Molecular Design, Spectroscopic, DFT, Pharmacological, and Molecular Docking Studies of Novel Ruthenium(III)–Schiff Base Complex: An Inhibitor of Progression in HepG2 Cells
Abstract
1. Introduction
2. Experimental
2.1. Materials and Instruments
2.2. Synthesis of 2-Chloro-5-Nitrophenyl-(4,6-Dimethylpyrimidinyl)methanimine Schiff Base; L
2.3. Synthesis of the (trichloro)(monoaquo)(2-Chloro-5-Nitrophenyl-(4,6-Dimethylpyrimidinyl)-Methanimine) ruthenium(III) Complex; RuL
2.4. Computational Details
2.5. In Vitro Studies
2.5.1. Cancer Cell Lines
2.5.2. Cytotoxicity Assay
2.5.3. Annexin V/Propidium Iodide Staining for Apoptosis Assessment
2.5.4. Cell Cycle Analysis
2.5.5. Expression Levels of Caspase 3, VEGF-A, mTOR, NF-kB, and SND1 by RT-PCR
2.5.6. Assay of Protein Levels of Caspase 3, VEGF-A, mTOR, NF-kB, and SND1
2.6. Molecular Docking Studies
2.7. Statistical Analysis
3. Result and Discussion
3.1. Spectroscopic Studies
3.2. Stereochemistry and Chemical Reactivity Prediction
3.3. In Vitro Studies
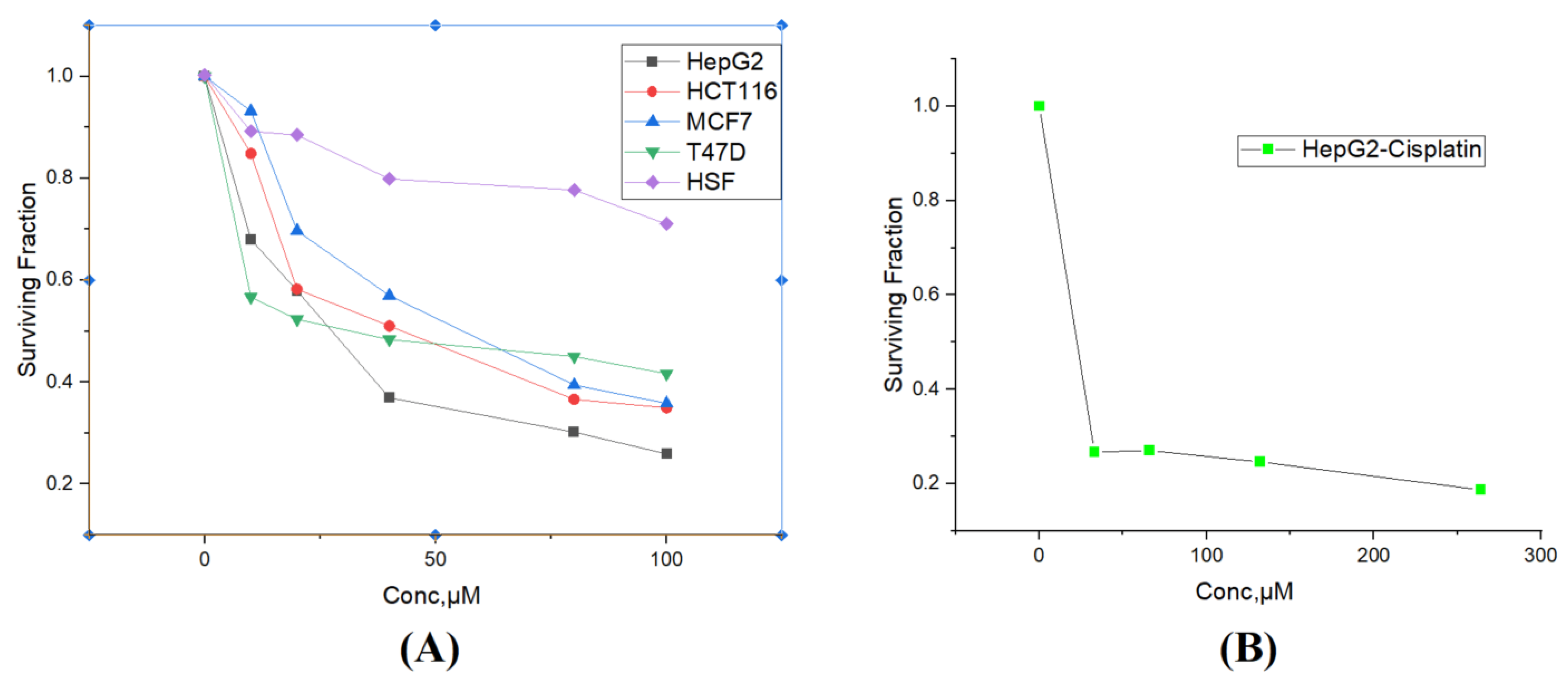
3.3.1. Cytotoxicity Screening on the Tested Human Cancer Cell Lines
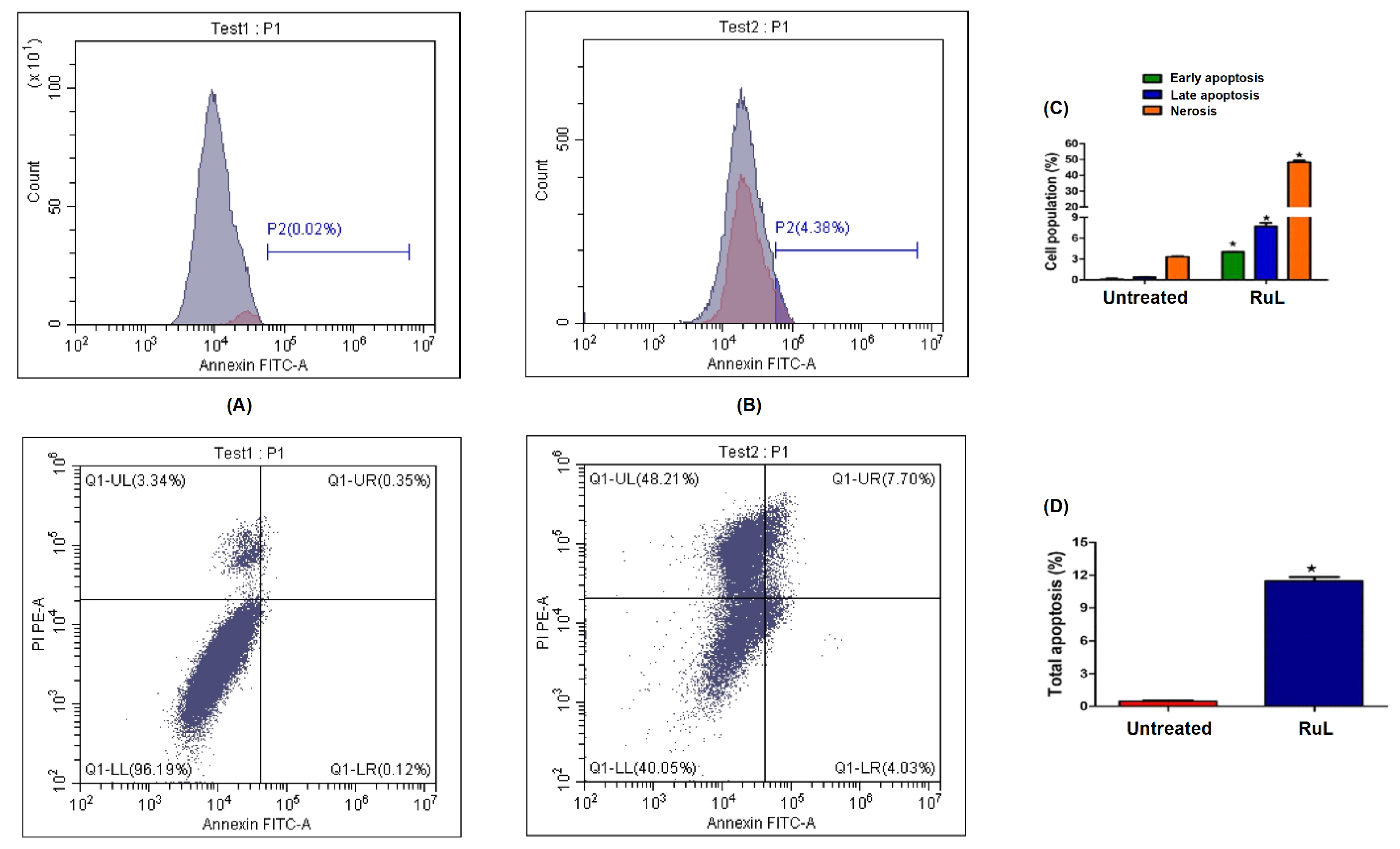
3.3.2. Annexin V/Propidium Iodide Staining for Apoptosis Assessment
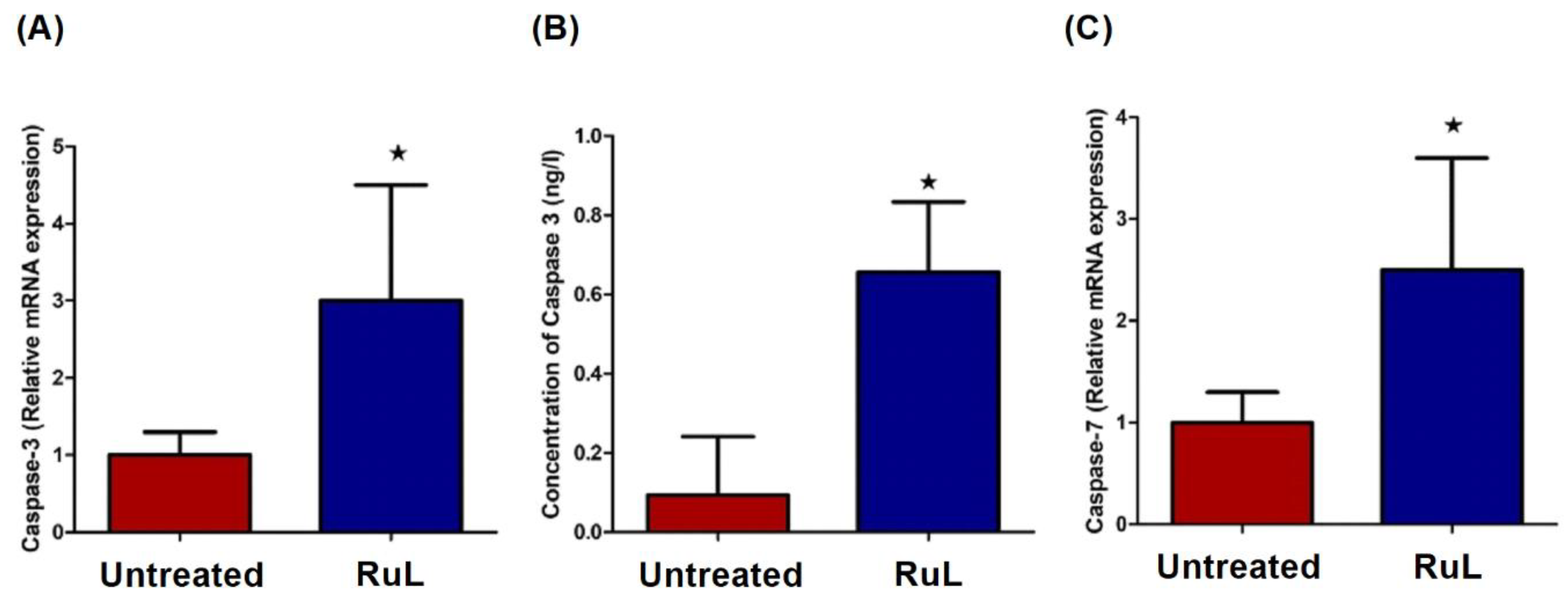
3.3.3. Expression Levels of Caspase 3 and 7
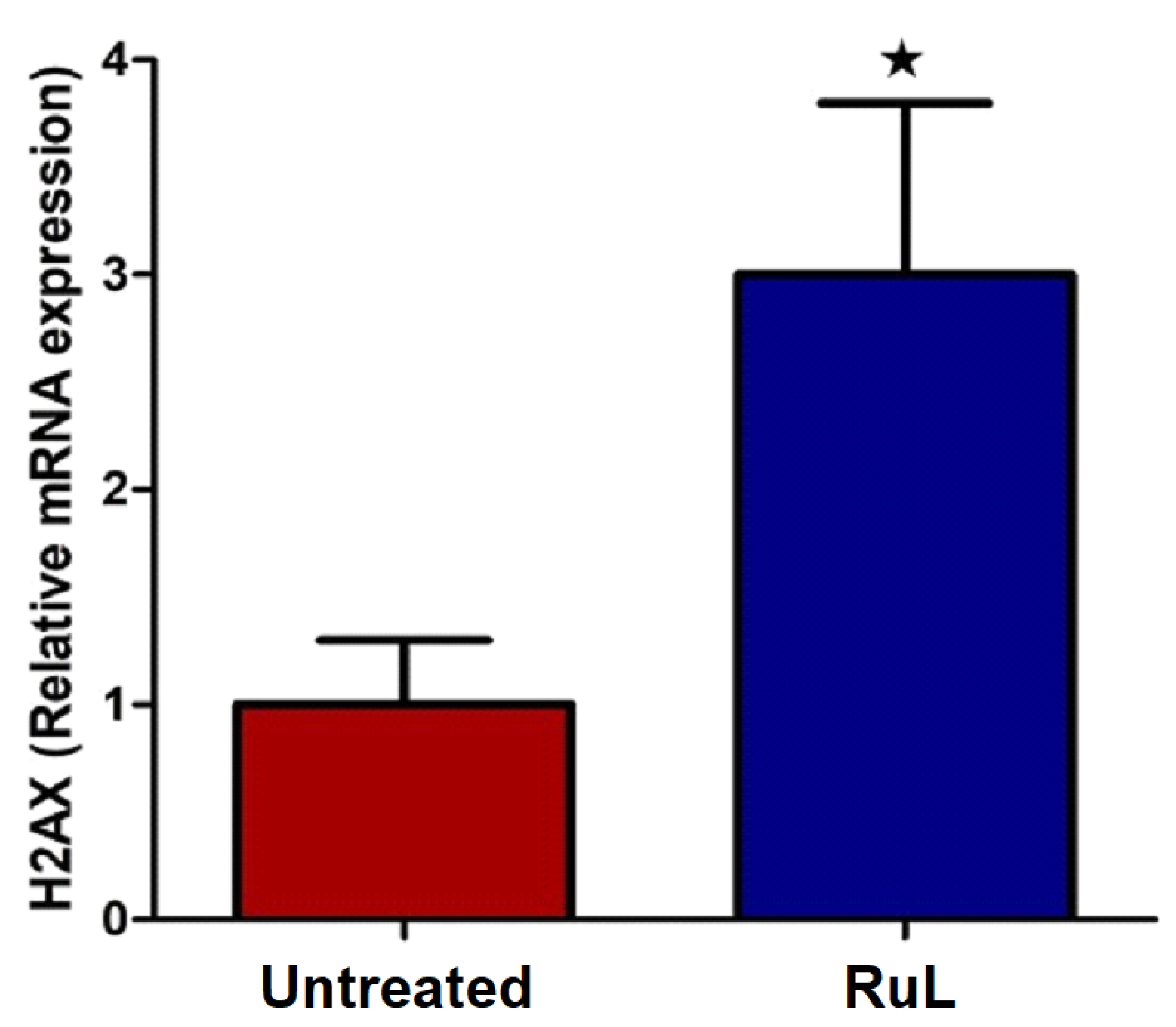
3.3.4. Effect of the RuL Complex on H2AX Expressions
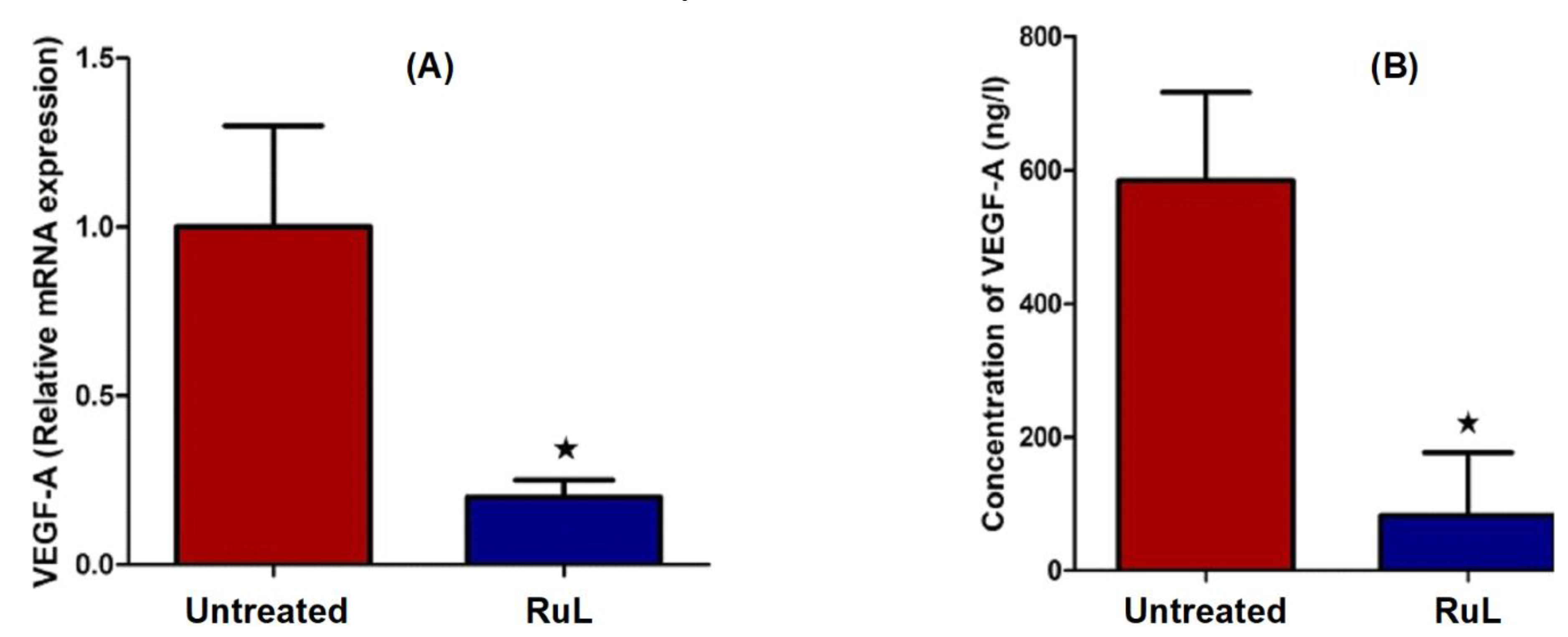
3.3.5. Effect of RuL Treatment on VEGF in HepG2 Cells
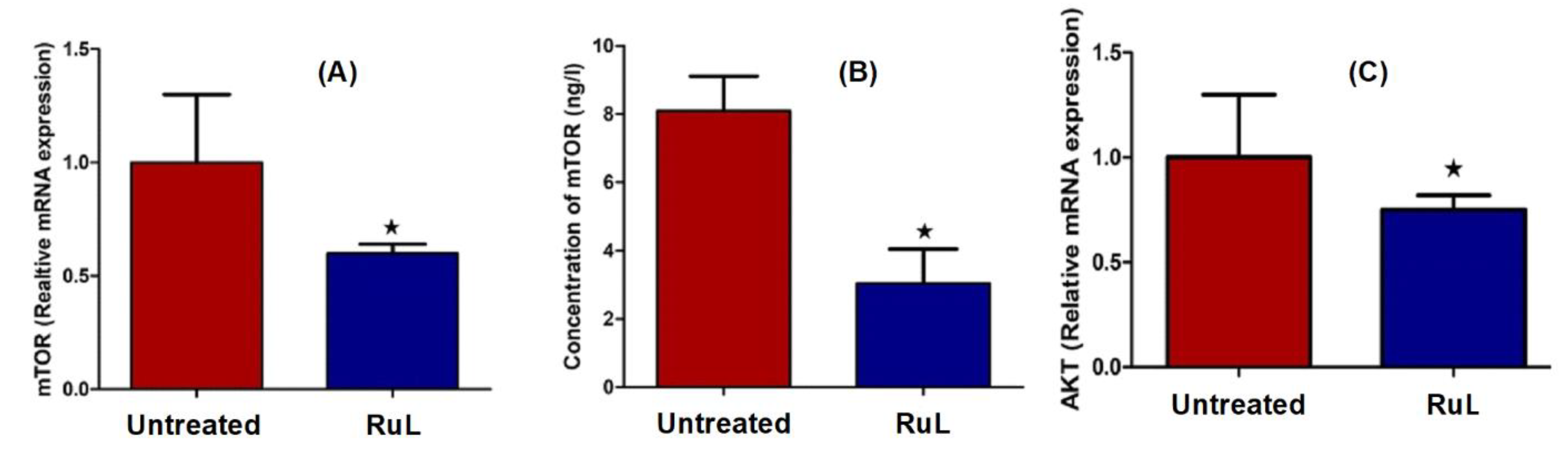
3.3.6. Effect of the RuL Complex on AKT and mTOR of HepG2 Cells
3.3.7. Effect of the RuL Complex on SND1 of HepG2 Cells
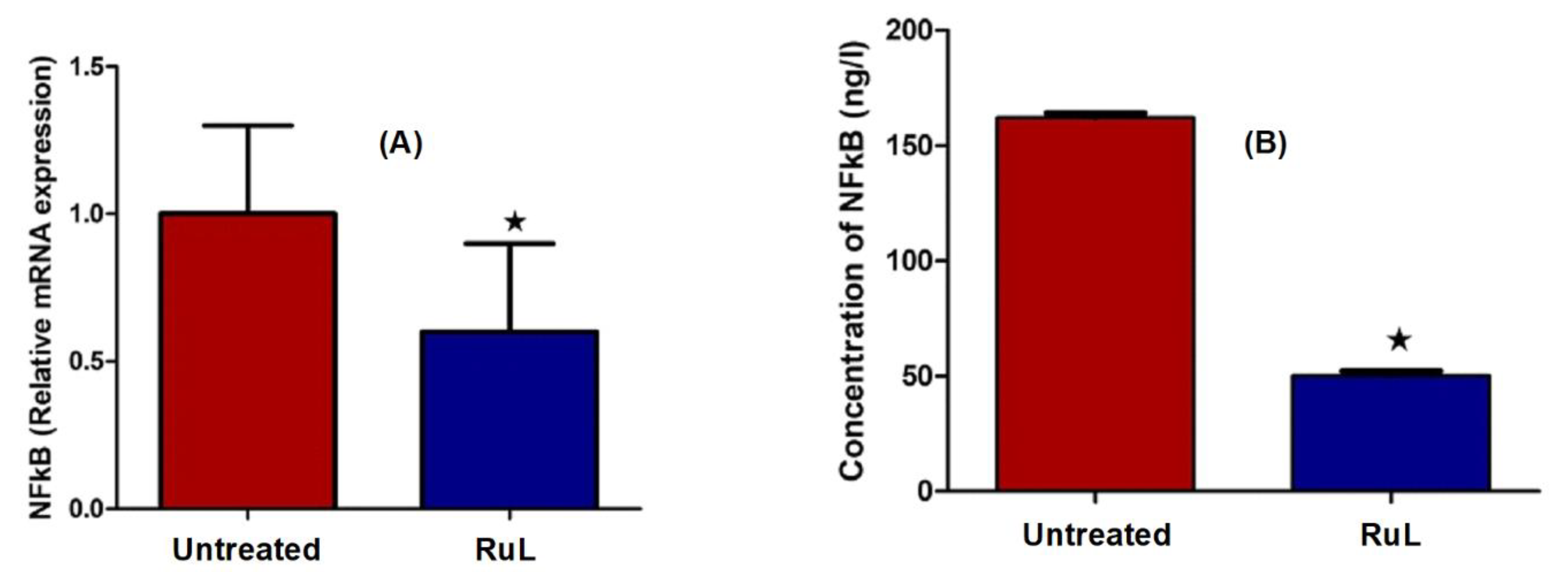
3.3.8. Effect of the RuL Complex on NF-kB in HepG2 Cells
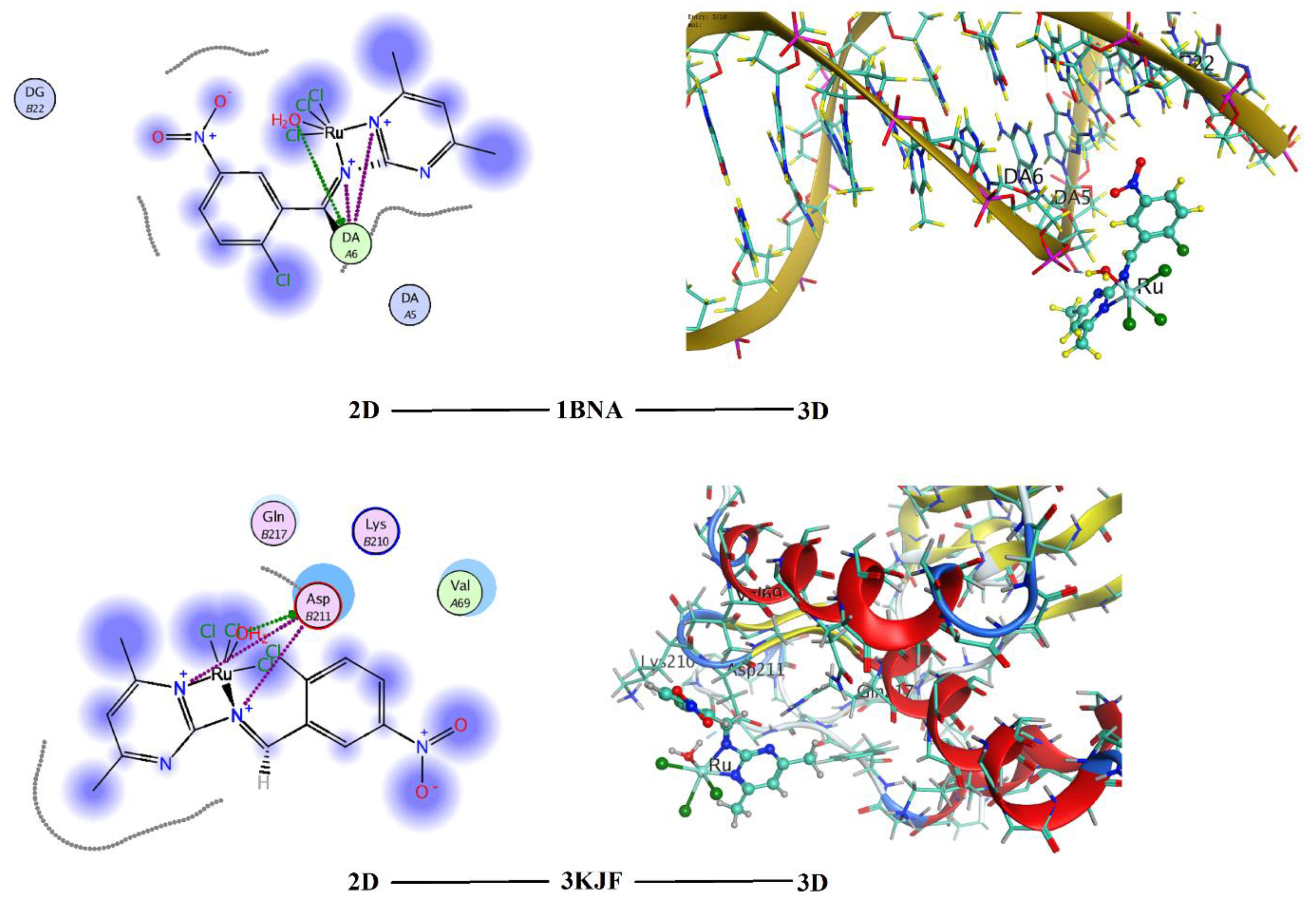
3.4. Molecular Docking of the Ruthenium (RuL) Complex
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Singal, A.G.; Lampertico, P.; Nahon, P. Epidemiology and surveillance for hepatocellular carcinoma: New trends. J. Hepatol. 2020, 72, 250–261. [Google Scholar] [CrossRef]
- Petrick, J.L.; Florio, A.A.; Znaor, A.; Ruggieri, D.; Laversanne, M.; Alvarez, C.S.; Ferlay, J.; Valery, P.C.; Bray, F.; McGlynn, K.A. International trends in hepatocellular carcinoma incidence, 1978–2012. Int. J. Cancer 2020, 147, 317–330. [Google Scholar] [CrossRef] [PubMed]
- Jafri, W.; Kamran, M. Hepatocellular carcinoma in Asia: A challenging situation. Euroasian J. Hepato Gastroenterol. 2019, 9, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Ohata, J.; Ball, Z.T. Rhodium at the chemistry–biology interface. Dalton Trans. 2018, 47, 14855–14860. [Google Scholar] [CrossRef]
- Lee, S.Y.; Kim, C.Y.; Nam, T.G. Ruthenium complexes as anticancer agents: A brief history and perspectives. Drug Des. Devel. Ther. 2020, 14, 5375–5392. [Google Scholar] [CrossRef] [PubMed]
