Assessing the Biodegradation of BTEX and Stress Response in a Bio-Permeable Reactive Barrier Using Compound-Specific Isotope Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Column Experimental Setup
2.3. Batch Experiments
2.4. Quantification of Benzene, Toluene, and TCE
2.5. CSIA Analysis
2.6. DNA Extraction and PCR Amplification
2.7. Quantitative Real-Time PCR (q-PCR)
2.8. Illumina High-Throughput Sequencing
2.9. Isotope Calculations
3. Results and Discussion
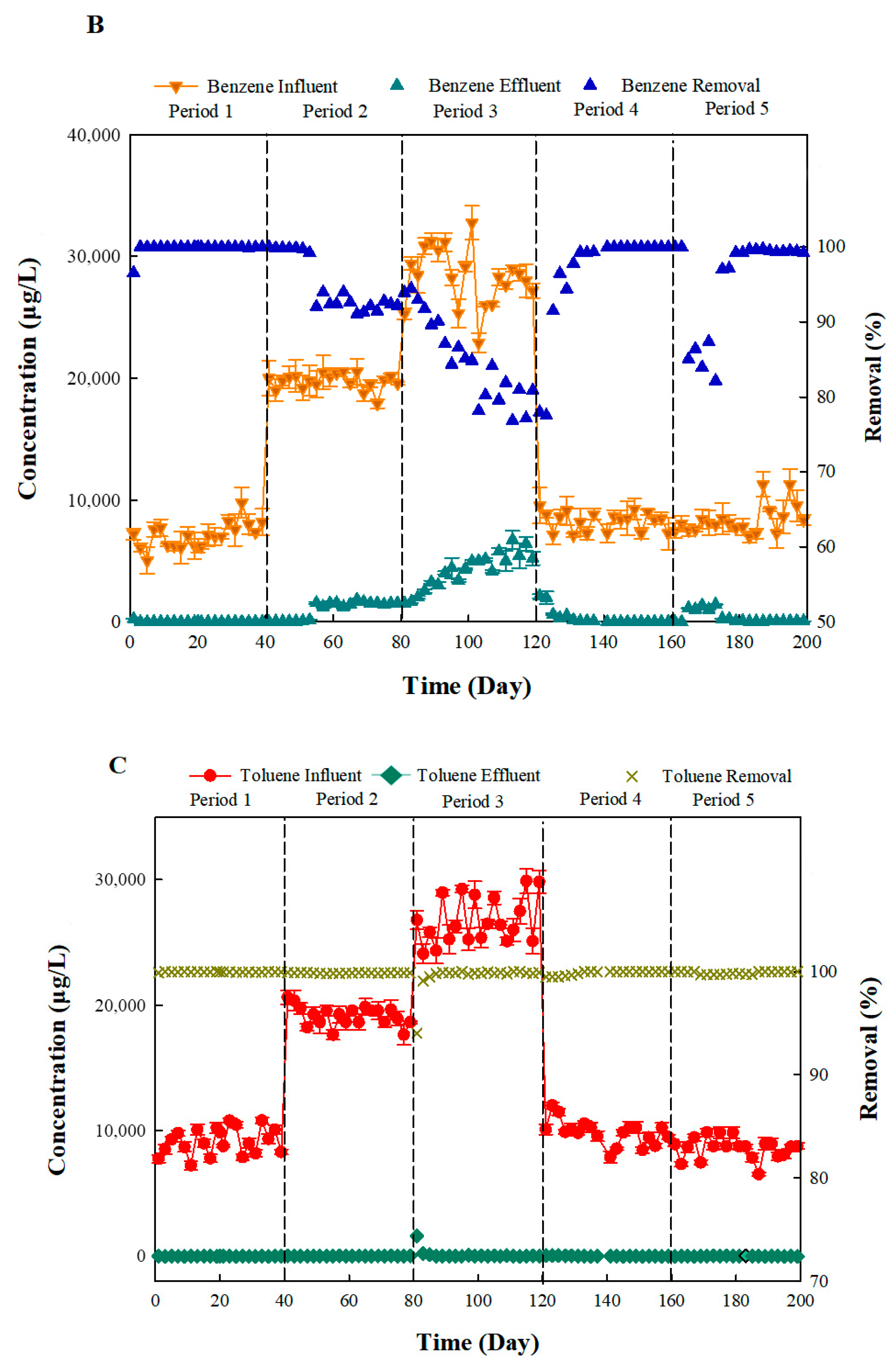
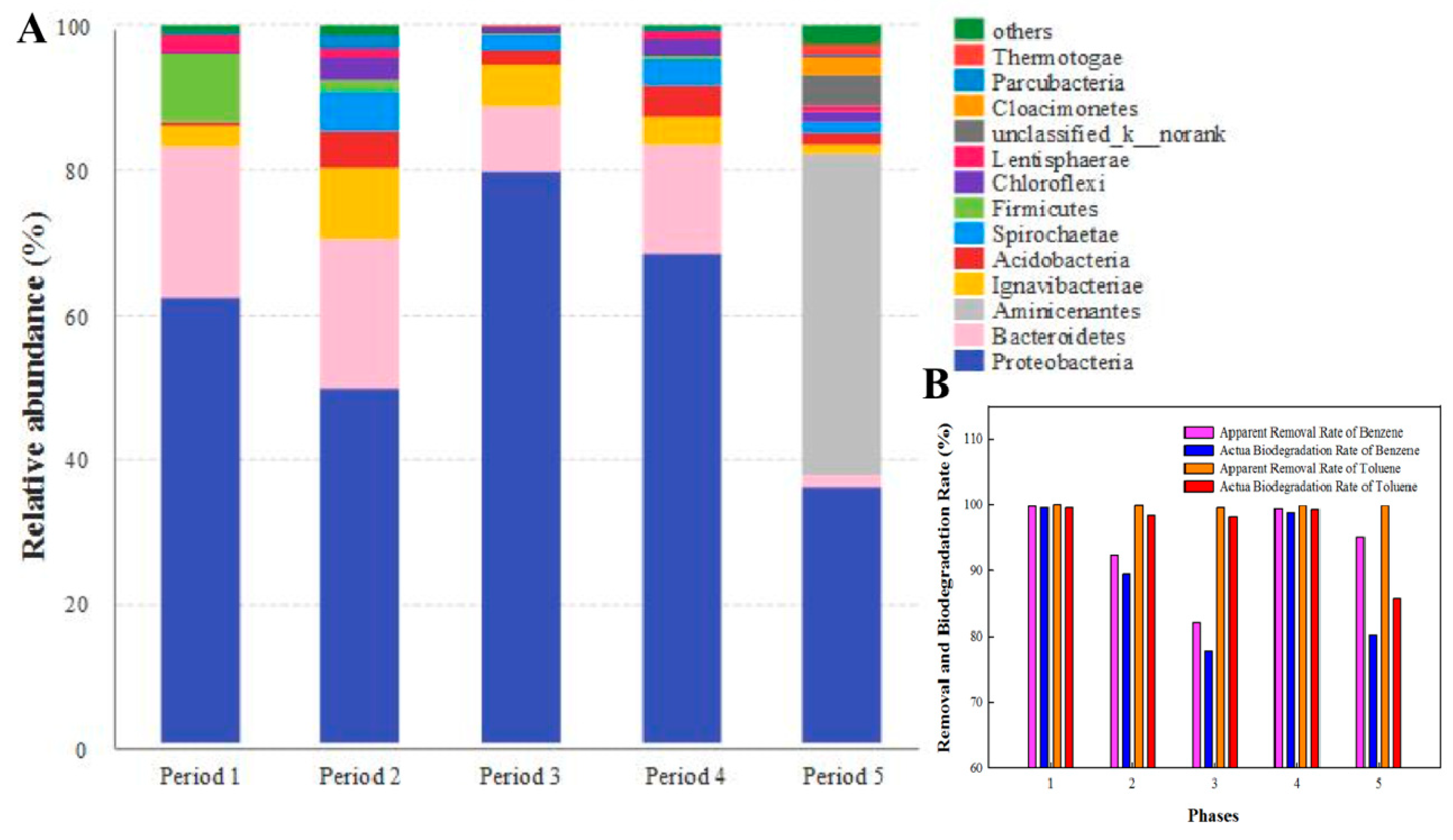
3.1. Apparent Removal Rate of Benzene and Toluene in Bio-PRB System
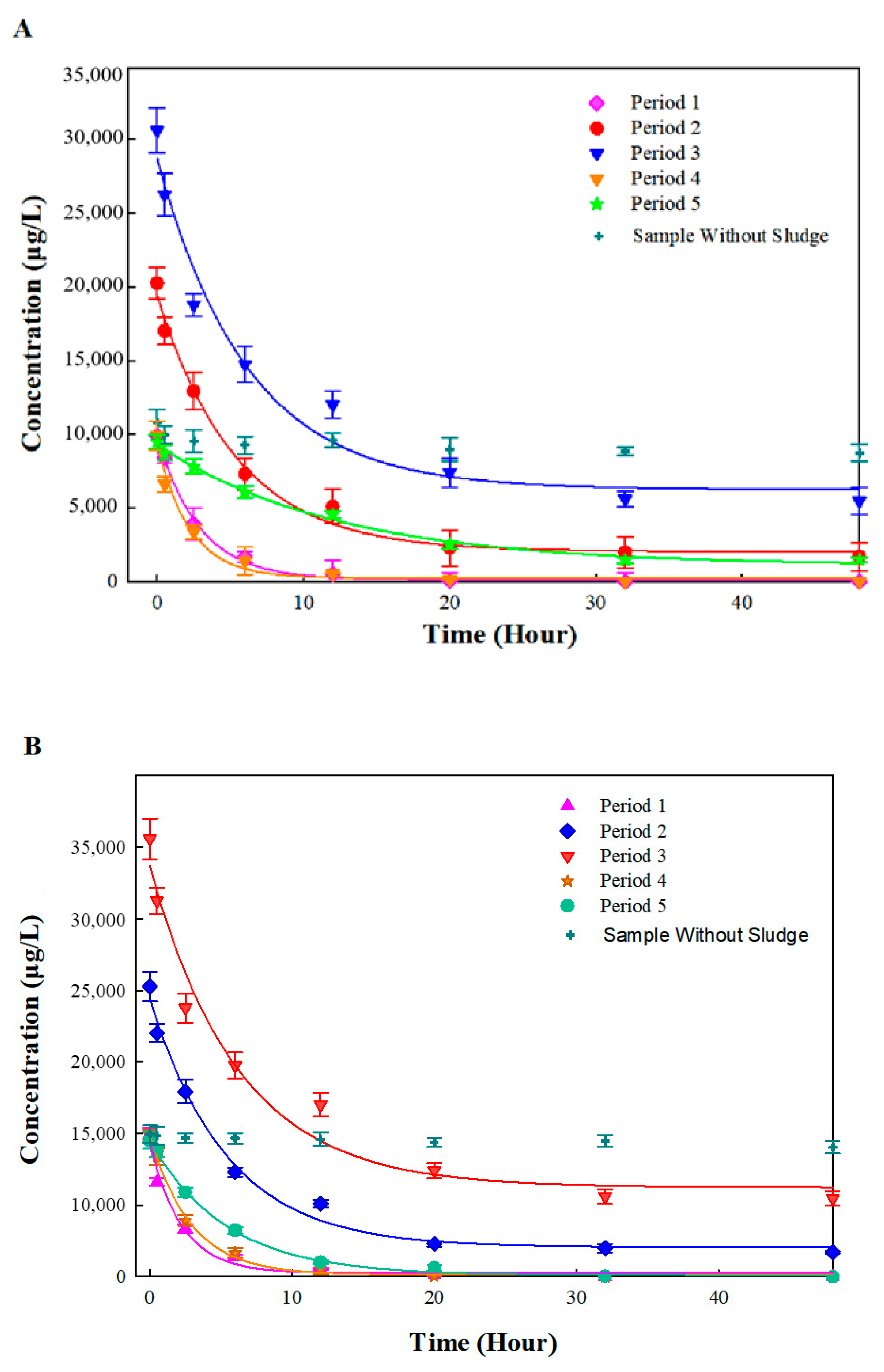
3.2. Batch Biodegradation Kinetics
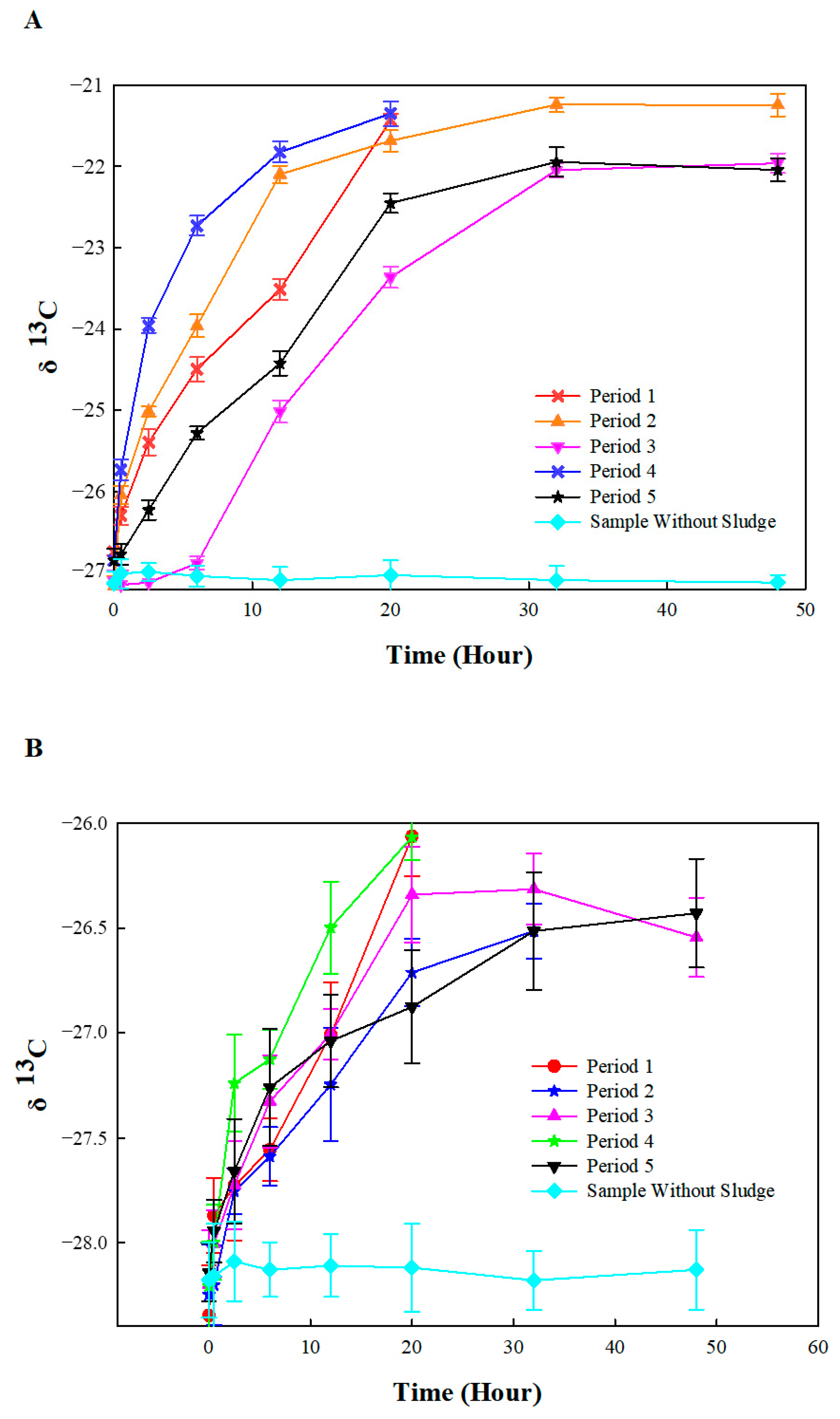
3.3. Compound-Specific Isotope Analysis
3.4. Calculation of Actual Biodegradation Rates of Benzene and Toluene in Bio-PRB
3.5. Functional Gene Copies in Bio-PRB System
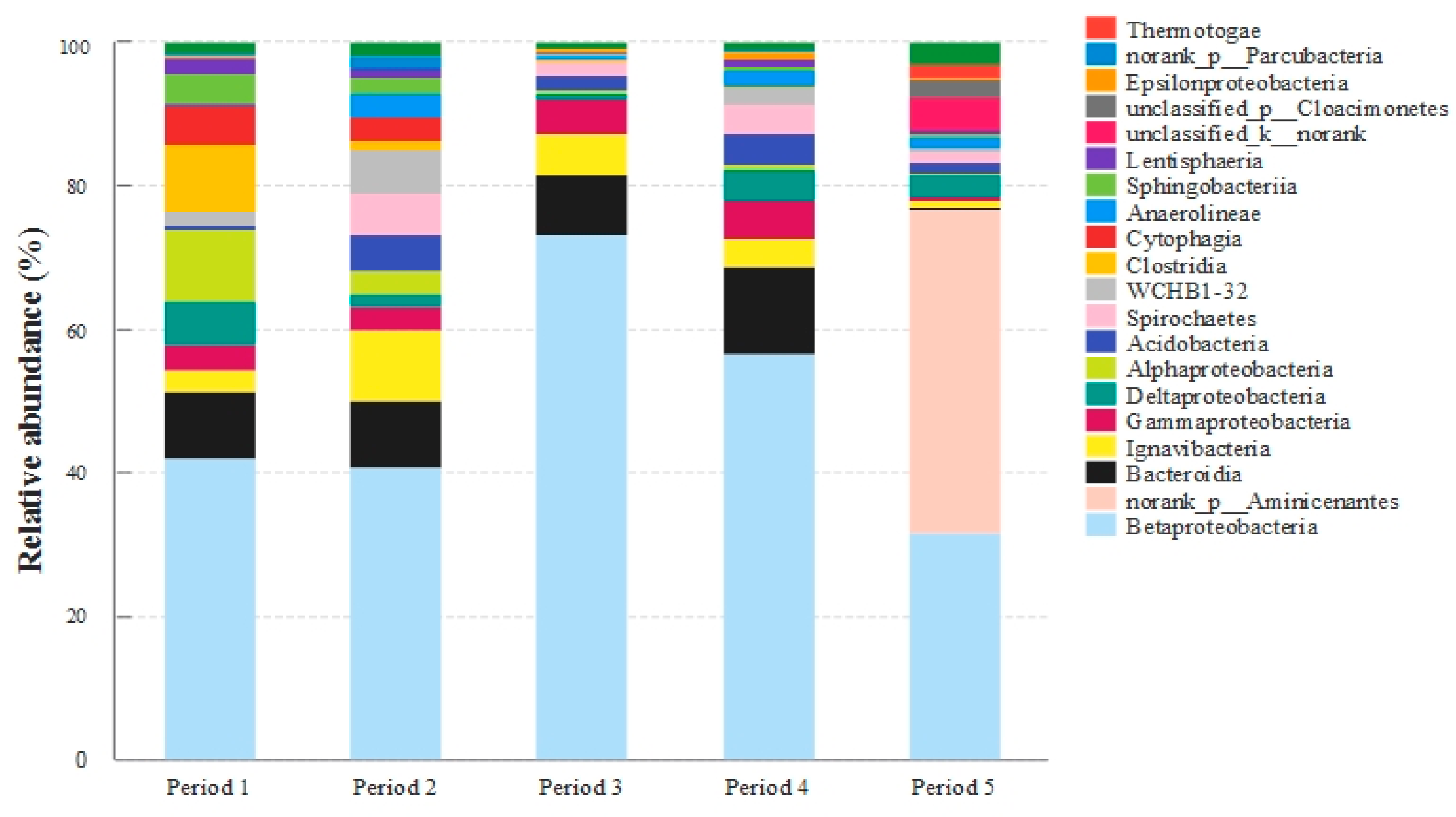
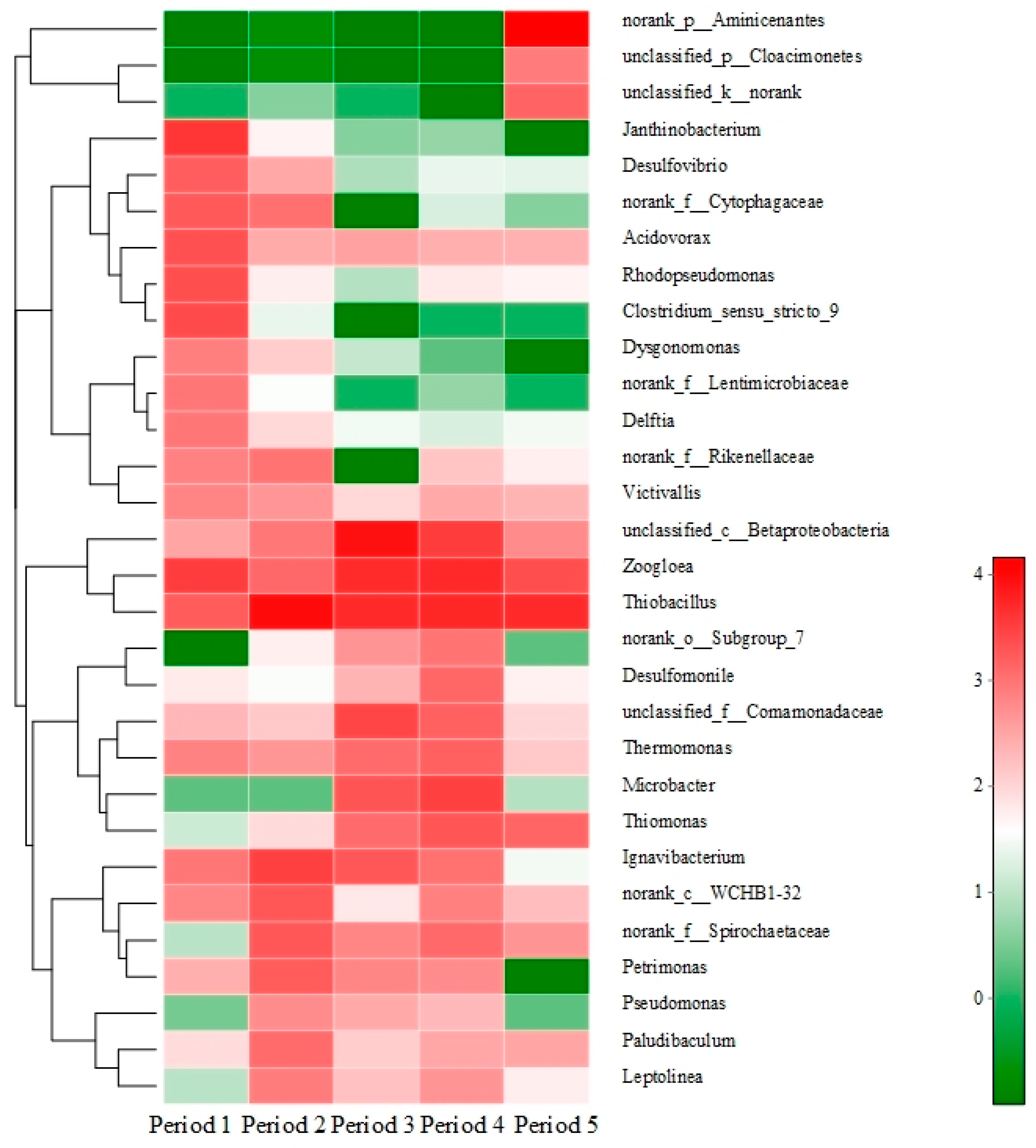
3.6. Bacterial Community Analyses
3.7. Conceptual Process of Benzene and Toluene Removal
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- An, Y.J. Toxicity of benzene, toluene, ethylbenzene, and xylene (BTEX) mixtures to Sorghum bicolor and Cucumis sativus. Bull. Environ. Contam. Toxicol. 2004, 72, 1006–1011. [Google Scholar] [CrossRef]
- Farhadian, M.; Duchez, D.; Vachelard, C.; Larroche, C. Monoaromatics removal from polluted water through bioreactors—A review. Water Res. 2008, 42, 1325–1341. [Google Scholar] [CrossRef]
