Infant Low Birth Weight Prediction Using Graph Embedding Features
Abstract
:1. Introduction
- A well-curated dataset obtained from 3453 patients with 41 important risk factors was used for infant BW prediction in the UAE.
- Experiments were performed using the five most commonly used ML classifiers on the original tabular dataset and graphs obtained from the original dataset.
- A detailed performance evaluation was performed using the original risk factors, graph features, and combinations of these features.
2. Materials and Methods
2.1. Dataset Collection and Data Preprocessing
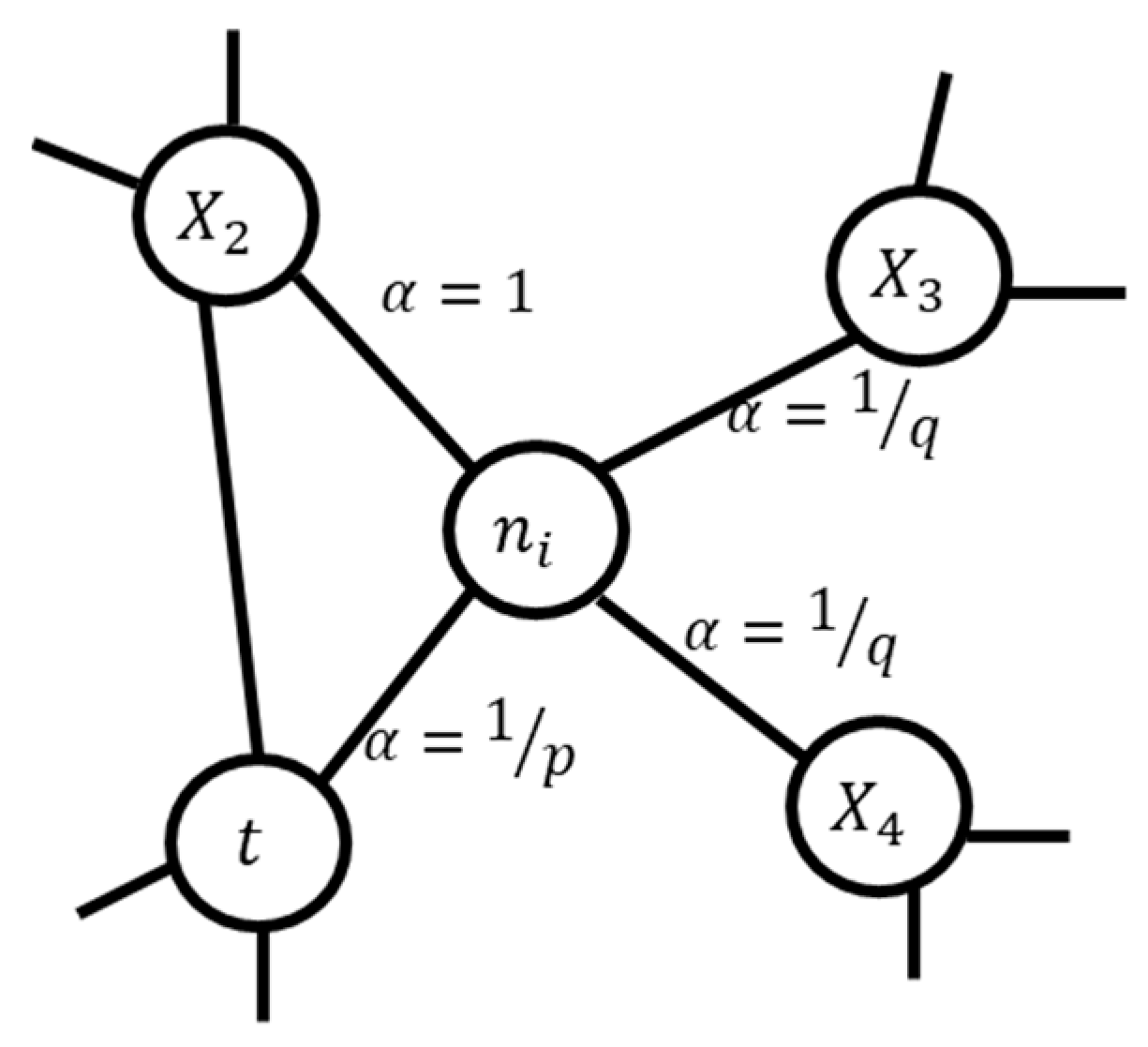
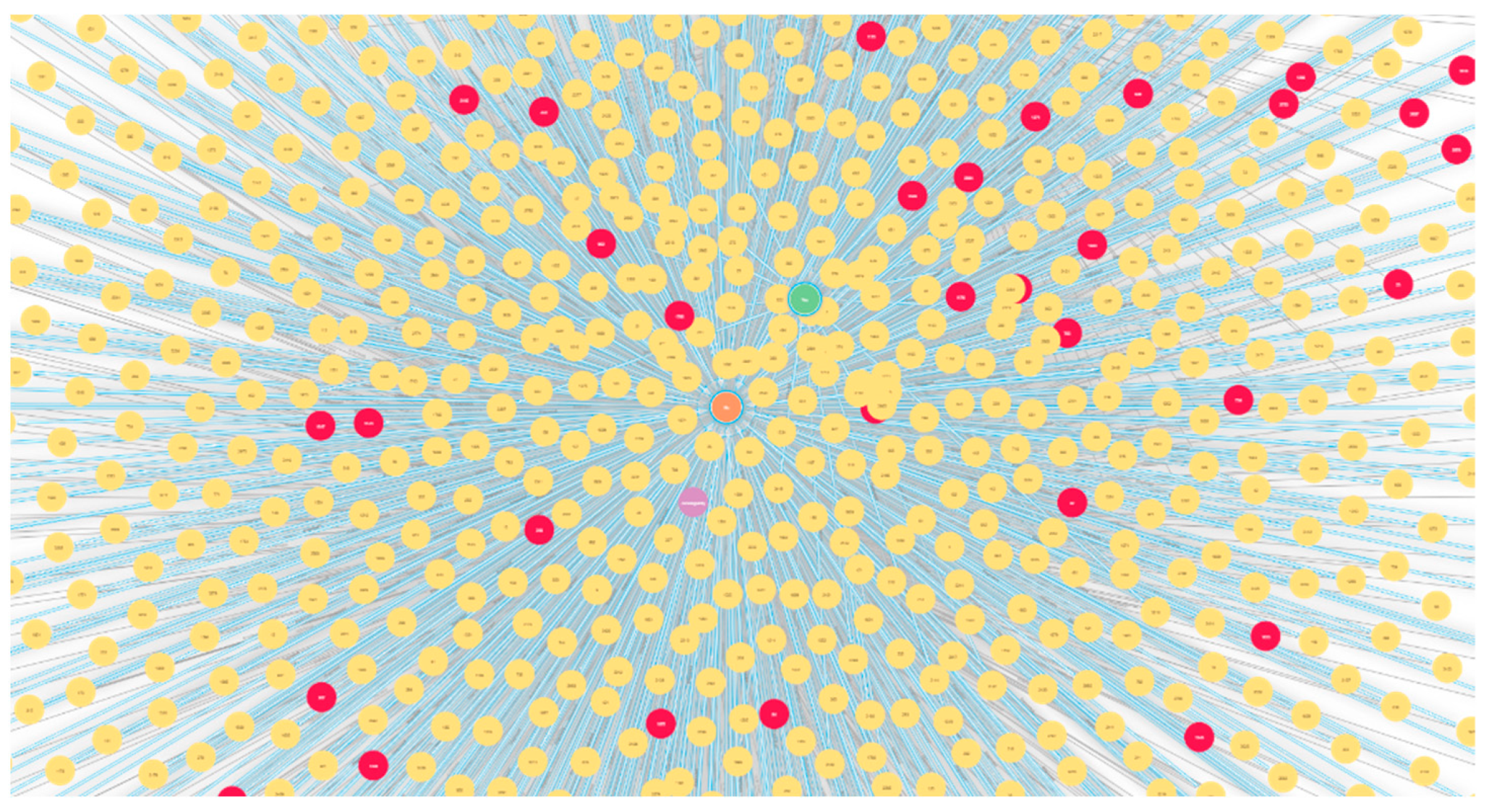
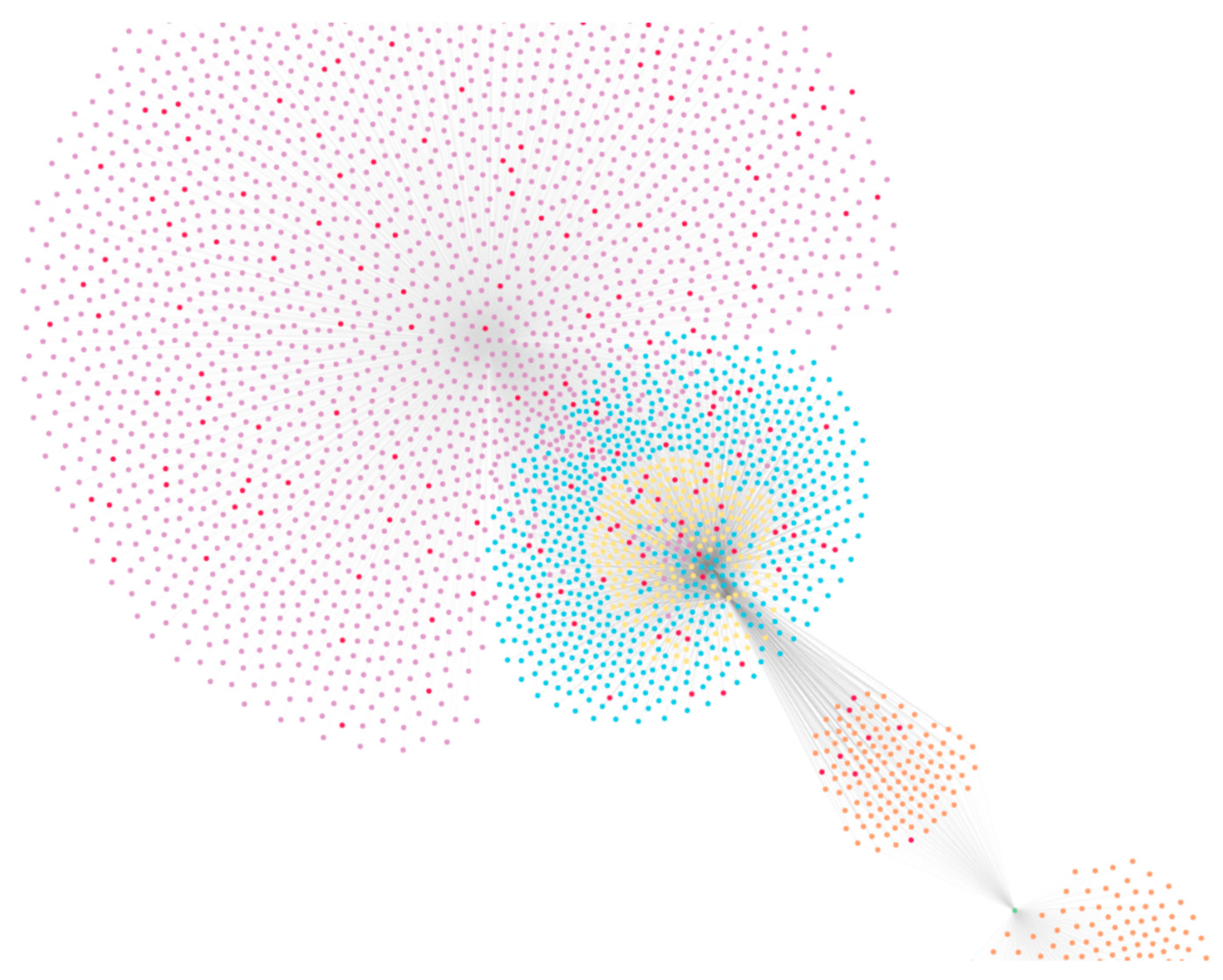
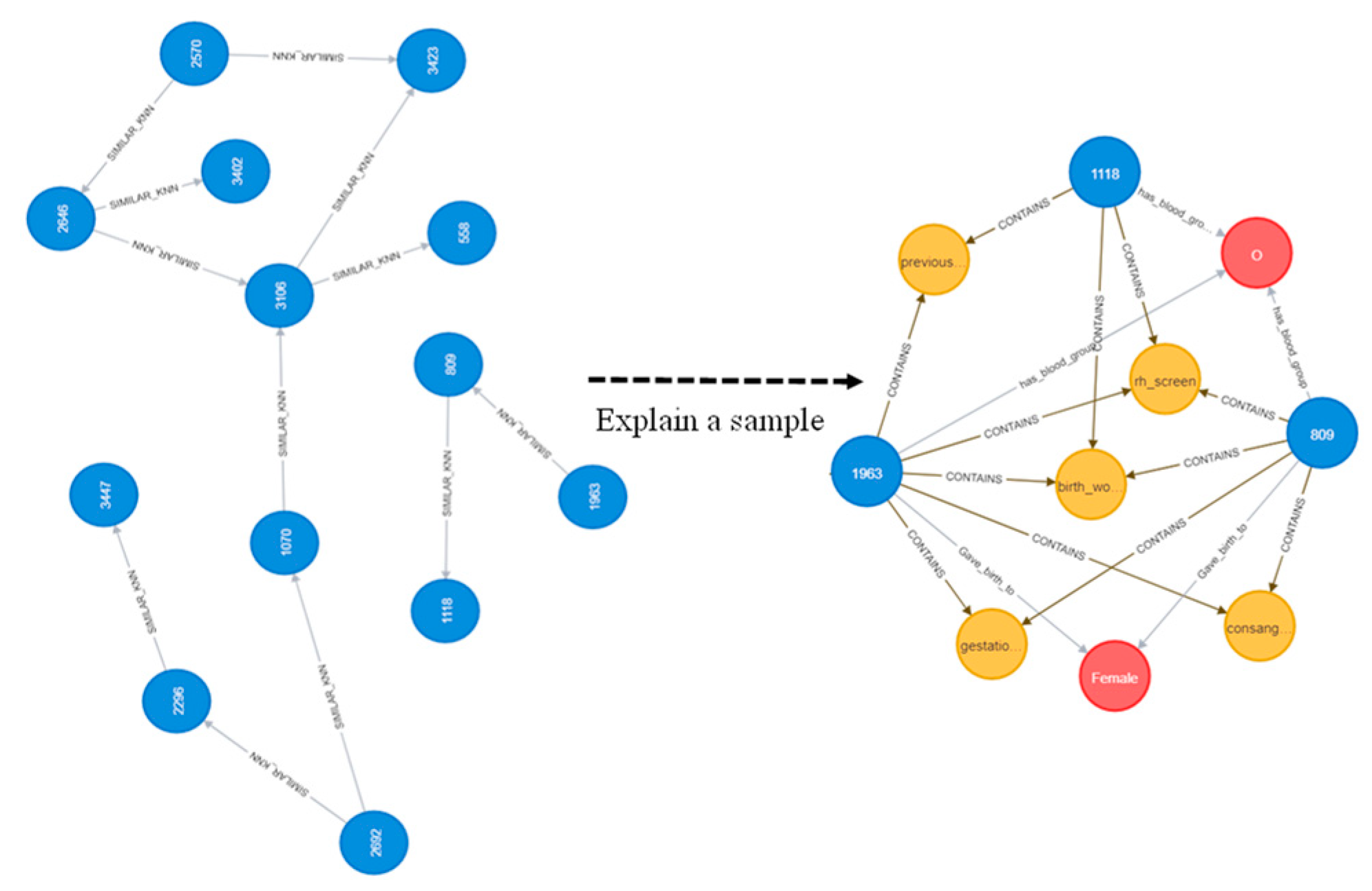
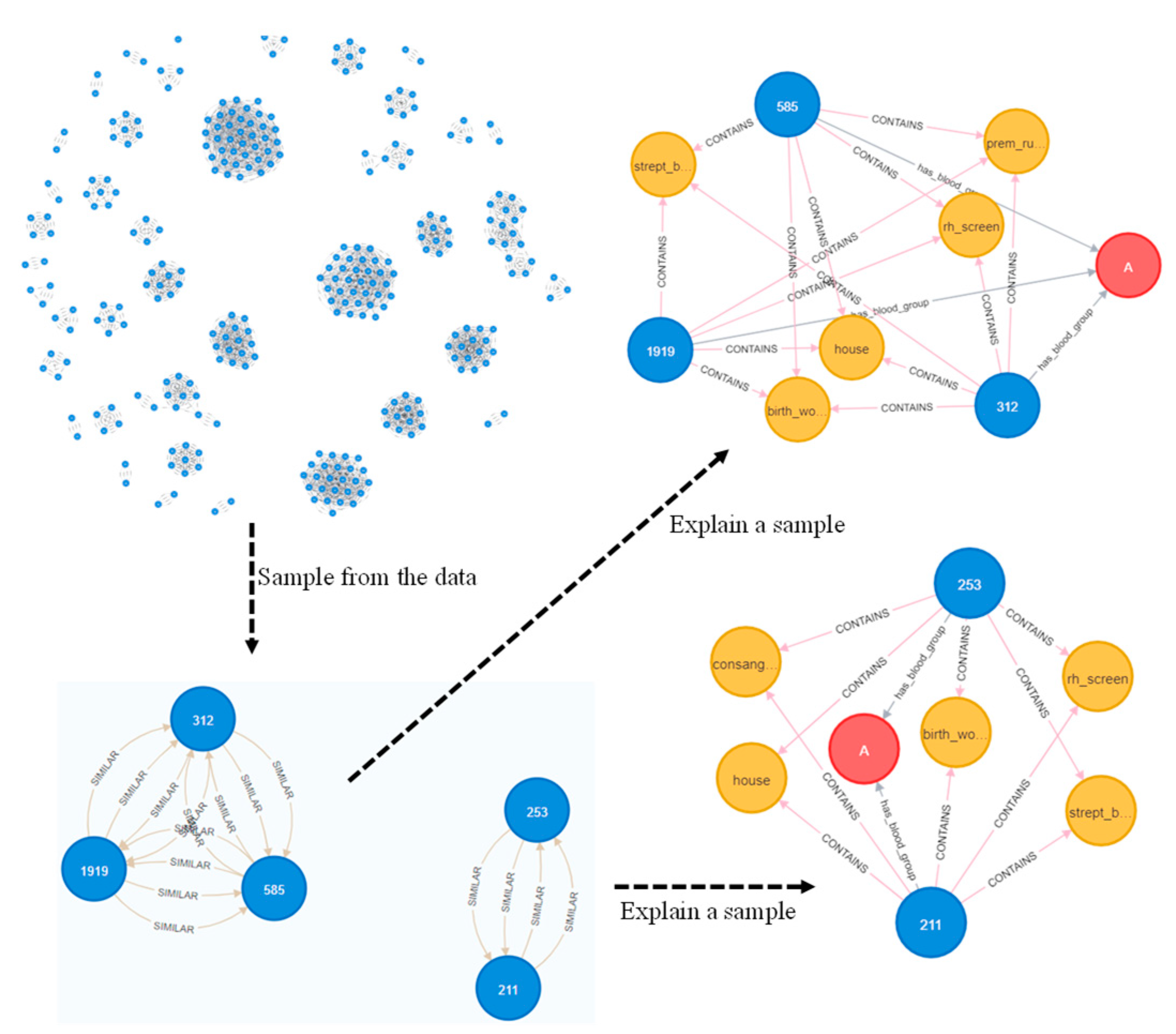
2.2. Problem Formulation and Knowledge Graph Construction
2.2.1. Node Embeddings
2.2.2. Graph Topological Features
- Graph degree;
- Closeness centrality;
- Betweenness centrality;
- Eigenvector centrality;
- Hub;
- Authority;
- PageRank;
- Clustering coefficient;
- K-nearest neighbors;
- Node similarity;
- Community detection;
2.2.3. Feature Combination for Classification
2.2.4. Machine Learning Models
2.2.5. Performance Metrics
3. Experiments and Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- WHO|World Health Organization. Available online: https://www.who.int/ (accessed on 21 December 2020).
- Khan, W.; Zaki, N.; Masud, M.M.; Ahmad, A.; Ali, L.; Ali, N.; Ahmed, L.A. Infant birth weight estimation and low birth weight classification in United Arab Emirates using machine learning algorithms. Sci. Rep. 2022, 12, 12110. [Google Scholar] [CrossRef]
- Jornayvaz, F.R.; Vollenweider, P.; Bochud, M.; Mooser, V.; Waeber, G.; Marques-Vidal, P. Low birth weight leads to obesity, diabetes and increased leptin levels in adults: The CoLaus study. Cardiovasc. Diabetol. 2016, 15, 73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reduction of Low Birth Weight: A South Asia Priority—PDF Free Download. Available online: https://docplayer.net/20755175-Reduction-of-low-birth-weight-a-south-asia-priority.html (accessed on 11 January 2021).
- Sitecontrol Low Birthweight UNICEF DATA. Available online: https://data.unicef.org/topic/nutrition/low-birthweight/ (accessed on 6 August 2022).