- Ramadan, R.M.; Noureldeen, A.F.H.; Abo-Aly, M.M.; El-Medani, S.M. Spectroscopic, DFT analysis, antimicrobial and cytotoxicity studies of three gold(III) complexes. Inorg. Nano-Met. Chem. 2022, 52, 213–225. [Google Scholar] [CrossRef]
- Southam, H.M.; Butler, J.A.; Chapman, J.A.; Poole, R.K. The microbiology of ruthenium complexes. Adv. Microb. Physiol. 2017, 71, 1–96. [Google Scholar]
- Mahmud, M.; Niloy, M.S.; Shakil, S.; Islam, A. Ruthenium Complexes: An Alternative to Platinum Drugs in Colorectal Cancer Treatment Kazi. Pharmaceutics 2021, 13, 1295. [Google Scholar] [CrossRef]
- Małecka, M.; Skoczyńska, A.; Goodman, D.M.; Hartinger, C.G.; Budzisz, E. Biological properties of ruthenium(II)/(III) complexes with flavonoids as ligands. Coord. Chem. Rev. 2021, 436, 213849. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, D.; Lv, F.; Yu, B.; Shen, Y.; Cong, H. Recent advances in ruthenium and platinum based supramolecular coordination complexes for antitumor therapy. Colloids Surf. B: Biointerfaces 2019, 182, 110373. [Google Scholar] [CrossRef]
- Mohamed, S.E.; Ramadan, R.M.; Aboelhasan, A.E.; Abdel Aziz, A.A. Design, synthesis, biomedical investigation, DFT calculation and molecular docking of novel Ru(II)-mixed ligand complexes. J. Biomol. Struct. Dyn. 2021, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Barman, P. Recent advances in Schiff base ruthenium metal complexes: Synthesis and applications. Top. Curr. Chem. 2021, 379, 1–71. [Google Scholar] [CrossRef] [PubMed]
- Bhat, A.R. Biological activity of pyrimidine derivativies: A review. Org. Med. Chem. Int. J. 2017, 2, 23–26. [Google Scholar]
- Festus, C.; Okafor, S.N.; Ekennia, A.C. Heteroleptic metal complexes of a pyrimidinyl based Schiff base ligand incorporating 2,2-bipyridine moiety: Synthesis, characterization, and biological studies. Front. Chem. 2019, 7, 862. [Google Scholar] [CrossRef] [PubMed]
- Al-Masoudi, N.A.; Abbas, A.; Al-Asadi, M.J.B. Synthesis, biological activity and modeling study of some thiopyrimidine derivatives and their platinum(II) and ruthenium(III) metal complexes. Z. Für Nat. B 2015, 70, 343–353. [Google Scholar] [CrossRef]
- Fandzloch, M.; Dobrzańska, L.; Jędrzejewski, T.; Jezierska, J.; Wiśniewska, J.; Łakomska, I. Synthesis, structure and biological evaluation of ruthenium(III) complexes of triazolopyrimidines with anticancer properties. J. Biol. Inorg. Chem. 2020, 25, 109–124. [Google Scholar] [CrossRef]
- Liu, J.; Lai, H.; Xiong, Z.; Chen, B.; Chen, T. Functionalization and cancer-targeting design of ruthenium complexes for precise cancer therapy. Chem. Commun. 2019, 55, 9904–9914. [Google Scholar] [CrossRef]
- Alessio, E.; Messori, L. NAMI-A and KP1019/1339, two iconic ruthenium anticancer drug candidates face-to-face: A case story in medicinal inorganic chemistry. Molecules 2019, 24, 1995. [Google Scholar] [CrossRef]