- Mehlman, M.A. Dangerous and cancer-causing properties of products and chemicals in the oil refining and petrochemical industry: Part I. Carcinogenicity of motor fuels: Gasoline. Toxicol. Ind. Health 1991, 7, 143–152. [Google Scholar] [CrossRef]
- Schaefer, C.E.; Yang, X.; Pelz, O.; Tsao, D.T.; Streger, S.H.; Steffan, R.J. Anaerobic biodegradation of iso-butanol and ethanol and their relative effects on BTEX biodegradation in aquifer materials. Chemosphere 2010, 81, 1111–1117. [Google Scholar] [CrossRef] [PubMed]
- Da Ifullah, A.; Girgis, B.S. Impact of surface characteristics of activated carbon on adsorption of BTEX. Colloids Surf. A Physicochem. Eng. Asp. 2003, 214, 181–193. [Google Scholar] [CrossRef]
- Mascolo, G.; Ciannarella, R.; Balest, L.; Lopez, A. Effectiveness of UV-based advanced oxidation processes for the remediation of hydrocarbon pollution in the groundwater: A laboratory investigation. J. Hazard. Mater. 2008, 152, 1138–1145. [Google Scholar] [CrossRef] [PubMed]
- Tiburtius, E.R.L.; Peralta-Zamora, P.; Emmel, A. Treatment of gasoline-contaminated waters by advanced oxidation processes. J. Hazard. Mater. 2005, 126, 86–90. [Google Scholar] [CrossRef]
- Weelink, S.A.; Van Eekert, M.H.; Stams, A.J. Degradation of BTEX by anaerobic bacteria: Physiology and application. Rev. Environ. Sci. Bio/Technol. 2010, 9, 359–385. [Google Scholar] [CrossRef]
- Yang, X.; Beckmann, D.; Fiorenza, S.; Niedermeier, C. Field Study of Pulsed Air Sparging for Remediation of Petroleum Hydrocarbon Contaminated Soil and Groundwater. Environ. Sci. Technol. 2005, 39, 7279–7286. [Google Scholar] [CrossRef]
