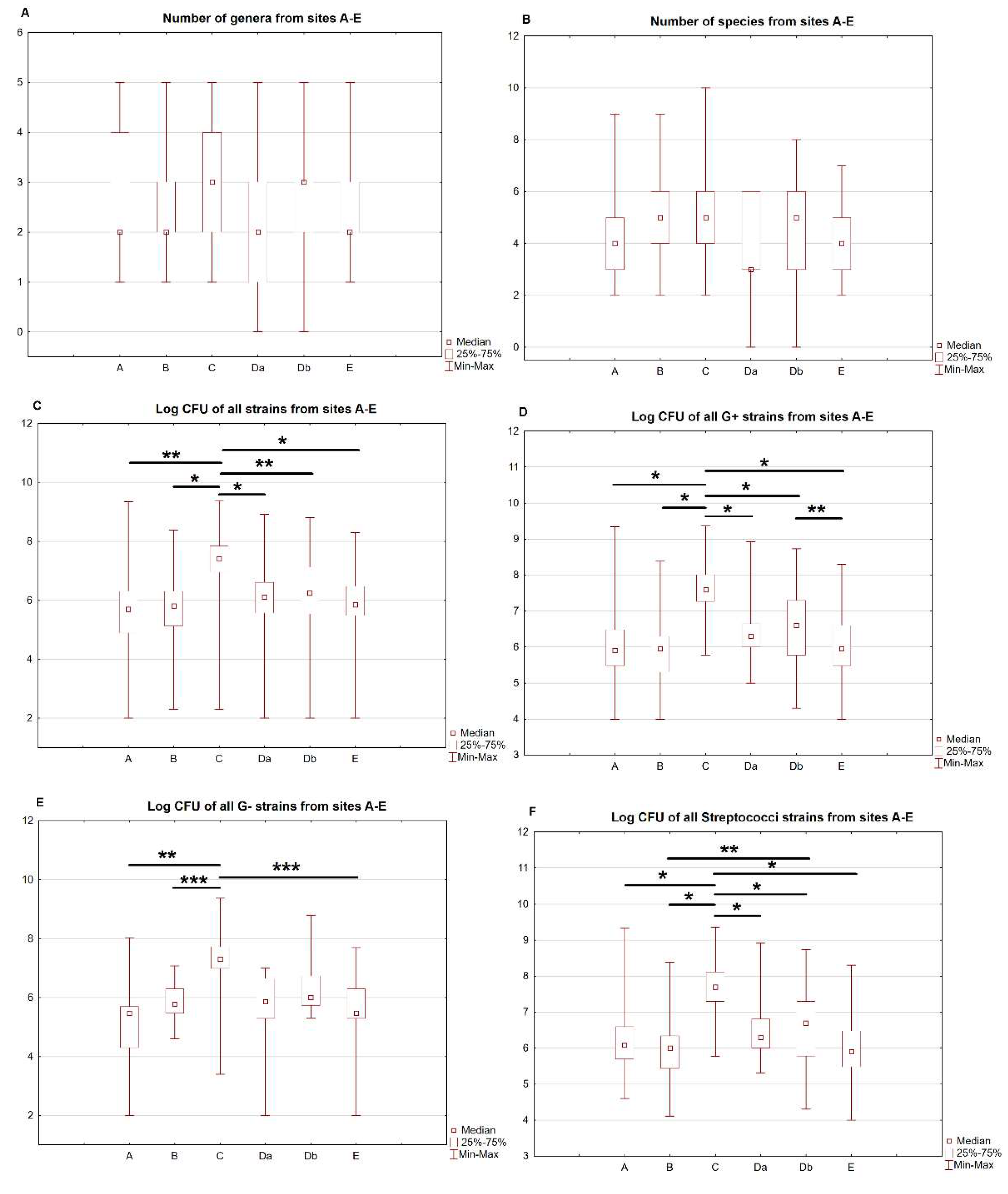
| Site | A | B | C | Da | Db | E | p-Value(A–E) | p-Value(A–E, exl. C) |
|---|---|---|---|---|---|---|---|---|
| No. of genera | 2 (1.75–4) | 2 (2–3) | 3 (2–4) | 2 (1–3) | 3 (2–3) | 2 (2–3) | 0.299 | - |
| No. of species | 4 (3–5) | 5 (4–6) | 5 (3.75–6.25) | 3 (2.75–6) | 5 (3–6) | 4 (3–5) | 0.124 | - |
| No. of samples [N], CFU [CFU/mL] | ||||||||
| Overall & | 100 4.34 × 107 (2.40 × 108) 5.00 × 105 (7.48 × 104–2.00 × 106) | 112 8.25 × 106 (3.56 × 107) 6.50 × 105 (1.28 × 105–2.00 × 106) | 114 1.05 × 108 (3.17 × 108) 2.50 × 107 (8.75 × 106–7.00 × 107) | 80 2.08 × 107 (1.09 × 108) 1.30 × 106 (3.55 × 105–4.00 × 107) | 96 2.57 × 107 (8.93 × 107) 1.75 × 106 (3.25 × 105–1.43 × 107) | 89 7.17 × 106 (2.43 × 107) 7.00 × 105 (3.00 × 105–3.00 × 106) | <0.001 # | 0.001 * |
| Gram-positive & | 72 5.85 × 107 (2.82 × 108) 8.00 × 105 (3.00 × 105–3.00 × 106) | 82 1.09 × 107 (4.13 × 107) 9.00 × 105 (2.00 × 105–2.00 × 106) | 76 1.08 × 108 (2.76 × 108) 3.90 × 107 (1.83 × 107–1.06 × 108) | 52 3.12 × 107 (1.34 × 108) 2.00 × 106 (1.00 × 106–4.75 × 106) | 63 2.73 × 107 (7.77 × 107) 4.00 × 106 (6.00 × 105–2.00 × 107) | 67 8.20 × 106 (2.73 × 107) 9.00 × 105 (3.00 × 105–4.00 × 106) | <0.001 $ | 0.001 φ |
| Staphylococci & | 4 2.03 × 105 (1.75 × 105) 1.80 × 105 (5.50 × 104 –3.50 × 105) | 5 2.06 × 105 (2.28 × 105) 1.00 × 105 (2.00 × 104–4.00 × 105) | 7 5.00 × 107 (7.76 × 107) 2.00 × 107 (1.00 × 106–6.00 × 107) | 2 1.75 × 106 (3.54 × 105) 1.75 × 106 (1.50 × 106–2.00 × 106) | 4 4.25 × 107 (2.25 × 107) 4.10 × 107 (2.50 × 107–6.00 × 107) | 6 2.47 × 106 (2.63 × 106) 1.70 × 106 (4.00 × 105–4.00 × 106) | 0.005 φ | 0.03 φ |