- Kapitza, S.; Pongratz, M.; Jakupec, M.A.; Heffeter, P.; Berger, W.; Lackinger, L.; Keppler, B.K.; Marian, B. Heterocyclic complexes of ruthenium (III) induce apoptosis in colorectal carcinoma cells. J. Cancer Res. Clin. Oncol. 2005, 131, 101–110. [Google Scholar] [CrossRef]
- Lentz, F.; Drescher, A.; Lindauer, A.; Henke, M.; Hilger, R.A.; Hartinger, C.G.; Scheulenb, M.E.; Dittrichd, C.; Kepplerc, B.K.; Jaehdea, U. In collaboration with Central European Society for Anticancer Drug Research-EWIV. Pharmacokinetics of a novel anticancer ruthenium complex (KP1019, FFC14A) in a phase I dose-escalation study. Anti-Cancer Drugs 2009, 20, 97–103. [Google Scholar] [CrossRef]
- Ramadan, R.M.; Abu Al-Nasr, K.; Noureldeen, A.F.H. Synthesis, spectroscopic studies, antimicrobial activities and antitumor of a new monodentate V-shaped Schiff base and its transition metal complexes. Spectrochim. Acta Part A 2014, 132, 417–422. [Google Scholar] [CrossRef] [PubMed]
- Ramadan, R.M.; Elsheemy, W.M.; Hassan, N.S.; Abdel Aziz, A.A. Synthesis, spectroscopic characterization, thermal behavior and in vitro antimicrobial and anticancer activities of novel ruthenium tricarbonyl complexes containing monodentate V-shaped Schiff bases. Appl. Organometal. Chem. 2018, 32, e4180. [Google Scholar] [CrossRef]
- Frisch, M.J.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; Nakatsuji, H.; et al. Official Gaussian 09 Literature Citation. Gaussian Inc.: Wallingford, CT, USA, 2009; Available online: https://gaussian.com/glossary/g09/ (accessed on 4 August 2022).
- Skehan, P.; Storeng, R.; Scudiero, D.; Monks, A.; McMahon, J.; Vistica, D.; Warren, J.T.; Bokesch, H.; Kenney, S.; Boyd, M.R. New colorimetric cytotoxicity assay for anticancer-drug screening. JNCI 1990, 82, 1107–1112. [Google Scholar] [CrossRef] [PubMed]
- Drew, H.R.; Wing, R.M.; Takano, T.; Broka, C.; Tanaka, S.; Itakura, K.; Dickerson, R.E. Structure of a B-DNA dodecamer: Conformation and dynamics. Proc. Natl. Acad. Sci. USA 1981, 78, 2179–2183. [Google Scholar] [CrossRef]
- Wang, Z.; Watt, W.; Brooks, N.A.; Harris, M.S.; Urban, J.; Boatman, D.; McMillan, M.; Kahn, M.; Heinrikson, R.L.; Finzel, B.C.; et al. Kinetic and structural characterization of caspase-3 and caspase-8 inhibition by a novel class of irreversible inhibitors. Biochim. Biophys. Acta 2010, 1804, 1817–1831. [Google Scholar] [CrossRef] [PubMed]
- Taher, M.A.; Jarelnabbi, S.E.; Al-Sehemi, A.G.M.; El-Medani, S.M.; Ramadan, R.M. Synthesis and spectroscopic studies of some new metal carbonyl derivatives of 1-(2-pyridylazo)-2-naphthol. J. Coord. Chem. 2009, 62, 1293–1301. [Google Scholar] [CrossRef]
- Ramadan, R.M.; El-Medani, S.M.; Makhlouf, A.A.; Moustafa, H.; Afifi, M.A.; Haukka, M.; Abdel Aziz, A. Spectroscopic, density functional theory, nonlinear optical properties and in vitro biological studies of Co(II), Ni(II), and Cu(II) complexes of hydrazide Schiff base derivatives. Appl. Organometal. Chem. 2021, 35, e6246. [Google Scholar] [CrossRef]