- Kao, C.M.; Huang, W.Y.; Chang, L.J.; Chen, T.Y.; Chien, H.Y.; Hou, F. Application of monitored natural attenuation to remediate a petroleum-hydrocarbon spill site. Water Sci. Technol. J. Int. Assoc. Water Pollut. Res. 2006, 53, 321–328. [Google Scholar] [CrossRef]
- Lueders, T. The ecology of anaerobic degraders of BTEX hydrocarbons in aquifers. FEMS Microbiol. Ecol. 2017, 93, 220. [Google Scholar] [CrossRef]
- Zein, M.M.; Pinto, P.X.; Garcia-Blanco, S.; Suidan, M.T.; Venosa, A.D. Treatment of groundwater contaminated with PAHs, gasoline hydrocarbons, and methyl tert-butyl ether in a laboratory biomass-retaining bioreactor. Biodegradation 2006, 17, 57–69. [Google Scholar] [CrossRef]
- Farhadian, M.; Vachelard, C.; Duchez, D.; Larroche, C. In situ bioremediation of monoaromatic pollutants in groundwater: A review. Bioresour. Technol. 2008, 99, 5296–5308. [Google Scholar] [CrossRef]
- Atteia, O.; Franceschi, M. Kinetics of natural attenuation of BTEX: Review of the critical chemical conditions and measurements at bore scale. Sci. World J. 2014, 2, 1338–1346. [Google Scholar] [CrossRef][Green Version]
- Deeb, R.A.; Alvarez-Cohen, L. Temperature effects and substrate interactions during the aerobic biotransformation of BTEX mixtures by toluene-enriched consortia and Rhodococcus rhodochrous. Biotechnol. Bioeng. 2015, 62, 526–536. [Google Scholar] [CrossRef]
- Abbai, N.S.; Pillay, B. Analysis of Hydrocarbon-Contaminated Groundwater Metagenomes as Revealed by High-Throughput Sequencing. Mol. Biotechnol. 2013, 54, 900–912. [Google Scholar] [CrossRef]
