Optimizing the Retrieval of the Vital Status of Cancer Patients for Health Data Warehouses by Using Open Government Data in France
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Sources and Architecture
2.2. Organisation of the Study
2.3. Matching Algorithms
2.3.1. Exact Matching Algorithm
2.3.2. Fuzzy Matching Algorithm
2.4. Evaluation
2.4.1. False Negatives
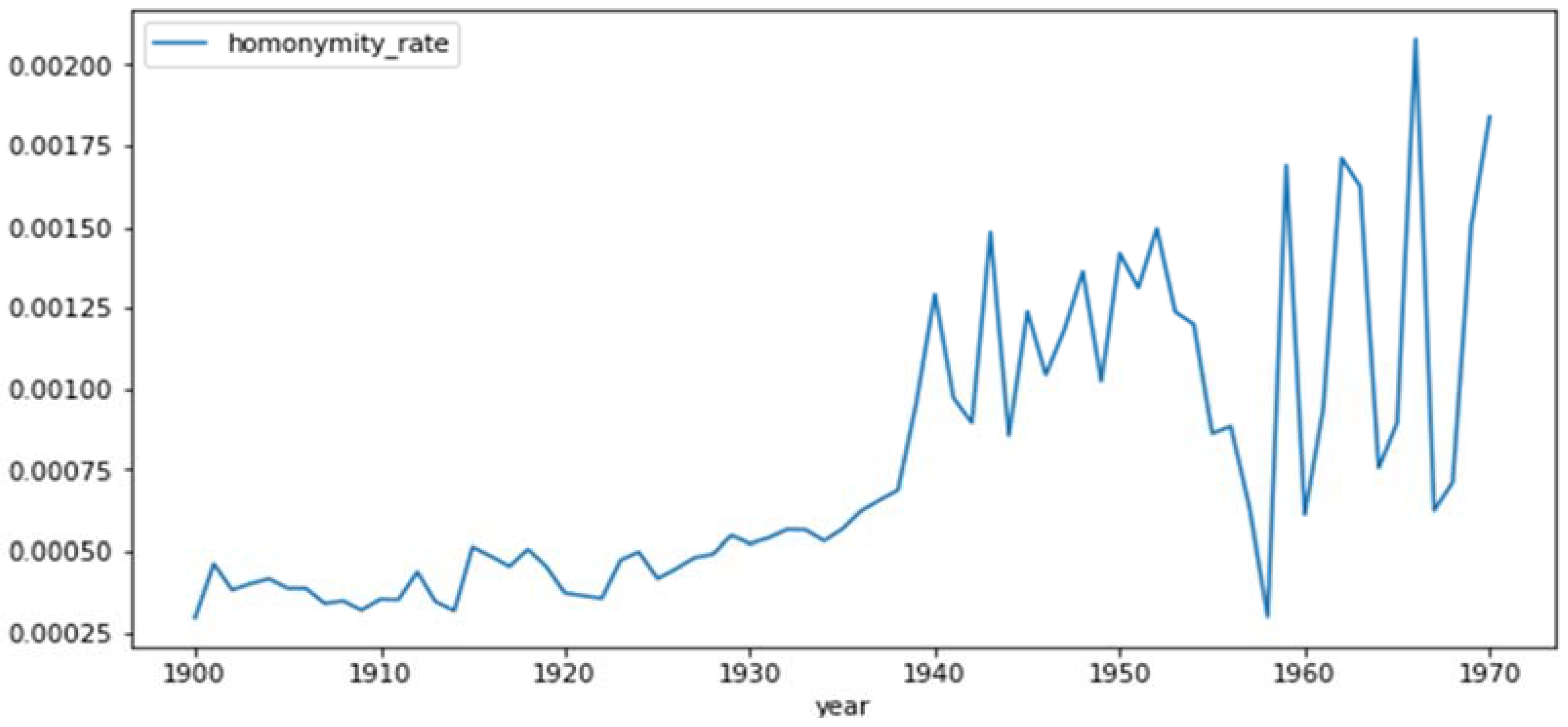
2.4.2. Homonymity Rates
2.4.3. False Positives
3. Results
3.1. Homonyms
3.2. Matching Results
3.3. False Negatives
3.4. Patients with Different Dates of Death
4. Discussion
4.1. False Negatives
4.2. Patients with Different Dates of Death
4.3. Estimated Precision
4.4. Homonyms
4.5. Probabilistic Linkage
4.6. Strengths and Limitations of the Study
5. Conclusions
Author Contributions
Funding
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Foran, D.J.; Chen, W.; Chu, H.; Sadimin, E.; Loh, D.; Riedlinger, G.; Goodell, L.A.; Ganesan, S.; Hirshfield, K.; Rodriguez, L.; et al. Roadmap to a comprehensive clinical data warehouse for precision medicine applications in oncology. Cancer Inform. 2017, 16, 1176935117694349. [Google Scholar] [CrossRef] [PubMed]
- Eschrich, S.; Teer, J.; Reisman, P.; Siegel, E.; Challa, C.; Lewis, P.; Fellows, K.; Malpica, E.; Carvajal, R.; Gonzalez, G.; et al. Enabling Precision Medicine in Cancer Care Through a Molecular Data Warehouse: The Moffitt Experience. JCO Clin. Cancer Inform. 2021, 5, 561–569. [Google Scholar] [CrossRef]
- Boyd, M.; Fulcher, N.; Annavarapu, S.; Aguilar, K.; Frytak, J.; Robert, N.; Espirito, J. PCN1 Concordance of death date assessments between the social security death master file and electronic health records in a US community oncology setting. Cancer Clin. Outcomes Value Health 2020, 23 (Suppl. S1), S22. [Google Scholar] [CrossRef]
- The American College of Obstetricians and Gynecologists, The Importance of Vital Records and Statistics for the Obstetrician–Gynecologist. Available online: https://www.acog.org/clinical/clinical-guidance/committee-opinion/articles/2018/08/the-importance-of-vital-records-and-statistics-for-the-obstetriciangynecologist (accessed on 20 January 2022).
- Driscoll, J.J.; Rixe, O. Overall survival: Still the gold standard: Why overall survival remains the definitive end point in cancer clinical trials. Cancer J. 2009, 15, 401–405. [Google Scholar] [CrossRef]
- Zhuang, S.H.; Xiu, L.; Elsayed, Y.A. Overall survival: A gold standard in search of a surrogate: The value of progression-free survival and time to progression as end points of drug efficacy. Cancer J. 2009, 15, 395–400. [Google Scholar] [CrossRef]