- Yıldız, M.; Karpuz, Ö.; Zeyrek, C.T.; Boyacıoğlu, B.; Dal, H.; Demir, N.; Yıldırım, N.; Ünver, H. Synthesis, biological activity, DNA binding and anion sensors, molecular structure and quantum chemical studies of a novel bidentate Schiff base derived from 3, 5-bis (triflouromethyl) aniline and salicylaldehyde. J. Mol Struct. 2015, 1094, 148–160. [Google Scholar] [CrossRef]
- Hassen, S.; Chebbi, H.; Zid, M.F.; Arfaoui, Y. Assembly and weak interactions in the crystal structure of 2-amino-4-(3-bromophenyl)-1, 3, 5-triazinobenzimidazolium chloride studied by X-ray diffraction, vibrational spectroscopy, Hirshfeld surface analysis and DFT calculations. J. Mol. Struct. 2019, 1179, 678–684. [Google Scholar] [CrossRef]
- El-Medani, S.M.; Makhlouf, A.A.; Moustafa, H.; Afifi, M.A.; Haukka, M.; Ramadan, R.M. Spectroscopic, crystal structural, theoretical and biological studies of phenylacetohydrazide Schiff base derivatives and their copper complexes. J. Mol. Struct. 2020, 1208, 127860. [Google Scholar] [CrossRef]
- Raja, N.; Ramesh, R.; Liu, Y. Paramagnetic ruthenium (III) complexes bearing O, O chelating ligands: Synthesis, spectra, molecular structure and electron transfer properties. Polyhedron 2012, 31, 196–201. [Google Scholar] [CrossRef]
- Aziz, A.A.A.; Elantabli, F.M.; Moustafa, H.; El-Medani, S.M. Spectroscopic, DNA binding ability, biological activity, DFT calculations and nonlinear optical properties (NLO) of novel Co (II), Cu (II), Zn (II), Cd (II) and Hg (II) complexes with ONS Schiff base. J. Mol. Struct. 2017, 1141, 563–576. [Google Scholar] [CrossRef]
- Junker, A.; Renn, C.; Dobelmann, C.; Namasivayam, V.; Jain, S.; Losenkova, K.; Irjala, H.; Duca, S.; Balasubramanian, R.; Chakraborty, S.; et al. Structure-Activity Relationship of Purine and Pyrimidine Nucleotides as Ecto-5′-Nucleotidase (CD73) Inhibitors. J. Med. Chem. 2019, 62, 3677–3695. [Google Scholar] [CrossRef] [PubMed]
- Wan, D.; Tang, B.; Wang, Y.J.; Guo, B.H.; Yin, H.; Yi, Q.Y.; Liu, Y.J. Synthesis and anticancer properties of ruthenium (II) complexes as potent apoptosis inducers through mitochondrial disruption. Eur. J. Med. Chem. 2017, 139, 180–190. [Google Scholar] [CrossRef] [PubMed]
- Ryu, H.; Nam, K.Y.; Kim, J.S.; Hwang, S.G.; Song, J.Y.; Ahn, J. The small molecule AU14022 promotes colorectal cancer cell death via p53-mediated G2/M-phase arrest and mitochondria-mediated apoptosis. J. Cell. Physiol. 2018, 233, 4666–4676. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.W.; Hu, J.J.; Fu, R.Q.; Liu, X.; Zhang, Y.H.; Li, J.; Liu, L.; Li, Y.N.; Deng, Q.; Luo, Q.S.; et al. Flavonoids inhibit cell proliferation and induce apoptosis and autophagy through downregulation of PI3Kgamma mediated PI3K/AKT/mTOR/p70S6K/ULK signaling pathway in human breast cancer cells. Sci. Rep. 2018, 8, 11255. [Google Scholar] [CrossRef]
