Sink Drains in a Neonatal Intensive Care Unit: A Retrospective Risk Assessment and Evaluation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Clinical Setting
2.2. Multidrug-Resistant Bacteria Screening and Infection Control Measures
2.3. Microbiological Sample Processing
2.4. Surveillance of Breastmilk
2.5. Environmental Surveillance and Infection Control Measures
2.6. Whole-Genome Sequencing-Based Typing
2.7. Ethics Statement
3. Results
3.1. Environmental, Patient, and Breastmilk Testing
3.2. Whole-Genome Sequencing-Based Typing
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Volling, C.; Ahangari, N.; Bartoszko, J.J.; Coleman, B.L.; Garcia-Jeldes, F.; Jamal, A.J.; Johnstone, J.; Kandel, C.; Kohler, P.; Maltezou, H.C.; et al. Are Sink Drainage Systems a Reservoir for Hospital-Acquired Gammaproteobacteria Colonization and Infection? A Systematic Review. Open Forum Infect. Dis. 2021, 8, ofaa590. [Google Scholar] [CrossRef] [PubMed]
- Carling, P.C. Wastewater drains: Epidemiology and interventions in 23 carbapenem-resistant organism outbreaks. Infect. Control Hosp. Epidemiol. 2018, 39, 972–979. [Google Scholar] [CrossRef] [PubMed]
- Salm, F.; Deja, M.; Gastmeier, P.; Kola, A.; Hansen, S.; Behnke, M.; Gruhl, D.; Leistner, R. Prolonged outbreak of clonal MDR Pseudomonas aeruginosa on an intensive care unit: Contaminated sinks and contamination of ultra-filtrate bags as possible route of transmission? Antimicrob. Resist. Infect. Control 2016, 5, 53. [Google Scholar] [CrossRef]
- Kossow, A.; Kampmeier, S.; Willems, S.; Berdel, W.E.; Groll, A.H.; Burkhardt, B.; Rossig, C.; Groth, C.; Idelevich, E.A.; Kipp, F.; et al. Control of Multidrug-Resistant Pseudomonas aeruginosa in Allogeneic Hematopoietic Stem Cell Transplant Recipients by a Novel Bundle Including Remodeling of Sanitary and Water Supply Systems. Clin. Infect. Dis. 2017, 65, 935–942. [Google Scholar] [CrossRef] [PubMed]
- Catho, G.; Martischang, R.; Boroli, F.; Chraïti, M.N.; Martin, Y.; Koyluk Tomsuk, Z.; Renzi, G.; Schrenzel, J.; Pugin, J.; Nordmann, P.; et al. Outbreak of Pseudomonas aeruginosa producing VIM carbapenemase in an intensive care unit and its termination by implementation of waterless patient care. Crit. Care 2021, 25, 301. [Google Scholar] [CrossRef]
- Starlander, G.; Melhus, A. Minor outbreak of extended-spectrum beta-lactamase-producing Klebsiella pneumoniae in an intensive care unit due to a contaminated sink. J. Hosp. Infect. 2012, 82, 122–124. [Google Scholar] [CrossRef]
- Lowe, C.; Willey, B.; O’Shaughnessy, A.; Lee, W.; Lum, M.; Pike, K.; Larocque, C.; Dedier, H.; Dales, L.; Moore, C.; et al. Outbreak of extended-spectrum beta-lactamase-producing Klebsiella oxytoca infections associated with contaminated handwashing sinks. Emerg. Infect. Dis. 2012, 18, 1242–1247. [Google Scholar] [CrossRef]
- Nurjadi, D.; Scherrer, M.; Frank, U.; Mutters, N.T.; Heininger, A.; Späth, I.; Eichel, V.M.; Jabs, J.; Probst, K.; Müller-Tidow, C.; et al. Genomic Investigation and Successful Containment of an Intermittent Common Source Outbreak of OXA-48-Producing Enterobacter cloacae Related to Hospital Shower Drains. Microbiol. Spectr. 2021, 9, e0138021. [Google Scholar] [CrossRef]