- Clement, T.P.; Johnson, C.D.; Sun, Y.; Klecka, G.M.; Bartlett, C. Natural attenuation of chlorinated ethane compounds: Model development and field-scale application a. J. Contam. Hydrol. 2000, 42, 113–140. [Google Scholar] [CrossRef]
- Jung, I.G.; Park, C.H. Characteristics of Rhodococcus pyridinovorans PYJ-1 for the Biodegradation of Benzene, Toluene, m-Xylene (BTX), and Their Mixtures. J. Biosci. Bioeng. 2004, 97, 429–431. [Google Scholar] [CrossRef]
- Kim, D.; Kim, Y.S.; Kim, S.K.; Kim, S.W.; Zylstra, G.J.; Kim, Y.M.; Kim, E. Monocyclic Aromatic Hydrocarbon Degradation by Rhodococcus sp. Strain DK17. Appl. Environ. Microbiol. 2002, 68, 3270–3278. [Google Scholar] [CrossRef]
- Morasch, B.; Richnow, H.H.; Schink, B.; Vieth, A.; Meckenstock, R.U. Carbon and Hydrogen Stable Isotope Fractionation during Aerobic Bacterial Degradation of Aromatic Hydrocarbons. Appl. Environ. Microbiol. 2002, 68, 5191–5194. [Google Scholar] [CrossRef]
- Nielsen, D.R.; Mclellan, P.J.; Daugulis, A.J. Direct estimation of the oxygen requirements of Achromobacter xylosoxidans for aerobic degradation of monoaromatic hydrocarbons (BTEX) in a bioscrubber. Biotechnol. Lett. 2006, 28, 1293–1298. [Google Scholar] [CrossRef]
- Parales, R.E.; Ditty, J.L.; Harwood, C.S. Toluene-Degrading Bacteria Are Chemotactic towards the Environmental Pollutants Benzene, Toluene, and Trichloroethylene. Appl. Environ. Microbiol. 2000, 66, 4098–4104. [Google Scholar] [CrossRef]