- de Carvalho, N.C.; Neves, S.P.; Dias, R.B.; Valverde, L.D.F.; Sales, C.B.S.; Rocha, C.A.G.; Soares, M.B.P.; Santos, E.R.D.; Oliveira, R.M.M.; Carlos, R.M.; et al. A novel ruthenium complex with xanthoxylin induces S-phase arrest and causes ERK1/2-mediated apoptosis in HepG2 cells through a p53-independent pathway. Cell Death Dis. 2018, 9, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Jan, R.; Chaudhry, G.E. Understanding apoptosis and apoptotic pathways targeted cancer therapeutics. Adv. Pharm. Bull. 2019, 9, 205–218. [Google Scholar] [CrossRef]
- Yadav, P.; Yadav, R.; Jain, S.; Vaidya, A. Caspase-3: A primary target for natural and synthetic compounds for cancer therapy. Chem. Biol. Drug Des. 2021, 98, 144–165. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.L.; Shen, W.Y.; Chen, Z.F.; Zhao, L.F.; Qin, Q.P.; Yu, Y.C.; Liang, H. Oxoaporphine metal complexes (CoII, NiII, ZnII) with high antitumor activity by inducing mitochondria-mediated apoptosis and S-phase arrest in HepG2. Sci. Rep. 2017, 7, 46056. [Google Scholar] [CrossRef]
- Sahyon, H.A.; El-Bindary, A.A.; Shoair, A.F.; Abdellatif, A.A. Synthesis and characterization of ruthenium(III) complex containing 2-aminomethyl benzimidazole, and its anticancer activity of in vitro and in vivo models. J. Mol. Liq. 2018, 255, 122–134. [Google Scholar] [CrossRef]
- Li, Y.; Wu, Q.; Yu, G.; Li, L.; Zhao, X.; Huang, X.; Mei, W. Polypyridyl Ruthenium(II) complex-induced mitochondrial membrane potential dissipation activates DNA damage-mediated apoptosis to inhibit liver cancer. Eur. J. Med. Chem. 2019, 164, 282–291. [Google Scholar] [CrossRef] [PubMed]
- Neves, S.P.; De Carvalho, N.C.; Da Silva, M.M.; Rodrigues, A.C.B.; Bomfim, L.M.; Dias, R.B.; Sales, C.B.S.; Rocha, C.A.G.; Soares, M.B.P.; Batista, A.A.; et al. Ruthenium complexes containing heterocyclic thioamidates trigger caspase-mediated apoptosis through MAPK signaling in human hepatocellular carcinoma cells. Front. Oncol. 2019, 9, 562. [Google Scholar] [CrossRef] [PubMed]
- Bourhis, M.; Palle, J.; Galy-Fauroux, I.; Terme, M. Direct and indirect modulation of T cells by VEGF-A counteracted by anti-angiogenic treatment. Front. Immunol. 2021, 12, 616837. [Google Scholar] [CrossRef] [PubMed]
- Hicklin, D.J.; Ellis, L.M. Role of the vascular endothelial growth factor pathway in tumor growth and angiogenesis. J. Clin. Oncol. 2005, 23, 1011–1027. [Google Scholar] [CrossRef]
- Kwong, W.L.; Lam, K.Y.; Lok, C.N.; Lai, Y.T.; Lee, P.Y.; Che, C.M. A Macrocyclic Ruthenium (III) Complex Inhibits Angiogenesis with Down-Regulation of Vascular Endothelial Growth Factor Receptor-2 and Suppresses Tumor Growth In Vivo. Angew. Chem. 2016, 128, 13722–13726. [Google Scholar] [CrossRef]
- Brindell, M.; Gurgul, I.; Janczy-Cempa, E.; Gajda-Morszewski, P.; Mazuryk, O. Moving Ru polypyridyl complexes beyond cytotoxic activity towards metastasis inhibition. J. Inorg. Biochem. 2022, 226, 111652. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Zhao, C.; Wang, L.; Cao, X.; Li, J.; Huang, R.; Lao, Q.; Yu, H.; Li, Y.; Du, H.; et al. A VEGFR1 antagonistic peptide inhibits tumor growth and metastasis through VEGFR1-PI3K-AKT signaling pathway inhibition. Am. J. Cancer Res. 2015, 5, 3149–3161. [Google Scholar] [PubMed]