- Taha, Z.; Hassan, A.A.; Wikkeling-Scott, L.; Papandreou, D. Factors Associated with Preterm Birth and Low Birth Weight in Abu Dhabi, the United Arab Emirates. Int. J. Environ. Res. Public Health 2020, 17, 1382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Faruk, A.; Cahyono, E.S.; Eliyati, N.; Arifieni, I. Prediction and classification of low birth weight data using machine learning techniques. Indones. J. Sci. Technol. 2018, 3, 18–28. [Google Scholar] [CrossRef] [Green Version]
- Feng, M.; Wan, L.; Li, Z.; Qing, L.; Qi, X. Fetal Weight Estimation via Ultrasound Using Machine Learning. IEEE Access 2019, 7, 87783–87791. [Google Scholar] [CrossRef]
- Lu, Y.; Zhang, X.; Fu, X.; Chen, F.; Wong, K.K.L. Ensemble machine learning for estimating fetal weight at varying gestational age. Proc. AAAI Conf. Artif. Intell. 2019, 33, 9522–9527. [Google Scholar] [CrossRef] [Green Version]
- Campos Trujillo, O.; Perez-Gonzalez, J.; Medina-Bañuelos, V. Early Prediction of Weight at Birth Using Support Vector Regression. In IFMBE Proceedings; Springer: Berlin/Heidelberg, Germany, 2020; Volume 75, pp. 37–41. [Google Scholar] [CrossRef]
- Pollob, S.M.A.I.; Abedin, M.M.; Islam, M.T.; Islam, M.M.; Maniruzzaman, M. Predicting risks of low birth weight in Bangladesh with machine learning. PLoS ONE 2022, 17, e0267190. [Google Scholar] [CrossRef]
- Do, H.J.; Moon, K.M.; Jin, H.-S. Machine Learning Models for Predicting Mortality in 7472 Very Low Birth Weight Infants Using Data from a Nationwide Neonatal Network. Diagnostics 2022, 12, 625. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.-T.; Wu, T.-Y.; Chen, Y.-J.; Chang, Y.-S.; Lin, C.-H.; Lin, Y.-J. Predicting in-hospital length of stay for very-low-birth-weight preterm infants using machine learning techniques. J. Formos. Med. Assoc. 2022, 121, 1141–1148. [Google Scholar] [CrossRef]
- Abdulrazzaq, Y.M.; Bener, A.; Dawodu, A.; Kappel, I.; Surouri, F.A.; Varady, E.; Liddle, L.; Varghese, M.; Cheema, M.Y. Obstetric risk factors affecting incidence of low birth weight in live-born infants. Biol. Neonate 1995, 67, 160–166. [Google Scholar] [CrossRef]
- Nasir, B.; Zaman, J.; Alqemzi, N.; Musavi, N.; Adbullah, T.; Shaikh, B. Prevalence and Factors Related to Low Birth Weight in a Tertiary Hospital in Ajman UAE. GMJ 2014, 5–6, 45–51. [Google Scholar]
- Dawodu, A.; Abdulrazzaq, Y.M.; Bener, A.; Kappel, I.; Liddle, L.; Varghese, M. Biologic risk factors for low birthweight in Al Ain, United Arab Emirates. Am. J. Hum. Biol. Off. J. Hum. Biol. Counc. 1996, 8, 341–345. [Google Scholar] [CrossRef]
- Oprescu, A.M.; Miró-amarante, G.; García-Díaz, L.; Beltrán, L.M.; Rey, V.E.; Romero-Ternero, M. Artificial Intelligence in Pregnancy: A Scoping Review. IEEE Access 2020, 8, 181450–181484. [Google Scholar] [CrossRef]
- Wu, Z.; Pan, S.; Chen, F.; Long, G.; Zhang, C.; Yu, P.S. A Comprehensive Survey on Graph Neural Networks. IEEE Trans. Neural Netw. Learn. Syst. 2021, 32, 4–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zaki, N. From Tabulated Data to Knowledge Graph: A Novel Way of Improving the Performance of the Classification Models in the Healthcare Data. medRxiv 2021. [Google Scholar] [CrossRef]
- Tsuang, M. Schizophrenia: Genes and environment. Biol. Psychiatry 2000, 47, 210–220. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Semerci, M.; Yener, B.; Zaki, M.J. Effective graph classification based on topological and label attributes. Stat. Anal. Data Min. ASA Data Sci. J. 2012, 5, 265–283. [Google Scholar] [CrossRef]
- Chami, I.; Abu-El-Haija, S.; Perozzi, B. Machine Learning on Graphs: A Model and Comprehensive Taxonomy. J. Mach. Learn. Res. 2022, 23, 1–64. [Google Scholar]
- Bean, D.M.; Wu, H.; Iqbal, E.; Dzahini, O.; Ibrahim, Z.M.; Broadbent, M.; Stewart, R.; Dobson, R.J.B. Knowledge graph prediction of unknown adverse drug reactions and validation in electronic health records. Sci. Rep. 2017, 7, 16416. [Google Scholar] [CrossRef] [Green Version]
- Francis, N.; Paris-Est Alastair Green Neo, U.; Guagliardo, P.; Libkin, L.; Lindaaker Neo, T.; Marsault, V.; Plantikow Neo, S.; Selmer Neo, P.; Taylor Neo, A.; Green, A.; et al. Cypher: An Evolving Query Language for Property Graphs. In Proceedings of the 2018 International Conference on Management of Data, Houston, TX, USA, 10–15 June 2018; Volume 13. [Google Scholar] [CrossRef]
- Zaki, N.; Singh, H.; Mohamed, E.A. Identifying Protein Complexes in Protein-Protein Interaction Data Using Graph Convolutional Network. IEEE Access 2021, 9, 123717–123726. [Google Scholar] [CrossRef]
- Yuan, H.; Deng, W. Doctor recommendation on healthcare consultation platforms: An integrated framework of knowledge graph and deep learning. Internet Res. 2021, 32, 454–476. [Google Scholar] [CrossRef]
- Malik, K.M.; Krishnamurthy, M.; Alobaidi, M.; Hussain, M.; Alam, F.; Malik, G. Automated domain-specific healthcare knowledge graph curation framework: Subarachnoid hemorrhage as phenotype. Expert Syst. Appl. 2020, 145, 113120. [Google Scholar] [CrossRef]
- Zhang, Y.; Sheng, M.; Zhou, R.; Wang, Y.; Han, G.; Zhang, H.; Xing, C.; Dong, J. HKGB: An Inclusive, Extensible, Intelligent, Semi-auto-constructed Knowledge Graph Framework for Healthcare with Clinicians’ Expertise Incorporated. Inf. Process. Manag. 2020, 57, 102324. [Google Scholar] [CrossRef]
- Chawla, N.V.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. SMOTE: Synthetic Minority Over-sampling Technique. J. Artif. Intell. Res. 2011, 16, 321–357. [Google Scholar] [CrossRef]
- Grover, A.; Leskovec, J. node2vec: Scalable Feature Learning for Networks. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 855–864. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Luo, Y. Degree Centrality, Betweenness Centrality, and Closeness Centrality in Social Network. In Proceedings of the 2017 2nd International Conference on Modelling, Simulation and Applied Mathematics (MSAM2017), Bangkok, Thailand, 26–27 March 2017; pp. 300–303. [Google Scholar] [CrossRef] [Green Version]
- Perez, C.; Germon, R. Chapter 7—Graph Creation and Analysis for Linking Actors: Application to Social Data. In Automating Open Source Intelligence; Layton, R., Watters, P.A., Eds.; Syngress: Boston, MA, USA, 2016; pp. 103–129. [Google Scholar] [CrossRef]
- Golbeck, J. Chapter 3—Network Structure and Measures. In Analyzing the Social Web; Golbeck, J., Ed.; Morgan Kaufmann: Boston, MA, USA, 2013; pp. 25–44. ISBN 978-0-12-405531-5. [Google Scholar]
- Berlingerio, M.; Coscia, M.; Giannotti, F.; Monreale, A.; Pedreschi, D. The pursuit of hubbiness: Analysis of hubs in large multidimensional networks. J. Comput. Sci. 2011, 2, 223–237. [Google Scholar] [CrossRef]
- The Web as a Graph: Measurements, Models, and Methods. SpringerLink. Available online: https://link.springer.com/chapter/10.1007/3-540-48686-0_1 (accessed on 9 August 2022).