- Prenafeta-Boldú, F.X.; Ballerstedt, H.; Gerritse, J.; Grotenhuis, J.T.C. Bioremediation of BTEX hydrocarbons: Effect of soil inoculation with the toluene-growing fungus Cladophialophora sp. strain T1. Biodegradation 2004, 15, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Shim, H.; Hwang, B.; Lee, S.S.; Kong, S.H. Kinetics of BTEX biodegradation by a coculture of Pseudomonas putida and Pseudomonas fluorescens under hypoxic conditions. Biodegradation 2005, 16, 319–327. [Google Scholar] [CrossRef]
- Batlle-Aguilar, J.; Morasch, B.; Hunkeler, D.; Brouyère, S. Benzene Dynamics and Biodegradation in Alluvial Aquifers Affected by River Fluctuations. Groundwater 2013, 52, 388–398. [Google Scholar] [CrossRef]
- Chen, L.; Liu, Y.; Liu, F.; Jin, S. Treatment of co-mingled benzene, toluene and TCE in groundwater. J. Hazard. Mater. 2014, 275, 116–120. [Google Scholar] [CrossRef]
- Firmino, P.I.M.; Farias, R.S.; Buarque, P.M.; Costa, M.C.; Rodríguez, E.; Lopes, A.C.; dos Santos, A.B. Engineering and microbiological aspects of BTEX removal in bioreactors under sulfate-reducing conditions. Chem. Eng. J. 2015, 260, 503–512. [Google Scholar] [CrossRef]
- Xin, B.P.; Wu, C.H.; Wu, C.H.; Lin, C.W. Bioaugmented remediation of high concentration BTEX-contaminated groundwater by permeable reactive barrier with immobilized bead. J. Hazard. Mater. 2013, 244–245, 765–772. [Google Scholar] [CrossRef]
- Yeh, C.H.; Lin, C.W.; Wu, C.H. A permeable reactive barrier for the bioremediation of BTEX-contaminated groundwater: Microbial community distribution and removal efficiencies. J. Hazard. Mater. 2010, 178, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Barbara, H.; Winnie, D.; Folkert, F.; Wesley, B.; Leen, B.; Willy, V.; Top, E.M.; Dirk, S. PCR-DGGE method to assess the diversity of BTEX mono-oxygenase genes at contaminated sites. Fems Microbiol. Ecol. 2010, 2, 262–273. [Google Scholar]