- Husain, A.; Khadka, A.; Ehrlicher, A.; Saint-Geniez, M.; Krishnan, R. Substrate stiffening promotes VEGF-A functions via the PI3K/Akt/mTOR pathway. Biochem. Biophys. Res. Commun. 2022, 586, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Ferrín, G.; Guerrero, M.; Amado, V.; Rodríguez-Perálvarez, M.; De la Mata, M. Activation of MTOR Signaling Pathway in Hepatocellular Carcinoma. Int. J. Mol. Sci. 2020, 21, 1266. [Google Scholar] [CrossRef]
- Zinatizadeh, M.R.; Schock, B.; Chalbatani, G.M.; Zarandi, P.K.; Jalali, S.A.; Miri, S.R. The Nuclear Factor Kappa B (NF-kB) signaling in cancer development and immune diseases. Genes Dis. 2021, 8, 287–297. [Google Scholar] [CrossRef]
- Duan, Z.; Zhao, X.; Fu, X.; Su, C.; Xin, L.; Saarikettu, J.; Yang, X.; Yao, Z.; Silvennoinen, O.; Wei, M.; et al. Tudor-SN, a novel coactivator of peroxisome proliferator-activated receptor gamma protein, is essential for adipogenesis. J Biol Chem. 2014, 289, 8364–8374. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Liu, X.; Cui, K.; Di, Y.; Xin, L.; Sun, X.; Zhang, W.; Yang, X.; Wei, M.; Yao, Z.; et al. SND1 acts downstream of TGFbeta1 and upstream of Smurf1 to promote breast cancer metastasis. Cancer Res. 2015, 75, 1275–1286. [Google Scholar] [CrossRef] [PubMed]
- Su, C.; Zhang, C.; Tecle, A.; Fu, X.; He, J.; Song, J.; Zhang, W.; Sun, X.; Ren, Y.; Silvennoinen, O.; et al. Tudor staphylococcal nuclease(Tudor-SN), a novel regulator facilitating G1/S phase transition, acting as a co-activator of E2F-1 in cell cycle regulation. J. Biol. Chem. 2015, 290, 7208–7220. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Zhang, C.; Meng, H.; Zhang, K.; Shi, L.; Cao, C.; Wang, Y.; Su, C.; Xin, L.; Ren, Y.; et al. Oncoprotein Tudor-SN is a key determinant providing survival advantage under DNA damaging stress. Cell Death Differ. 2018, 25, 1625–1637. [Google Scholar] [CrossRef]
- Santhekadur, P.K.; Akiel, M.; Emdad, L.; Gredler, R.; Srivastava, J.; Rajasekaran, D.; Robertson, C.L.; Mukhopadhyay, N.D.; Fisher, P.B.; Sarkar, D. Staphylococcal nuclease domain containing-1 (SND1) promotes migration and invasion via angiotensin II type 1 receptor (AT1R) and TGFβ signaling. FEBS Open Bio. 2014, 4, 353–361. [Google Scholar] [CrossRef]
- Gutierrez-Beltran, E.; Denisenko, T.V.; Zhivotovsky, B.; Bozhkov, P.V. Tudor staphylococcal nuclease: Biochemistry and functions. Cell Death Differ. 2016, 23, 1739–1748. [Google Scholar] [CrossRef]
- Zhou, M.; Wang, A.; Yin, B.; Wu, D.; Han, S.; Zhang, W.; Liu, J.; Sun, K. SND1 promotes the proliferation of osteosarcoma cells by upregulating COX 2/PGE2 expression via activation of NF κB. Oncol. Rep. 2019, 41, 579–589. [Google Scholar] [CrossRef]