- Lakdawalla, D.N.; Shafrin, J.; Hou, N.; Peneva, D.; Vine, S.; Park, J.; Zhang, J.; Brookmeyer, R.; Figlin, R.A. Predicting Real-World Effectiveness of Cancer Therapies Using Overall Survival and Progression-Free Survival from Clinical Trials: Empirical Evidence for the ASCO Value Framework. Value Health 2017, 20, 866–875. [Google Scholar] [CrossRef]
- Newman, T.B.; Brown, A.N. Use of commercial record linkage software and vital statistics to identify patient deaths. J. Am. Med. Inform. Assoc. 1997, 4, 233–237. [Google Scholar] [CrossRef]
- Zingmond, D.S.; Ye, Z.; Ettner, S.L.; Liu, H. Linking hospital discharge and death records—accuracy and sources of bias. J. Clin. Epidemiol. 2004, 57, 21–29. [Google Scholar] [CrossRef]
- Fournel, I.; Schwarzinger, M.; Binquet, C.; Benzenine, E.; Hill, C.; Quantin, C. Contribution of record linkage to vital status determination in cancer patients. In Medical Informatics in a United and Healthy Europe; IOS Press: Amsterdam, The Netherlands, 2009; pp. 91–95. [Google Scholar]
- Jaro, M.A. Probabilistic linkage of large public health data files. Stat. Med. 1995, 14, 491–498. [Google Scholar] [CrossRef]
- Sylvestre, E.; Bouzille, G.; Breton, M.; Cuggia, M.; Campillo-Gimenez, B. Retrieving the vital status of patients with cancer using online obituaries. In Building Continents of Knowledge in Oceans of Data: The Future of Co-Created eHealth; IOS Press: Amsterdam, The Netherlands, 2018; pp. 571–575. [Google Scholar]
- Schnell, R.; Redlich, S. Web Scraping Online Newspaper Death Notices for the Estimation of the Local Number of Deaths. In Proceedings of the 12th International Joint Conference on Biomedical Engineering Systems and Technologies, Lisbon, Portugal, 12–15 January 2019; pp. 319–325. [Google Scholar]
- Doidge, J.C.; Harron, K.L. Reflections on modern methods: Linkage error bias. Int. J. Epidemiol. 2019, 48, 2050–2060. [Google Scholar] [CrossRef]
- Harron, K.; Dibben, C.; Boyd, J.; Hjern, A.; Azimaee, M.; Barreto, M.L.; Goldstein, H. Challenges in administrative data linkage for research. Big Data Soc. 2017, 4, 2053951717745678. [Google Scholar] [CrossRef] [PubMed]
- Doidge, J.C.; Harron, K. Demystifying probabilistic linkage: Common myths and misconceptions. Int. J. Popul. Data Sci. 2018, 3, 410. [Google Scholar] [CrossRef] [PubMed]
- Newcombe, H.B.; Kennedy, J.M.; Axford, S.J.; James, A.P. Automatic linkage of vital records. Science 1959, 130, 954–959. [Google Scholar] [CrossRef] [PubMed]
- Fellegi, I.P.; Sunter, A.B. A theory for record linkage. J. Am. Stat. Assoc. 1969, 64, 1183–1210. [Google Scholar] [CrossRef]