| Streptococci & | 61 6.90 × 107 (3.05 × 108) 1.20 × 106 (5.00 × 105 –4.00 × 106) | 67 1.32 × 107 (4.54 × 107) 1.00 × 106 (2.80 × 105–2.20 × 106) | 58 1.27 × 108 (3.12 × 108) 4.95 × 107 (2.00 × 107–1.30 × 108) | 44 3.66 × 107 (1.46 × 108) 2.00 × 106 (1.00 × 106–6.50 × 106) | 51 2.99 × 107 (1.46 × 108) 5.00 × 106 (6.0 × 105–2.00 × 107) | 51 8.66 × 106 (3.06 × 107) 8.00 × 105 (3.00 × 105–3.00 × 106) | <0.001 ψ | 0.001 § |
| Gram-negative & | 18 6.64 × 106 (2.58 × 107) 3.00 × 105 (1.88 × 104–5.50 × 105) | 19 1.62 × 106 (2.87 × 106) 6.00 × 105 (3.00 × 105–2.00 × 106) | 19 5.01 × 107 (5.31 × 108) 2.00 × 107 (1.00 × 107–4.00 × 107) | 12 2.59 × 106 (3.42 × 106) 7.50 × 105 (2.00 × 105–4.75 × 106) | 20 3.71 × 107 (1.40 × 108) 1.00 × 106 (5.25 × 105–5.75 × 106) | 19 4.55 × 106 (1.17 × 107) 3.00 × 105 (2.00 × 105–2.00 × 106) | <0.001% | 0.054 |
| Candida spp. | 5 9.00 × 102 (8.33 × 102) 5.00 × 102 (2.00 × 102–1.80 × 103) | 4 4.50 × 102 (2.38 × 102) 4.50 × 102 (2.25 × 102–6.75 × 102) | 9 1.52 × 103 (1.84 × 103) 1.20 × 103 (2.00 × 103–1.95 × 103) | 9 1.64 × 105 (4.28 × 105) 4200 (2.70 × 103–7.50 × 104) | 9 2.86 × 104 (7.2 × 104) 2.80 × 103 (4.50 × 102–1.38 × 104) | 0 | 0.22 | 0.23 |
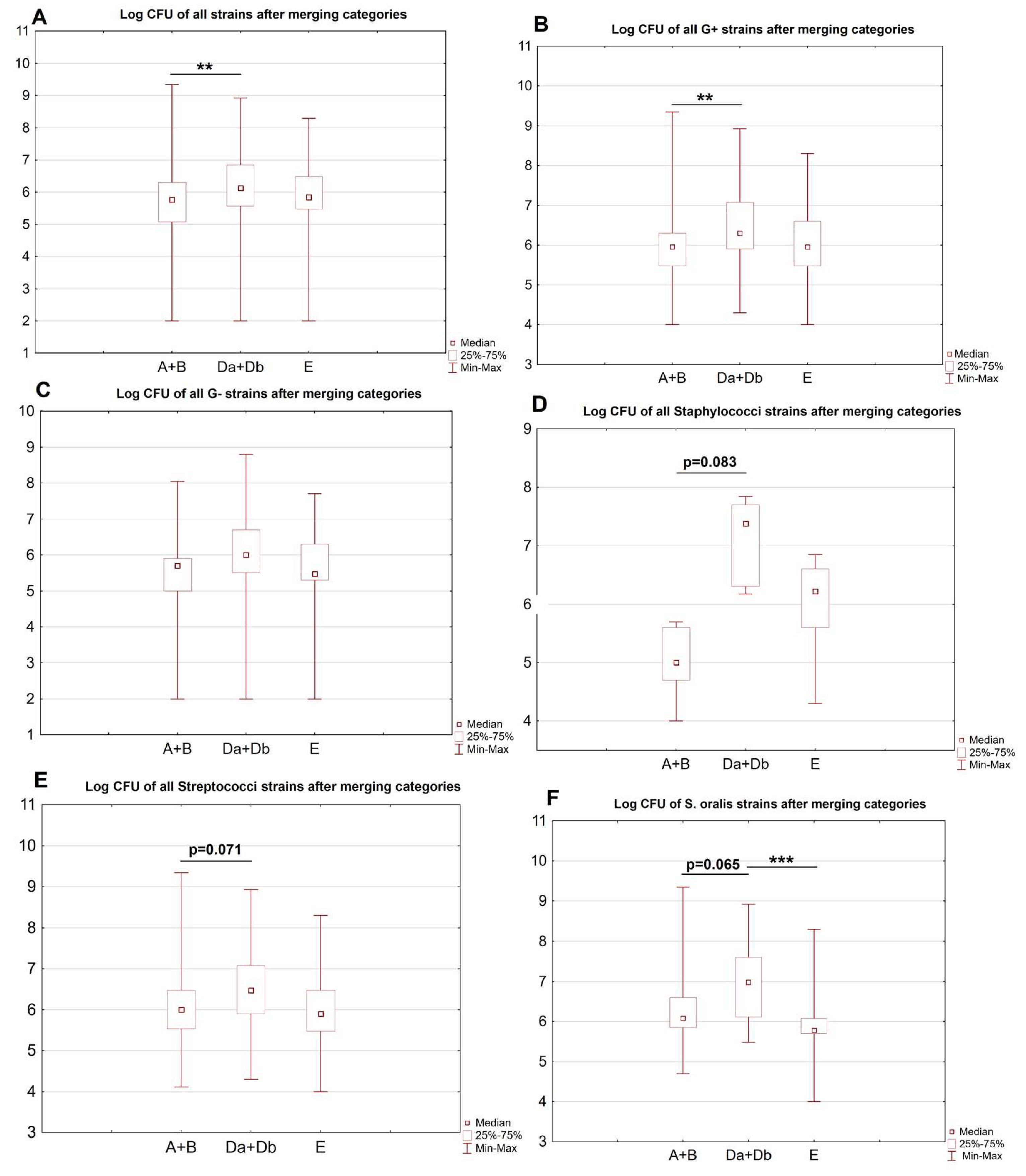
| Site | A + B | Da + Db | E | p-Value |
|---|---|---|---|---|
| No. of cultures [N], CFU counts [CFU/mL] | ||||
| Overall # | 212 2.48 × 107 (1.67 × 108) 6.00 × 105 (1.20 × 105–2.00 × 106) | 176 2.34 × 107 (9.85 × 107) 1.35 × 106 (3.70 × 105–7.00 × 106) | 89 7.17 × 106 (2.43 × 107) 7.00 × 105 (3.00 × 105–3.00 × 106 | 0.006 $ |