- Jariwala, N.; Rajasekaran, D.; Mendoza, R.G.; Shen, X.N.; Siddiq, A.; Akiel, M.A.; Robertson, C.L.; Subler, M.A.; Windle, J.J.; Fisher, P.B.; et al. Oncogenic role of SND1 in development and progression of hepatocellular carcinoma. Cancer Res. 2017, 77, 3306–3316. [Google Scholar] [CrossRef]
- Cui, X.; Zhao, C.; Yao, X.; Qian, B.; Su, C.; Ren, Y.; Yao, Z.; Gao, X.; Yang, J. SND1 acts as an anti-apoptotic factor via regulating the expression of lncRNA UCA1 in hepatocellular carcinoma. RNA Biol. 2018, 15, 1364–1375. [Google Scholar] [CrossRef]
- Chidambaranathan-Reghupaty, S.; Mendoza, R.; Fisher, P.B.; Sarkar, D. The multifaceted oncogene SND1 in cancer: Focus on hepatocellular carcinoma. Hepatoma Res. 2018, 4, 32. [Google Scholar] [CrossRef] [PubMed]
- Song, W.; Mazzieri, R.; Yang, T.; Gobe, G.C. Translational significance for tumor metastasis of tumor-associated macrophages and epithelial-mesenchymal transition. Front Immunol. 2017, 8, 1106. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Shen, S.; Verma, I.M. NF-kappaB, an active player in human cancers. Cancer Immunol. Res. 2014, 2, 823–830. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Luo, Q.; Qin, Y.; Wu, Y.; Liu, X.-W. DNA interaction, DNA photocleavage, photocytotoxicity in vitro, and molecular docking of naphthyl-appended ruthenium complexes. Molecules 2022, 27, 3676. [Google Scholar] [CrossRef] [PubMed]
- Balou, S.; Zarkadoulas, A.; Koukouvitaki, M.; Marchiò, L.; Efthimiadou, E.K.; Mitsopoulou, C.A. Synthesis, DNA-binding, anticancer evaluation, and molecular docking studies of bishomoleptic and trisheteroleptic Ru-diimine complexes bearing 2-(2-pyridyl)-quinoxaline. Bioinorg. Chem. Appl. 2021, 2021, 5599773. [Google Scholar] [CrossRef]
- Liu, X.-W.; Liu, N.-Y.; Deng, Y.-Q.; Wang, S.; Liu, T.; Tang, Y.-C.; Chen, Y.-D.; Lu, J.-L. Anticancer activity, topoisomerase I inhibition, DNA ‘light switch’ behavior and molecular docking of two ruthenium complexes containing phenazine ring. J. Biomol. Struct. Dyn. 2021, 39, 5953–5962. [Google Scholar] [CrossRef] [PubMed]
- Tang, B.; Shen, F.; Wan, D.; Guo, B.-H.; Wang, Y.-J.; Yi, Q.-Y.; Liu, Y.-J. DNA-binding, molecular docking studies and biological activity studies of ruthenium(II) polypyridyl complexes. RSC Adv. 2017, 7, 34945–34958. [Google Scholar] [CrossRef]
- Tweedy, B.G. Plant Extracts with Metal Ions as Potential Antimicrobial Agents. Phytopathology 1964, 55, 910. [Google Scholar]
- Benos, D.J.; Deamer, D.W.; Kleinzeller, A.; Fambrough, D.M. (Eds.) Membrane Permeability: 100 Years since Ernest Overton; Current Topics in Membranes; Academic Press: San Diego, CA, USA, 1999; p. 48. [Google Scholar]
- Smith, N.A.; Zhang, P.; Greenough, S.E.; Horbury, M.D.; Clarkson, G.J.; McFeely, D.; Habtemariam, A.; Salassa, L.; Stavros, V.G.; Dowson, C.G.; et al. Combatting AMR: Photoactivatable ruthenium(II)-isoniazid complex exhibits rapid selective antimycobacterial activ-ity. Chem. Sci. 2017, 8, 395–404. [Google Scholar] [CrossRef] [PubMed]