- Brin, S.; Page, L. The anatomy of a large-scale hypertextual Web search engine. Comput. Netw. ISDN Syst. 1998, 30, 107–117. [Google Scholar] [CrossRef]
- Que, X.; Checconi, F.; Petrini, F.; Gunnels, J.A. Scalable Community Detection with the Louvain Algorithm. In Proceedings of the 2015 IEEE International Parallel and Distributed Processing Symposium, Hyderabad, India, 25–29 May 2015; pp. 28–37. [Google Scholar] [CrossRef]
- Khan, W.; Phaisangittisagul, E.; Ali, L.; Gansawat, D.; Kumazawa, I. Combining features for RGB-D object recognition. In Proceedings of the 2017 International Electrical Engineering Congress (iEECON), Pattaya, Thailand, 8–10 March 2017. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Hearst, M.A.; Dumais, S.T.; Osuna, E.; Platt, J.; Scholkopf, B. Support vector machines. IEEE Intell. Syst. Their Appl. 1998, 13, 18–28. [Google Scholar] [CrossRef] [Green Version]
- Hosmer, D.W., Jr.; Lemeshow, S.; Sturdivant, R.X. Applied Logistic Regression; John Wiley & Sons: New York, NY, USA, 2013; ISBN 978-0-470-58247-3. [Google Scholar]
- Desiani, A.; Primartha, R.; Arhami, M.; Orsalan, O. Naive Bayes classifier for infant weight prediction of hypertension mother. Proc. J. Phys. Conf. Ser. 2019, 1282, 012005. [Google Scholar] [CrossRef]
- Gardner, M.W.; Dorling, S.R. Artificial neural networks (the multilayer perceptron)—A review of applications in the atmospheric sciences. Atmos. Environ. 1998, 32, 2627–2636. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. XGBoost: A Scalable Tree Boosting System. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar] [CrossRef] [Green Version]
- Ke, G.; Meng, Q.; Finley, T.; Wang, T.; Chen, W.; Ma, W.; Ye, Q.; Liu, T.-Y. LightGBM: A Highly Efficient Gradient Boosting Decision Tree. In Advances in Neural Information Processing Systems; 2017; Volume 30, Available online: https://proceedings.neurips.cc/paper/2017/hash/6449f44a102fde848669bdd9eb6b76fa-Abstract.html (accessed on 9 December 2022).
- Powers, D.M.W. Evaluation: From precision, recall and F-measure to ROC, informedness, markedness and correlation. arXiv 2020, arXiv:2010.16061. [Google Scholar] [CrossRef]
- Davis, J.; Goadrich, M. The relationship between Precision-Recall and ROC curves. In Proceedings of the 23rd International Conference on Machine Learning—ICML ’06, Pittsburgh, PA, USA, 25–29 June 2006; pp. 233–240. [Google Scholar] [CrossRef] [Green Version]
- Neo4j Graph Data Platform—The Leader in Graph Databases. Available online: https://neo4j.com/ (accessed on 22 November 2022).