| Gram-positive # | 154 3.32 × 107 (1.96 × 108) 9.00 × 105 (3.00 × 105–2.00 × 106) | 115 2.91 × 107 (1.07 × 108) 2.00 × 106 (8.00 × 105–1.20 × 107) | 67 8.20 × 106 (2.73 × 107) 9.00 × 105 (3.00 × 105–4.00 × 106) | 0.002% |
| Gram-negative # | 37 4.06 × 106 (1.80 × 107) 5.00 × 105 (1.00 × 105–8.00 × 105) | 32 2.42 × 107 (1.11 × 108) 1.00 × 106 (3.20 × 105–5.00 × 106) | 19 4.55 × 106 (1.17 × 107) 3.00 × 105 (2.00 × 105–2.00 × 106) | 0.083 |
| Veillonella # | 9 1.02 × 106 (5.15 × 106) 5.00 × 105 (3.00 × 105–9.00 × 105) | 6 8.12 × 106 (1.52 × 106) 1.45 × 106 (6.00 × 105–5.00 × 106) | 4 3.38 × 106 (1.57 × 107) 1.15 × 106 (2.50 × 105–6.50 × 106) | 0.42 |
| Neisseria # | 21 6.63 × 106 (2.38 × 107) 4.00 × 105 (8.00 × 104–2.00 × 106) | 15 4.59 × 107 (1.62 × 108) 9.00 × 105 (3.00 × 105–4.80 × 106) | 2 2.75 × 107 (3.18 × 107) 2.75 × 107 (5.00 × 106–5.00 × 107) | 0.14 |
| Staphylococci # | 9 2.04 × 105 (1.94 × 105) 1.00 × 105 (5.00 × 104–4.00 × 106) | 6 2.89 × 107 (2.73 × 107) 2.50 × 107 (2.00 × 105–4.00 × 106) | 9 2.47 × 106 (2.63 × 106) 1.70 × 106 (4.00 × 105–5.00 × 107) | 0.016 & |
| Actinomyces # | 8 7.14 × 105 (8.17 × 105) 4.00 × 105 (1.50 × 105–1.30 × 106) | 9 1.57 × 106 (1.11 × 106) 1.00 × 106 (1.00 × 106–2.00 × 106) | 3 2.77 × 106 (2.36 × 106) 3.0 0 × 106 (3.00 × 105–5.00 × 106) | 0.2 |
| Streptococci # | 128 3.98 × 107 (2.14 × 108) 1.00 × 106 (3.50 × 105–3.00 × 106) | 95 3.30 × 107 (1.17 × 108) 3.00 × 106 (8.00 × 105–1.20 × 107) | 51 8.66 × 106 (3.06 × 107) 8.00 × 105 (3.00 × 105–3.00 × 106) | 0.034 & |