- Wilson, D.R. Beyond probabilistic record linkage: Using neural networks and complex features to improve genealogical record linkage. In Proceedings of the 2011 International Joint Conference on Neural Networks, San Jose, CA, USA, 31 July–5 August 2011; pp. 9–14. [Google Scholar]
- Goldstein, H.; Harron, K.; Wade, A. The analysis of record-linked data using multiple imputation with data value priors. Stat. Med. 2012, 31, 3481–3493. [Google Scholar] [CrossRef]
- Goldstein, H.; Harron, K. Record linkage: A missing data problem. In Methodological Developments in Data Linkage; Wiley: London, UK, 2015; Chapter 6; pp. 109–124. [Google Scholar]
- Tancredi, A.; Liseo, B. A hierarchical Bayesian approach to record linkage and population size problems. Ann. Appl. Stat. 2011, 5, 1553–1585. [Google Scholar] [CrossRef]
- Richard, E.G.; Nancy, A.D. The National Center for Biotechnology Information, Registries for Evaluating Patient Outcomes: A User’s Guide; Report No.: 13(14)-EHC111; Leavy, M.B., Ed.; AHRQ Methods for Effective Health Care: Rockville, MD, USA, 2014. [Google Scholar]
- Data Gouv, National Institute of Statistics and Economic Studies, France. 2022. Available online: https://www.data.gouv.fr/fr/organizations/institut-national-de-la-statistique-et-des-etudes-economiques-insee/ (accessed on 20 January 2022).
- Baghdadi, Y.; Gallay, A.; Caserio-Schönemann, C.; Fouillet, A. Evaluation of the French reactive mortality surveillance system supporting decision making. Eur. J. Public Health 2019, 29, 601–607. [Google Scholar] [CrossRef]
- The National Center for Biotechnology Information, Files of Deceased Persons Since 1970, France. 2021. Available online: https://www.insee.fr/fr/information/4190491 (accessed on 20 January 2022).
- Van Eycken, E.; Haustermans, K.; Buntinx, F.; Ceuppens, A. Evaluation of the encryption procedure and record linkage in the Belgian national cancer registry. Arch. Public Health 2000, 58, 285–287. [Google Scholar]
- Young, B.; Faris, T.; Armogida, L. Levenshtein distance as a measure of accuracy and precision in forensic PCR-MPS methods. Forensic Sci. Int. Genet. 2021, 55, 102594. [Google Scholar] [CrossRef]
- Guillen, L.C.; Domenico, J.; Camargo Jr Kenneth, R.P.; Coeli, C. Match quality of a linkage strategy based on the combined use of a statistical linkage key and the Levenshtein distance to link birth to death records in Brazil. Int. J. Popul. Data Sci. 2017, 1, 53. [Google Scholar] [CrossRef][Green Version]
- National Institute of Statistics and Economic Studies, Births from 1900 to 2019 [INSEE, Naissances de 1900 à 2019], France. 2022. Available online: https://www.insee.fr/fr/statistiques/4277635?sommaire=4318291#tableau-figure3 (accessed on 15 January 2022).