- Trautmann, M.; Lepper, P.M.; Haller, M. Ecology of Pseudomonas aeruginosa in the intensive care unit and the evolving role of water outlets as a reservoir of the organism. Am. J. Infect. Control 2005, 33, 41–49. [Google Scholar] [CrossRef]
- Podschun, R.; Ullmann, U. Klebsiella spp. as nosocomial pathogens: Epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin. Microbiol. Rev. 1998, 11, 589–603. [Google Scholar] [CrossRef]
- Lewis, S.S.; Smith, B.A.; Sickbert-Bennett, E.E.; Weber, D.J. Water as a source for colonization and infection with multidrug-resistant pathogens: Focus on sinks. Infect. Control Hosp. Epidemiol. 2018, 39, 1463–1466. [Google Scholar] [CrossRef]
- Jones, L.D.; Mana, T.S.C.; Cadnum, J.L.; Jencson, A.L.; Silva, S.Y.; Wilson, B.M.; Donskey, C.J. Effectiveness of foam disinfectants in reducing sink-drain gram-negative bacterial colonization. Infect. Control Hosp. Epidemiol. 2020, 41, 280–285. [Google Scholar] [CrossRef]
- Kommission für Krankenhaushygiene und Infektprävention beim Robert Koch-Institut. Anforderungen der Hygiene an abwasserführende Systeme in medizinischen Einrichtungen. Bundesgesundheitsblatt Gesundheitsforschung Gesundheitsschutz 2020, 63, 484–501. [Google Scholar] [CrossRef]
- Kommission für Krankenhaushygiene und Infektprävention beim Robert Koch-Institut. Empfehlung zur Prävention nosokomialer Infektionen bei neonatologischen Intensivpflegepatienten mit einem Geburtsgewicht unter 1500 g. Bundesgesundheitsblatt Gesundheitsforschung Gesundheitsschutz 2007, 50, 1265–1303. [Google Scholar] [CrossRef]
- Kommission für Krankenhaushygiene und Infektprävention beim Robert Koch-Institut. Ergänzende Empfehlung (2011) zur Prävention nosokomialer Infektionen bei neonatologischen Intensivpflegepatienten mit einem Geburtsgewicht unter 1.500 g (2007). Epid. Bull. 2012, 2, 13–15. [Google Scholar]
- Simon, A.; Dame, C.; Christoph, J.; Eckmanns, T.; Gärtner, B.; Geffers, C.; Gille, C.; Haertel, C.; Haller, S.; Hartl, D.; et al. Risikocharakterisierung intensivmedizinisch behandelter Früh- und Neugeborener und Daten zur Ist-Situation in deutschen neonatologischen Intensivpflegestationen 2013—Fachliche Erläuterungen zu folgender Empfehlung: Praktische Umsetzung sowie krankenhaushygienische und infektionspräventive Konsequenzen des mikrobiellen Kolonisationsscreenings bei intensivmedizinisch behandelten Früh- und Neugeborenen Ergänzende Empfehlung der Kommission für Krankenhaushygiene und Infektionsprävention (KRINKO) beim Robert Koch-Institut, Berlin zur Implementierung der Empfehlungen zur Prävention nosokomialer Infektionen bei neonatologischen Intensivpflegepatienten mit einem Geburtsgewicht unter 1.500 g aus dem Jahr 2007 und 2012. Epid. Bull. 2013, 42, 1–52. [Google Scholar]
- Verordnung Über die Qualität von Wasser für den Menschlichen Gebrauch (Trinkwasserverordnung—TrinkwV) vom 20. Juni 2023 (BGBl. 2023 I Nr. 159, S. 2). TrinkwV–Verordnung über die Qualität von Wasser für den Menschlichen Gebrauch. Available online: gesetze-im-internet.de (accessed on 15 July 2023).