| S. vetibularis | 20 1.71 × 107 (4.57 × 107) 1.30 × 106 (4.50 × 105–3.80 × 106) | 10 1.64 × 107 (4.03 × 107) 2.50 × 106 (8.00 × 105–3.00 × 106) | 3 2.97 × 106 (3.52 × 106) 1.40 × 106 (5.00 × 105–7.00 × 106 | 0.38 |
| S. salivarius | 30 3.43 × 107 (1.57 × 108) 9.50 × 105 (3.00 × 105–2.00 × 106) | 10 7.25 × 106 (1.27 × 107) 3.00 × 106 (8.00 × 105–8.00 × 106) | 7 8.37 × 105 (1.05 × 106) 8.00 × 105 (4.00 × 104–1.00 × 106) | 0.22 |
| S. parapneumonie | 18 1.87 × 107 (6.10 × 107) 9.50 × 105 (5.00 × 105–2.20 × 106) | 7 5.83 × 106 (5.28 × 106) 5.00 × 106 (1.40 × 106–1.00 × 107) | 3 7.10 × 106 (6.85 × 106) 7.00 × 106 (3.00 × 105–1.4 0 × 107) | 0.72 |
| S. oralis | 26 1.28 × 107 (4.36 × 108) 1.20 × 106 (7.00 × 105–4.00 × 106) | 26 1.04 × 108 (1.89 × 108) 9.50 × 106 (1.30 × 106–4.00 × 107) | 17 6.93 × 107 (4.83 × 107) 6.00 × 105 (5.00 × 105–1.20 × 106) | <0.001 * |
| S. mitis | 19 1.42 × 107 (5.00 × 107) 1.10 × 106 (2.00 × 105–4.00 × 106) | 3 2.87 × 106 (3.60 × 106) 1.20 × 106 (4.00 × 105–7.00 × 106) | 9 4.79 × 106 (7.61 × 106) 9.00 × 105 (3.00 × 105–3.00 × 106) | 0.47 |
| Candida spp. | 9 7.00 × 102 (6.52 × 102) 5.00 × 102 (2.00 × 102–7.00 × 102) | 18 9.63 × 104 (3.06 × 105) 4.05 × 103 (1.50 × 103–1.8 × 104) | 0 | 0.015 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gregorczyk-Maga, I.; Fiema, M.; Kania, M.; Jachowicz-Matczak, E.; Romaniszyn, D.; Gerreth, K.; Klupa, T.; Wójkowska-Mach, J. Oral Microbiota—One Habitat or Diverse Niches? A Pilot Study of Sampling and Identification of Oral Bacterial and Fungal Biota in Patients with Type I Diabetes Mellitus Treated with Insulin Pump. Int. J. Environ. Res. Public Health 2023, 20, 2252. https://doi.org/10.3390/ijerph20032252