- Harron, K.; Goldstein, H.; Wade, A.; Muller-Pebody, B.; Parslow, R.; Gilbert, R. Linkage, evaluation and analysis of national electronic healthcare data: Application to providing enhanced blood-stream infection surveillance in paediatric intensive care. PLoS ONE 2013, 8, e85278. [Google Scholar] [CrossRef] [PubMed]
- Harron, K.; Wade, A.; Gilbert, R.; Muller-Pebody, B.; Goldstein, H. Evaluating bias due to data linkage error in electronic healthcare records. BMC Med. Res. Methodol. 2014, 14, 36. [Google Scholar] [CrossRef] [PubMed]
- Harron, K.; Doidge, J.C.; Knight, H.E.; Gilbert, R.; Goldstein, H.; Cromwell, D.A.; van der Meulen, J.H. A guide to evaluating linkage quality for the analysis of linked data. Int. J. Epidemiol. 2017, 46, 1699–1710. [Google Scholar] [CrossRef] [PubMed]
- Clark, D.E.; Hahn, D.R. Comparison of probabilistic and deterministic record linkage in the development of a statewide trauma registry. In Proceedings of the Annual Symposium on Computer Application in Medical Care, American Medical Informatics Association, New Orleans, LA, USA, 28 October–1 November 1995; p. 397. [Google Scholar]
- Gomatam, S.; Carter, R.; Ariet, M.; Mitchell, G. An empirical comparison of record linkage procedures. Stat. Med. 2002, 21, 1485–1496. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Matsuyama, Y.; Ohashi, Y.; Setoguchi, S. When to conduct probabilistic linkage vs. deterministic linkage? A simulation study. J. Biomed. Inform. 2015, 56, 80–86. [Google Scholar] [CrossRef] [PubMed]
- Porter, E.H.; Winkler, W.E. Approximate string comparison and its effect on an advanced record linkage system. In Advanced Record Linkage System; Research Report; US Bureau of the Census: Suitland-Silver Hill, MD, USA, 1997. [Google Scholar]
- DuVall, S.L.; Kerber, R.A.; Thomas, A. Extending the Fellegi–Sunter probabilistic record linkage method for approximate field comparators. J. Biomed. Inform. 2010, 43, 24–30. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Guttmann, A.; Cipière, S.; Maigne, L.; Demongeot, J.; Boire, J.Y.; Ouchchane, L. Implementation of an extended Fellegi-Sunter probabilistic record linkage method using the Jaro-Winkler string comparator. In Proceedings of the IEEE-EMBS International Conference on Biomedical and Health Informatics, Valencia, Spain, 1–4 June 2014; pp. 375–379. [Google Scholar]


| Patient Data | Death Register | Match | Score | ||
|---|---|---|---|---|---|
| First Name | Last Name | First Name | Last Name | ||
| Maxim | Dupond | Maxime | Dupond | YES | 0.5 |
| Maxime | Dupont | Maxime | Dupond | YES | 0.833 |
| Maxim | Dupont | Maxime | Dupond | NO | 1.333 |
| Exact Matching | Fuzzy Matching | |
|---|---|---|
| Total number of matches | 26,193 | 37,434 |
| Matches not in hospital data warehouse | 14,907 | 25,146 |
| Matches already present in hospital data warehouse | 11,286 | 12,288 |
| Matches with a difference of more than 62 days | 18 | 18 |
| Matches missing compared to the hospital data warehouse | 1381 | 379 |
| Estimated linkage precision (%) | 99.95 | 99.67 |
| Estimated number of false links | 13.86 | 122.83 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lauzanne, O.; Frenel, J.-S.; Baziz, M.; Campone, M.; Raimbourg, J.; Bocquet, F. Optimizing the Retrieval of the Vital Status of Cancer Patients for Health Data Warehouses by Using Open Government Data in France. Int. J. Environ. Res. Public Health 2022, 19, 4272. https://doi.org/10.3390/ijerph19074272
Lauzanne O, Frenel J-S, Baziz M, Campone M, Raimbourg J, Bocquet F. Optimizing the Retrieval of the Vital Status of Cancer Patients for Health Data Warehouses by Using Open Government Data in France. International Journal of Environmental Research and Public Health. 2022; 19(7):4272. https://doi.org/10.3390/ijerph19074272
Chicago/Turabian StyleLauzanne, Olivier, Jean-Sébastien Frenel, Mustapha Baziz, Mario Campone, Judith Raimbourg, and François Bocquet. 2022. "Optimizing the Retrieval of the Vital Status of Cancer Patients for Health Data Warehouses by Using Open Government Data in France" International Journal of Environmental Research and Public Health 19, no. 7: 4272. https://doi.org/10.3390/ijerph19074272
APA StyleLauzanne, O., Frenel, J.-S., Baziz, M., Campone, M., Raimbourg, J., & Bocquet, F. (2022). Optimizing the Retrieval of the Vital Status of Cancer Patients for Health Data Warehouses by Using Open Government Data in France. International Journal of Environmental Research and Public Health, 19(7), 4272. https://doi.org/10.3390/ijerph19074272