- Webber, J. A programmatic introduction to Neo4j. In Proceedings of the 3rd Annual Conference on Systems, Programming, and Applications: Software for Humanity, Tucson, AZ, USA, 19–26 October 2012; pp. 217–218. [Google Scholar] [CrossRef]
- Kipf, T.N.; Welling, M. Semi-Supervised Classification with Graph Convolutional Networks. arXiv 2016, arXiv:1609.02907. [Google Scholar]
- Yeh, H.-Y.; Chao, C.-T.; Lai, Y.-P.; Chen, H.-W. Predicting the Associations between Meridians and Chinese Traditional Medicine Using a Cost-Sensitive Graph Convolutional Neural Network. Int. J. Environ. Res. Public Health 2020, 17, 740. [Google Scholar] [CrossRef] [PubMed]
- Davahli, M.R.; Fiok, K.; Karwowski, W.; Aljuaid, A.M.; Taiar, R. Predicting the Dynamics of the COVID-19 Pandemic in the United States Using Graph Theory-Based Neural Networks. Int. J. Environ. Res. Public Health 2021, 18, 3834. [Google Scholar] [CrossRef] [PubMed]

| References | Method Used | Performance | Limitations |
|---|---|---|---|
| Faruk et al. [7] | LBW prediction using LR, RF. Basic data preprocessing was performed. | AUC of LR was 0.50, Accuracy of RF was 93%. | No other performance other than accuracy was shown for RF. A small set of features were used. |
| Feng et al. [8] | Fetal weight estimation and classification using ultrasound features. SMOTE [29] was used for data balancing. deep belief network (DBN) for estimation. SVM for classification. | DBN achieved better performance with an MAE of 198.55 g ± 158 g, MAPE of 6.09 ± 5.06%, | LBW and NBW samples were treated as the same class to predict High BW. |
| Lu et al. [9] | Fetal weight estimation using ensemble (RF, XGBoost, and LightGBM) models. | Accuracy of 64.3% and mean relative error of 7% which was improved by 12% and 3% respectively. | Performance needs further improvement. |
| Trujillo et al. [10] | Infant BW estimation using support vector regression. | Results show the SVR was able to predict BW with nearly 250 g. | Only one ML model was used for evaluation. |
| Pollob et al. [11] | LBW classification using LR and decision tree | Sensitivity, specificity, and AUC of 0.99, 0.18, and 0.59 was achieved using LR. | Low performance was achieved with a Specificity of 0.18 and an AUC of only 0.59. |
| Do et al. [12] | Mortality prediction in very LBW infants using ML (LR, ANN, KNN, RF, SVM) models. | ANN achieved an AUC of 0.845, a sensitivity, and specificity of 0.76 and 0.78, respectively. | A small set of features was used. The sensitivity and specificity need further improvement. |
| Lin et al. [13] | Prediction of in-hospital length of stay of very LBW infants. Six ML models (KNN, MLP, RF, LR etc) were used. | LR achieved the best performance with AUC of 0.72, precision, recall, and F-score of 0.76, 0.78. 0.744. | Performance needs further improvement. |
| Khan et al. [2] | BW estimation and LBW classification, SMOTE for data balancing, and multiple sets of features. | LR achieved the best classification performance with an accuracy of 0.90, precision, recall, and F-score of 0.88, 0.90, and 0.89, respectively. Important risk factors were highlighted. | Performance metric such as AUC and PR-value was not used. The classification performance of LBW samples was low. |
| Classifier | Parameter(s) |
|---|---|
| RF | Batch Size = 100, number of trees = 100, Break Ties Randomly = False, Maximum Depth = None, maximum features = “sqrt”, bootstrap = True, base estimator = DecisionTreeClassifier |
| SVM | , loss = squared hinge, maximum iterations = 1000. |
| Logistic Regression | Batch Size = 100, Ridge = 1.0 × 10−8, . |
| Naïve Bayes | Batch Size = 100, parameters = default. |
| MLP | Hidden layers = default, activation = relu, alpha = 0.001, learning rate = 0.001, maximum iterations = 200, |
| KNN | K = 3, distance measure = Euclidean |
| LightGBM | Batch Size = 100, learning rate = 0.01 |
| XGBoost | Number of estimators = 100, learning rate = 0.01, random state = 42, maximum features = number of features |
| CatBoost | Iterations = 20, learning rate = 0.01, loss function = cross entropy |
| Method | Precision (SD) | Recall (SD) | F-Score (SD) | AUC (SD) | PR LBW (SD) | PR Overall (SD) |
|---|---|---|---|---|---|---|
| Original | 0.843 (0.01) | 0.887 (0.001) | 0.864 (0.001) | 0.746 (0.002) | 0.306 (0.001) | 0.878 (0.01) |
| Node Embedding | 0.876 (0.05) | 0.886 (0.004 | 0.881 (0.004) | 0.767 (0.01) | 0.330 (0.01) | 0.886 (0.002) |
| Combination of Graph Features | 0.868 (0.01) | 0.887 (0.01) | 0.877 (0.01) | 0.777 (0.01) | 0.355 (0.01) | 0.888 (0.01) |
| Combination of all features | 0.877 (0.02) | 0.887 (0.01) | 0.882 (0.01) | 0.807 (0.01) | 0.401 (0.01) | 0.901 (0.01) |