Gregorczyk-Maga I, Fiema M, Kania M, Jachowicz-Matczak E, Romaniszyn D, Gerreth K, Klupa T, Wójkowska-Mach J. Oral Microbiota—One Habitat or Diverse Niches? A Pilot Study of Sampling and Identification of Oral Bacterial and Fungal Biota in Patients with Type I Diabetes Mellitus Treated with Insulin Pump. International Journal of Environmental Research and Public Health. 2023; 20(3):2252. https://doi.org/10.3390/ijerph20032252
Chicago/Turabian StyleGregorczyk-Maga, Iwona, Mateusz Fiema, Michal Kania, Estera Jachowicz-Matczak, Dorota Romaniszyn, Karolina Gerreth, Tomasz Klupa, and Jadwiga Wójkowska-Mach. 2023. "Oral Microbiota—One Habitat or Diverse Niches? A Pilot Study of Sampling and Identification of Oral Bacterial and Fungal Biota in Patients with Type I Diabetes Mellitus Treated with Insulin Pump" International Journal of Environmental Research and Public Health 20, no. 3: 2252. https://doi.org/10.3390/ijerph20032252
APA StyleGregorczyk-Maga, I., Fiema, M., Kania, M., Jachowicz-Matczak, E., Romaniszyn, D., Gerreth, K., Klupa, T., & Wójkowska-Mach, J. (2023). Oral Microbiota—One Habitat or Diverse Niches? A Pilot Study of Sampling and Identification of Oral Bacterial and Fungal Biota in Patients with Type I Diabetes Mellitus Treated with Insulin Pump. International Journal of Environmental Research and Public Health, 20(3), 2252. https://doi.org/10.3390/ijerph20032252