- Mercier, A.; Joulian, C.; Michel, C.; Auger, P.; Coulon, S.; Amalric, L.; Morlay, C.; Battaglia-Brunet, F. Evaluation of three activated carbons for combined adsorption and biodegradation of PCBs in aquatic sediment. Water Res. 2014, 59, 304–315. [Google Scholar] [CrossRef]
- Xiong, W.; Mathies, C.; Bradshaw, K.; Carlson, T.; Tang, K.; Wang, Y. Benzene removal by a novel modification of enhanced anaerobic biostimulation. Water Res. 2012, 46, 4721–4731. [Google Scholar] [CrossRef]
- Tong, Z.; Shao, M.F.; Lin, Y. 454 Pyrosequencing reveals bacterial diversity of activated sludge from 14 sewage treatment plants. ISME J. 2012, 6, 1137–1147. [Google Scholar]
- Zhou, J.; Jiang, Y.H.; Deng, Y.; Shi, Z.; Zhou, B.Y.; Xue, K.; Wu, L.; He, Z.; Yang, Y. Random Sampling Process Leads to Overestimation of β-Diversity of Microbial Communities. Mbio 2013, 4, e00324-13. [Google Scholar] [CrossRef]
- Dorer, C.; Höhener, P.; Hedwig, N.; Richnow, H.H.; Vogt, C. Rayleigh-Based Concept to Tackle Strong Hydrogen Fractionation in Dual Isotope Analysis-The Example of Ethylbenzene Degradation by Aromatoleum aromaticum. Environ. Sci. Technol. 2014, 48, 5788–5797. [Google Scholar] [CrossRef]
- Schmidt, T.C.; Jochmann, M.A. Origin and Fate of Organic Compounds in Water: Characterization by Compound-Specific Stable Isotope Analysis. Annu. Rev. Anal. Chem. 2012, 5, 133–155. [Google Scholar] [CrossRef]