- Souvorov, A.; Agarwala, R.; Lipman, D.J. SKESA: Strategic k-mer extension for scrupulous assemblies. Genome Biol. 2018, 19, 153. [Google Scholar] [CrossRef]
- Mellmann, A.; Bletz, S.; Böking, T.; Kipp, F.; Becker, K.; Schultes, A.; Prior, K.; Harmsen, D. Real-Time Genome Sequencing of Resistant Bacteria Provides Precision Infection Control in an Institutional Setting. J. Clin. Microbiol. 2016, 54, 2874–2881. [Google Scholar] [CrossRef]
- Tönnies, H.; Prior, K.; Harmsen, D.; Mellmann, A. Establishment and Evaluation of a Core Genome Multilocus Sequence Typing Scheme for Whole-Genome Sequence-Based Typing of Pseudomonas aeruginosa. J. Clin. Microbiol. 2021, 59, e01987-20. [Google Scholar] [CrossRef]
- Feldgarden, M.; Brover, V.; Haft, D.H.; Prasad, A.B.; Slotta, D.J.; Tolstoy, I.; Tyson, G.H.; Zhao, S.; Hsu, C.H.; McDermott, P.F.; et al. Validating the AMRFinder Tool and Resistance Gene Database by Using Antimicrobial Resistance Genotype-Phenotype Correlations in a Collection of Isolates. Antimicrob. Agents Chemother. 2019, 63, e00483-19. [Google Scholar] [CrossRef]
- Berberian, G.; Brizuela, M.; Rosanova, M.T.; Travaglianti, M.; Mastroiani, A.; Reijtman, V.; Fiorili, G.; Santa Cruz, D.; Castro, G. Multidrug resistant Gram-negative infections in neonatology. Arch. Argent. Pediatr. 2019, 117, 6–11. [Google Scholar] [CrossRef] [PubMed]
- Benjamin, D.K.; DeLong, E.; Cotten, C.M.; Garges, H.P.; Steinbach, W.J.; Clark, R.H. Mortality following blood culture in premature infants: Increased with Gram-negative bacteremia and candidemia, but not Gram-positive bacteremia. J. Perinatol. 2004, 24, 175–180. [Google Scholar] [CrossRef] [PubMed]
- Auriti, C.; Ronchetti, M.P.; Pezzotti, P.; Marrocco, G.; Quondamcarlo, A.; Seganti, G.; Bagnoli, F.; De Felice, C.; Buonocore, G.; Arioni, C.; et al. Determinants of nosocomial infection in 6 neonatal intensive care units: An Italian multicenter prospective cohort study. Infect. Control Hosp. Epidemiol. 2010, 31, 926–933. [Google Scholar] [CrossRef] [PubMed]