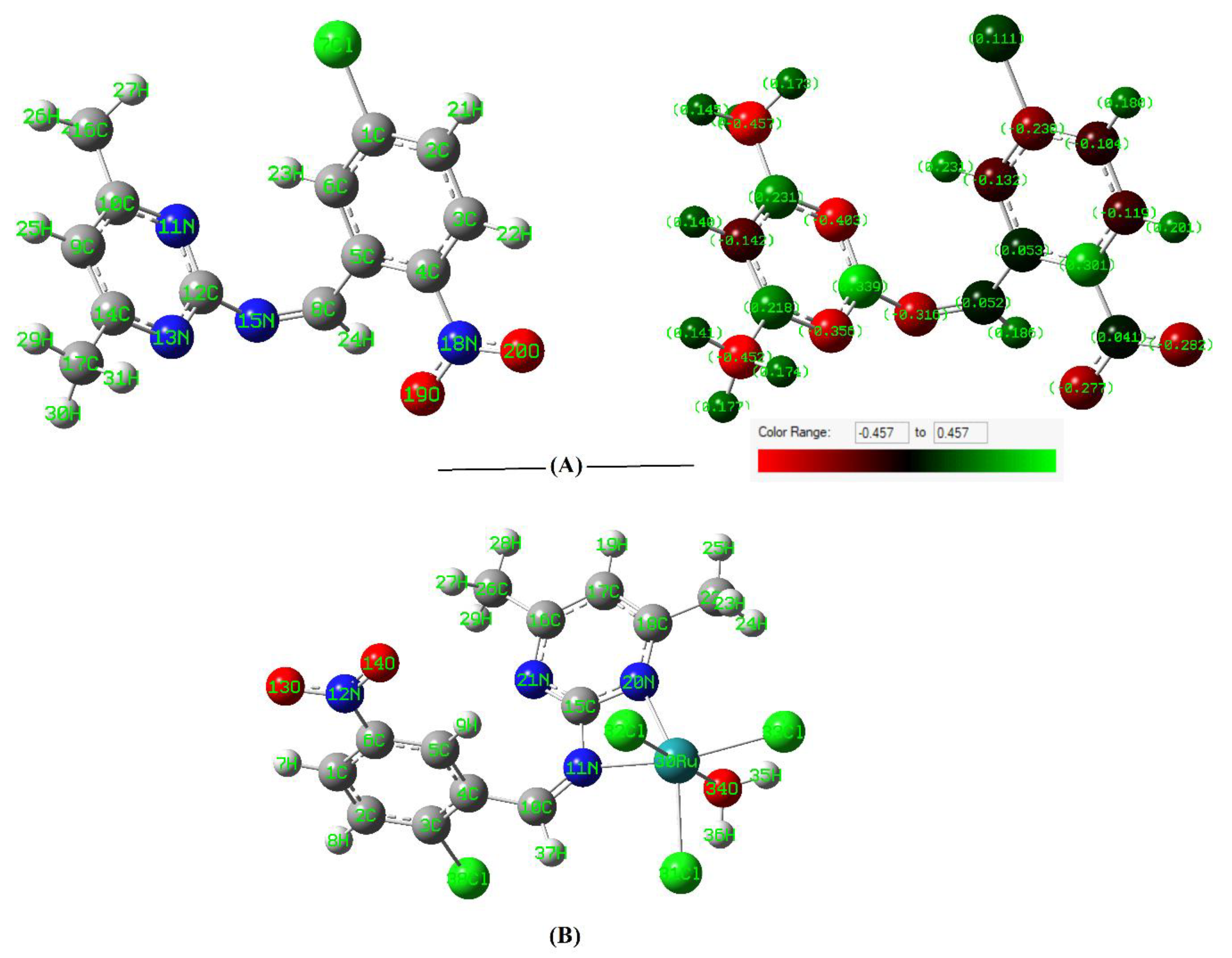
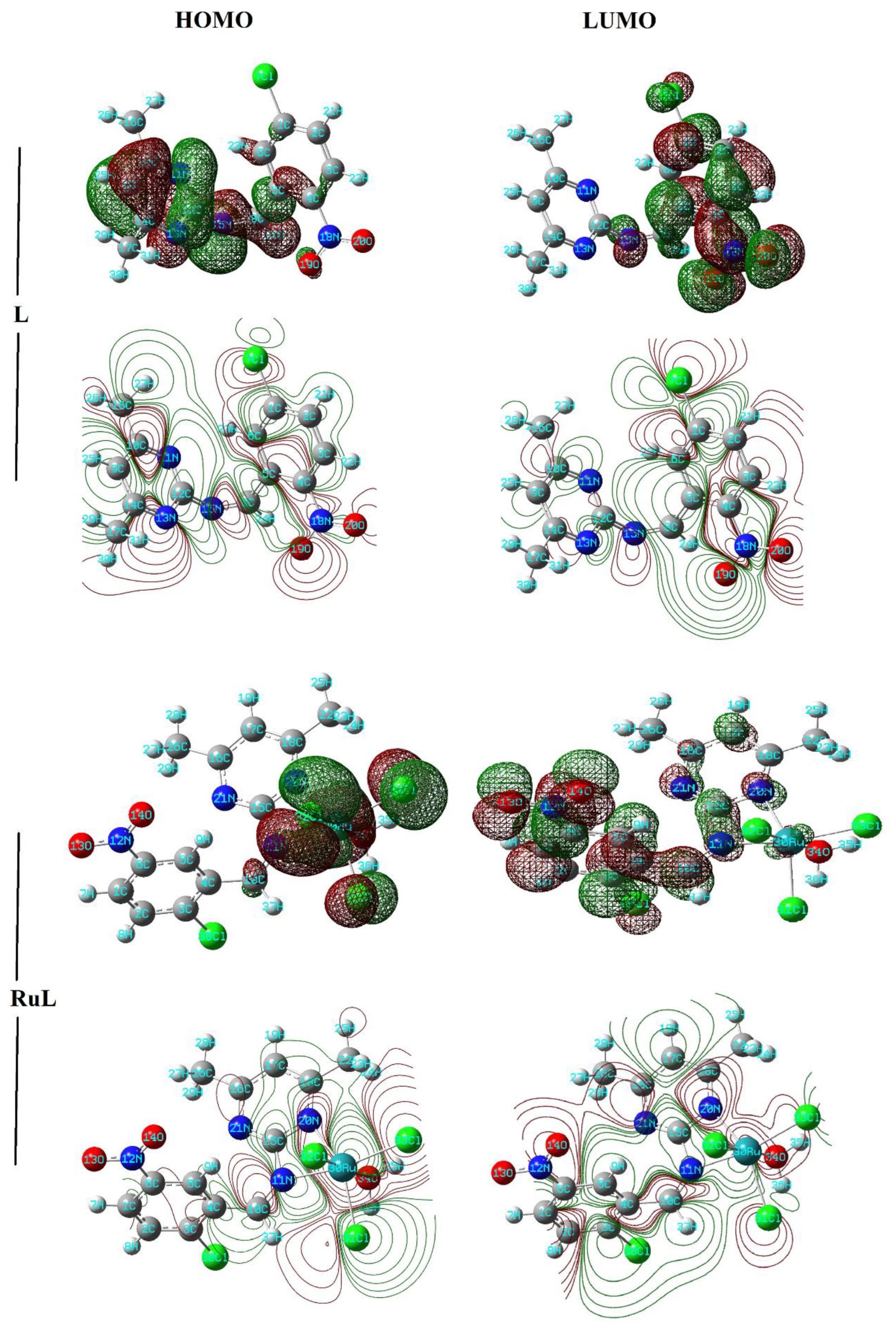


| L | RuL | |||||||
|---|---|---|---|---|---|---|---|---|
| Bond length (Ǻ) | C(12)-N(15) | 1.40 | C(8)-N(15) | 1.28 | C(15)-N(11) | 1.43 | C(10)-N(11) | 1.29 |
| C(12)-N(11) | 1.36 | C(12)-N(13) | 1.35 | Ru(30)-O(34) | 2.19 | Ru(30)-Cl(31) | 2.41 | |
| C(4)-N(18) | 1.47 | C(1)-Cl(7) | 1.82 | C(3)-Cl(38) | 1.81 | C(6)-N(12) | 1.48 | |
| Ru(30)-N(20) | 2.11 | Ru(30)-N(11) | 2.10 | |||||
| Bond angle (°) | C(8)-N(15)-C(12) | 131.0 | N(11)-C(12)-N(13) | 125.8 | C(10)-N(11)-C(15) | 123.6 | N(20)-Ru(30)-O(34) | 78.0 |
| C(2)-C(1)-Cl(7) | 118.9 | C(5)-C(8)-N(15) | 127.6 | N(11)-Ru(30)-N(20) | 64.2 | Cl(32)-Ru(30)-O(34) | 171.6 | |
| C(4)-N(18)-O(20) | 118.0 | C(6)-C(5)-C(8) | 118.5 | Cl(32)-Ru(30)-Cl(33) | 105.6 | N(11)-C(15)-N(20) | 103.5 | |
| N(11)-C(12)-N(15) | 123.2 | |||||||
| Parameter | L | RuL |
|---|---|---|
| DM (Debye) | 5.22 | 13.44 |
| HOMO (eV) | −6.61 | −6.47 |
| LUMO (eV) | −3.33 | −3.82 |
| ΔE (eV) | 3.28 | 2.64 |
| Χ (eV) | 4.97 | 5.15 |
| V (eV) | −4.97 | −5.15 |
| A (eV) | 3.33 | 3.82 |
| I (eV) | 6.61 | 6.47 |
| η (eV) | 1.64 | 1.32 |
| S (eV) | 0.82 | 0.66 |
| ω (eV) | 7.53 | 10.02 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Noureldeen, A.F.H.; Aziz, S.W.; Shouman, S.A.; Mohamed, M.M.; Attia, Y.M.; Ramadan, R.M.; Elhady, M.M. Molecular Design, Spectroscopic, DFT, Pharmacological, and Molecular Docking Studies of Novel Ruthenium(III)–Schiff Base Complex: An Inhibitor of Progression in HepG2 Cells. Int. J. Environ. Res. Public Health 2022, 19, 13624. https://doi.org/10.3390/ijerph192013624
Noureldeen AFH, Aziz SW, Shouman SA, Mohamed MM, Attia YM, Ramadan RM, Elhady MM. Molecular Design, Spectroscopic, DFT, Pharmacological, and Molecular Docking Studies of Novel Ruthenium(III)–Schiff Base Complex: An Inhibitor of Progression in HepG2 Cells. International Journal of Environmental Research and Public Health. 2022; 19(20):13624. https://doi.org/10.3390/ijerph192013624
Chicago/Turabian StyleNoureldeen, Amani F. H., Safa W. Aziz, Samia A. Shouman, Magdy M. Mohamed, Yasmin M. Attia, Ramadan M. Ramadan, and Mostafa M. Elhady. 2022. "Molecular Design, Spectroscopic, DFT, Pharmacological, and Molecular Docking Studies of Novel Ruthenium(III)–Schiff Base Complex: An Inhibitor of Progression in HepG2 Cells" International Journal of Environmental Research and Public Health 19, no. 20: 13624. https://doi.org/10.3390/ijerph192013624
APA StyleNoureldeen, A. F. H., Aziz, S. W., Shouman, S. A., Mohamed, M. M., Attia, Y. M., Ramadan, R. M., & Elhady, M. M. (2022). Molecular Design, Spectroscopic, DFT, Pharmacological, and Molecular Docking Studies of Novel Ruthenium(III)–Schiff Base Complex: An Inhibitor of Progression in HepG2 Cells. International Journal of Environmental Research and Public Health, 19(20), 13624. https://doi.org/10.3390/ijerph192013624