- Wang, Z.; Jian, L.; Hesham, E.L.; He, S.; Yu, Z.; Wang, Z.; Yang, M. Co-variations of bacterial composition and catabolic genes related to PAH degradation in a produced water treatment system consisting of successive anoxic and aerobic units. Sci. Total Environ. 2007, 373, 356–362. [Google Scholar] [CrossRef]
- Ye, L.; Zhang, T.; Wang, T.; Fang, Z. Microbial structures, functions, and metabolic pathways in wastewater treatment bioreactors revealed using high-throughput sequencing. Environ. Sci. Technol. 2012, 46, 13244–13252. [Google Scholar] [CrossRef]
- Ke, Y.; Tong, Z. Metagenomic and Metatranscriptomic Analysis of Microbial Community Structure and Gene Expression of Activated Sludge. PLoS ONE 2011, 7, e38183. [Google Scholar]
- Chee, G.J. Biodegradation analyses of trichloroethylene (TCE) by bacteria and its use for biosensing of TCE. Talanta 2011, 85, 1778–1782. [Google Scholar] [CrossRef]
- Chakraborty, R. Anaerobic Monoaromatic Hydrocarbon Degradation by a Novel Chlorate Reducing Organism, Dechloromonas Aromatica Strain RCB. Ph.D. Thesis, University of California, Berkeley, CA, USA, 4 December 2005. [Google Scholar]
- Kasai, Y.; Takahata, Y.; Manefield, M.; Watanabe, K. RNA-Based Stable Isotope Probing and Isolation of Anaerobic Benzene-Degrading Bacteria from Gasoline-Contaminated Groundwater. Appl. Environ. Microbiol. 2006, 72, 3586–3592. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.W.; La, H.J.; Lee, S.J. Microbial degradation of benzene, toluene, ethylbenzene and xylene isomers (BTEX) contaminated groundwater in Korea. Water Sci. Technol. J. Int. Assoc. Water Pollut. Res. 2001, 44, 165–171. [Google Scholar] [CrossRef]