- Barker, D.P.; Rutter, N. Exposure to invasive procedures in neonatal intensive care unit admissions. Arch. Dis. Child. Fetal Neonatal Ed. 1995, 72, F47–F48. [Google Scholar] [CrossRef] [PubMed]
- Hota, S.; Hirji, Z.; Stockton, K.; Lemieux, C.; Dedier, H.; Wolfaardt, G.; Gardam, M.A. Outbreak of multidrug-resistant Pseudomonas aeruginosa colonization and infection secondary to imperfect intensive care unit room design. Infect. Control Hosp. Epidemiol. 2009, 30, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Kotay, S.; Chai, W.; Guilford, W.; Barry, K.; Mathers, A.J. Spread from the Sink to the Patient: In Situ Study Using Green Fluorescent Protein (GFP)-Expressing Escherichia coli To Model Bacterial Dispersion from Hand-Washing Sink-Trap Reservoirs. Appl. Environ. Microbiol. 2017, 83, e03327-16. [Google Scholar] [CrossRef]
- Exner, M.; Kramer, A.; Lajoie, L.; Gebel, J.; Engelhart, S.; Hartemann, P. Prevention and control of health care-associated waterborne infections in health care facilities. Am. J. Infect. Control 2005, 33, S26–S40. [Google Scholar] [CrossRef]
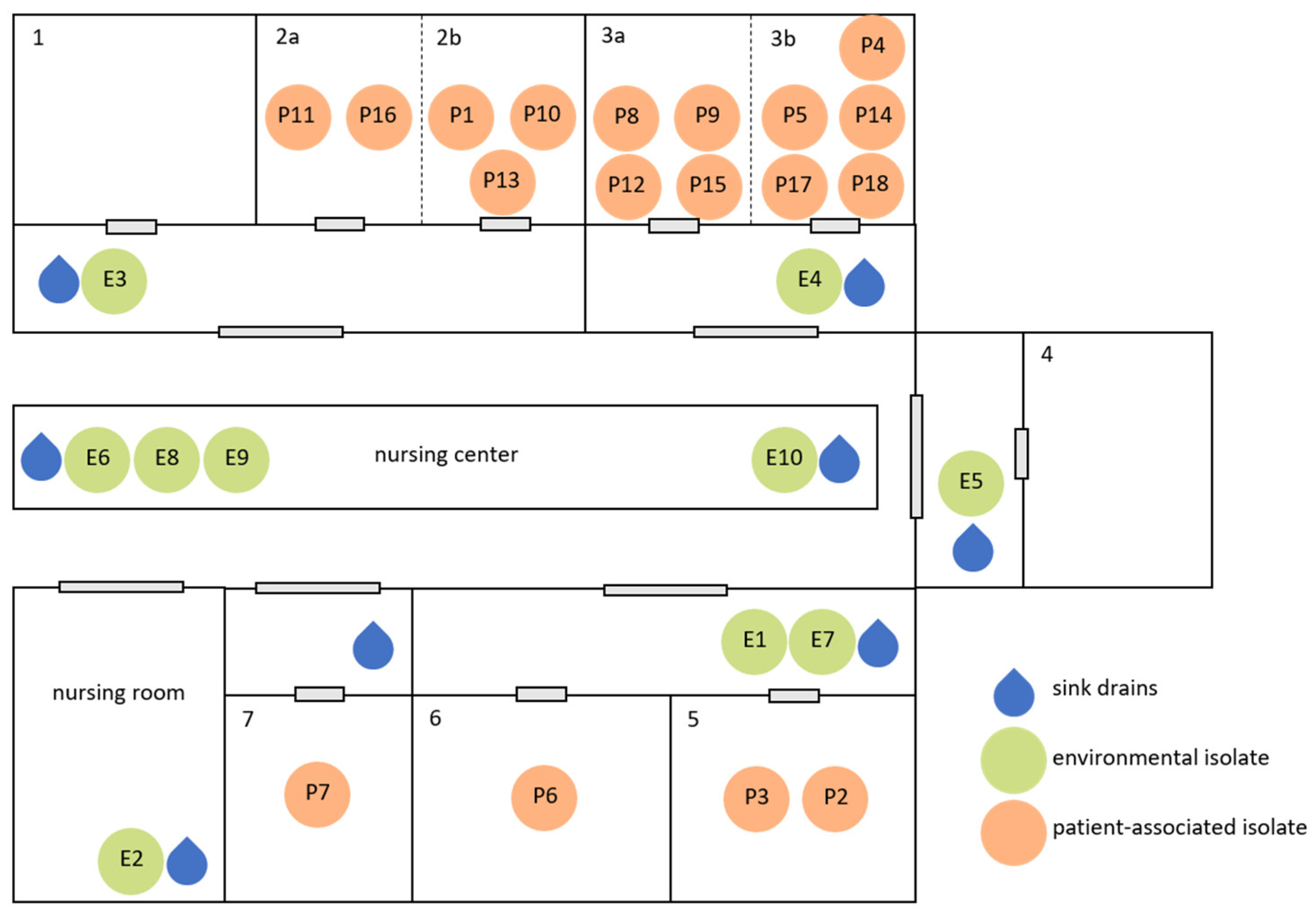
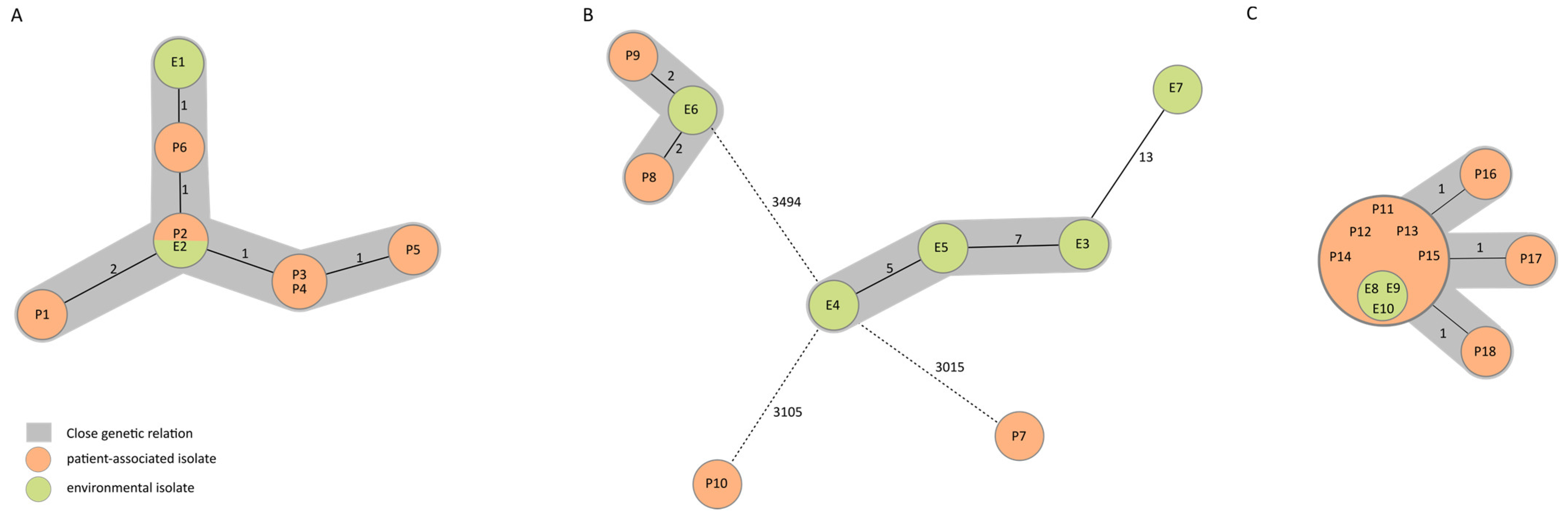
| Isolate no. | Isolation Date | Localization |
|---|---|---|
| P1 | 18/06/2017 | Throat swab |
| P2 | 25/06/2017 | Anal swab |
| P3 | 09/07/2017 | Anal swab |
| P4 | 17/07/2017 | Throat swab |
| P5 | 17/07/2017 | Anal swab |
| P6 | 24/07/2017 | Anal swab |
| E1 | 15/08/2017 | Washbasin |
| E2 | 15/08/2017 | Washbasin |
| Isolate no. | Isolation Date | Localization |
|---|---|---|
| P7 | 28/10/2021 | Throat swab |
| E3 | 29/10/2021 | Washbasin |
| E4 | 29/10/2021 | Washbasin |
| E5 | 29/10/2021 | Washbasin |