| Method | Precision (SD) | Recall (SD) | F-Score (SD) | AUC (SD) | PR LBW (SD) | PR Overall (SD) |
|---|---|---|---|---|---|---|
| Original | 0.840 (0.01) | 0.870 (0.01) | 0.855 (0.01) | 0.726 (0.01) | 0.260 (0.02) | 0.868 (0.01) |
| Node Embedding | 0.867 (0.01) | 0.889 (0.01) | 0.878 (0.01) | 0.803 (0.02) | 0.390 (0.02) | 0.902 (0.01) |
| Combination of Graph Features | 0.855 (0.02) | 0.866 (0.01) | 0.860 (0.01) | 0.779 (0.02) | 0.322 (0.01) | 0.889 (0.01) |
| Combination of all features | 0.862 (0.01) | 0.860 (0.01) | 0.861 (0.01) | 0.799 (0.02) | 0.346 (0.01) | 0.895 (0.01) |
| Method | Precision (SD) | Recall (SD) | F-Score (SD) | AUC (SD) | PR LBW (SD) | PR Overall (SD) |
|---|---|---|---|---|---|---|
| Original | 0.858 (0.003) | 0.888 (0.003) | 0.873 (0.005) | 0.754 (0.003) | 0.347 (0.003) | 0.884 (0.002) |
| Node Embedding | 0.872 (0.01) | 0.895 (0.01) | 0.883 (0.01) | 0.809 (0.008) | 0.419 (0.02) | 0.906 (0.004) |
| Combinations of Graph Features | 0.875 (0.01) | 0.895 (0.01) | 0.885 (0.01) | 0.814 (0.01) | 0.431 (0.02) | 0.908 (0.01) |
| Combination of all features | 0.870 (0.01) | 0.884 (0.01) | 0.877 (0.01) | 0.8189 (0.01) | 0.392 (0.01) | 0.909 (0.01) |
| Method | Precision (SD) | Recall (SD) | F-Score (SD) | AUC (SD) | PR LBW (SD) | PR Overall (SD) |
|---|---|---|---|---|---|---|
| Original | 0.803 (0.01) | 0.876 (0.02) | 0.838 (0.01) | 0.530 (0.02) | 0.127 (0.01) | 0.806 (0.01) |
| Node Embedding | 0.835 (0.01) | 0.873 (0.02) | 0.854 (0.01) | 0.600 (0.02) | 0.166 (0.02) | 0.824 (0.01) |
| Combinations of Graph Features | 0.821 (0.01) | 0.867 (0.03) | 0.843 (0.01) | 0.573 (0.03) | 0.149 (0.02) | 0.817 (0.01) |
| Combination of all features | 0.827 (0.02) | 0.876 (0.02) | 0.851 (0.01) | 0.530 (0.01) | 0.132 (0.01) | 0.806 (0.01) |
| Method | Precision (SD) | Recall (SD) | F-Score (SD) | AUC (SD) | PR LBW (SD) | PR Overall (SD) |
|---|---|---|---|---|---|---|
| Original | 0.831 (0.01) | 0.854 (0.01) | 0.842 (0.01) | 0.652 (0.02) | 0.239 (0.02) | 0.850 (0.01) |
| Node Embedding | 0.844 (0.01) | 0.860 (0.01) | 0.852 (0.01) | 0.7217 (0.01) | 0.286 (0.02) | 0.8734 (0.02) |
| Combinations of Graph Features | 0.848 (0.01) | 0.857 (0.01) | 0.852 (0.01) | 0.745 (0.01) | 0.307 (0.01) | 0.881 (0.01) |
| Combination of all features | 0.863 (0.01) | 0.876 (0.01) | 0.869 (0.01) | 0.787 (0.01) | 0.384 (0.02) | 0.897 (0.02) |
| Method | Precision (SD) | Recall (SD) | F-Score (SD) | AUC (SD) | PR LBW (SD) | PR Overall (SD) |
|---|---|---|---|---|---|---|
| Original | 0.858 (0.01) | 0.888 (0.01) | 0.873 (0.01) | 0.756 (0.01) | 0.329 (0.01) | 0.882 (0.01) |
| Node Embedding | 0.868 (0.01) | 0.911 (0.03) | 0.889 (0.01) | 0.807 (0.01) | 0.409 (0.01) | 0.905 (0.01) |
| Combination of Graph Features | 0.872 (0.01) | 0.889 (0.01) | 0.880 (0.01) | 0.811 (0.01) | 0.411 (0.01) | 0.905 (0.01) |
| Combination of all features | 0.878 (0.01) | 0.894 (0.01) | 0.886 (0.01) | 0.819 (0.02) | 0.459 (0.02) | 0.913 (0.02) |
| Method | Precision (SD) | Recall (SD) | F-Score (SD) | AUC (SD) | PR LBW (SD) | PR Overall (SD) |
|---|---|---|---|---|---|---|
| Original | 0.865 (0.01) | 0.891 (0.01) | 0.878 (0.01) | 0.762 (0.01) | 0.406 (0.01) | 0.891 (0.01) |
| Node Embedding | 0.870 (0.01) | 0.892 (0.01) | 0.881 (0.01) | 0.802 (0.01) | 0.410 (0.02) | 0.902 (0.01) |
| Combination of Graph Features | 0.872 (0.01) | 0.894 (0.01) | 0.883 (0.01) | 0.822 (0.01) | 0.440 (0.01) | 0.909 (0.01) |
| Combination of all features | 0.888 (0.01) | 0.898 (0.01) | 0.893 (0.01) | 0.834 (0.01) | 0.481 (0.02) | 0.916 (0.01) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khan, W.; Zaki, N.; Ahmad, A.; Bian, J.; Ali, L.; Mehedy Masud, M.; Ghenimi, N.; Ahmed, L.A. Infant Low Birth Weight Prediction Using Graph Embedding Features. Int. J. Environ. Res. Public Health 2023, 20, 1317. https://doi.org/10.3390/ijerph20021317
Khan W, Zaki N, Ahmad A, Bian J, Ali L, Mehedy Masud M, Ghenimi N, Ahmed LA. Infant Low Birth Weight Prediction Using Graph Embedding Features. International Journal of Environmental Research and Public Health. 2023; 20(2):1317. https://doi.org/10.3390/ijerph20021317
Chicago/Turabian StyleKhan, Wasif, Nazar Zaki, Amir Ahmad, Jiang Bian, Luqman Ali, Mohammad Mehedy Masud, Nadirah Ghenimi, and Luai A. Ahmed. 2023. "Infant Low Birth Weight Prediction Using Graph Embedding Features" International Journal of Environmental Research and Public Health 20, no. 2: 1317. https://doi.org/10.3390/ijerph20021317
APA StyleKhan, W., Zaki, N., Ahmad, A., Bian, J., Ali, L., Mehedy Masud, M., Ghenimi, N., & Ahmed, L. A. (2023). Infant Low Birth Weight Prediction Using Graph Embedding Features. International Journal of Environmental Research and Public Health, 20(2), 1317. https://doi.org/10.3390/ijerph20021317