| E6 | 29/10/2021 | Washbasin 1 |
| E7 | 29/10/2021 | Washbasin |
| BM1 (P8) | 06/11/2021 | Breast milk |
| BM2 (P9) | 07/11/2021 | Breast milk |
| P10 | 17/01/2022 | Throat swab |
| Isolate no. | Isolation Date | Localization |
|---|---|---|
| P11 | 23/01/2023 | Anal swab |
| P12 | 23/01/2023 | Anal swab |
| P13 | 23/01/2023 | Throat swab |
| P14 | 31/01/2023 | Anal swab |
| P15 | 31/01/2023 | Anal swab |
| P16 | 31/01/2023 | Anal swab |
| P17 | 31/01/2023 | Throat swab |
| P18 | 02/02/2023 | Anal swab |
| E8 | 01/02/2023 | Washbasin 1 |
| E9 | 01/02/2023 | Washbasin 1 |
| E10 | 01/02/2023 | Washbasin 2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schneider, J.S.; Froböse, N.J.; Kuczius, T.; Schwierzeck, V.; Kampmeier, S. Sink Drains in a Neonatal Intensive Care Unit: A Retrospective Risk Assessment and Evaluation. Int. J. Environ. Res. Public Health 2023, 20, 6692. https://doi.org/10.3390/ijerph20176692
Schneider JS, Froböse NJ, Kuczius T, Schwierzeck V, Kampmeier S. Sink Drains in a Neonatal Intensive Care Unit: A Retrospective Risk Assessment and Evaluation. International Journal of Environmental Research and Public Health. 2023; 20(17):6692. https://doi.org/10.3390/ijerph20176692
Chicago/Turabian StyleSchneider, Julia S., Neele J. Froböse, Thorsten Kuczius, Vera Schwierzeck, and Stefanie Kampmeier. 2023. "Sink Drains in a Neonatal Intensive Care Unit: A Retrospective Risk Assessment and Evaluation" International Journal of Environmental Research and Public Health 20, no. 17: 6692. https://doi.org/10.3390/ijerph20176692
APA StyleSchneider, J. S., Froböse, N. J., Kuczius, T., Schwierzeck, V., & Kampmeier, S. (2023). Sink Drains in a Neonatal Intensive Care Unit: A Retrospective Risk Assessment and Evaluation. International Journal of Environmental Research and Public Health, 20(17), 6692. https://doi.org/10.3390/ijerph20176692