- Fischer, A.; Theuerkorn, K.; Stelzer, N.; Gehre, M.; Thullner, M.; Richnow, H.H. Applicability of Stable Isotope Fractionation Analysis for the Characterization of Benzene Biodegradation in a BTEX-contaminated Aquifer. Environ. Sci. Technol. 2007, 41, 3689–3696. [Google Scholar] [CrossRef]
- Greenberg, A.E.; Clesceri, L.S.; Eaton, A.D. Standard Methods for the Examination of Water and Wastewater, 18th ed.; Amer Public Health Assn: Washington, DC, USA, 1992. [Google Scholar]
- Fischer, A.; Bauer, J.; Meckenstock, R.U.; Stichler, W.; Griebler, C.; Maloszewski, P.; Kästner, M.; Richnow, H. A multitracer test proving the reliability of Rayleigh equation-based approach for assessing biodegradation in a BTEX contaminated aquifer. Environ. Sci. Technol. 2006, 40, 4245–4252. [Google Scholar] [CrossRef]
- Mancini, S.A.; Ulrich, A.C.; Lacrampe-Couloume, G.; Sleep, B.; Edwards, E.A.; Lollar, B.S. Carbon and Hydrogen Isotopic Fractionation during Anaerobic Biodegradation of Benzene. Appl. Environ. Microbiol. 2003, 69, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Baldwin, B.R.; Peacock, A.D.; Park, M.; Ogles, D.M.; Istok, J.D.; Mckinley, J.P.; Resch, C.T.; White, D.C. Multilevel Samplers as Microcosms to Assess Microbial Response to Biostimulation. Ground Water 2008, 46, 295–304. [Google Scholar] [CrossRef]
- Beller, H.R.; Kane, S.R.; Legler, T.C.; Alvarez, P.J.J. A real-time polymerase chain reaction method for monitoring anaerobic, hydrocarbon-degrading bacteria based on a catabolic gene. Environ. Sci. Technol. 2002, 36, 3977–3984. [Google Scholar] [CrossRef]
- Stults, J.R.; Snoeyenbos-West, O.; Methe, B.; Lovley, D.R.; Chandler, D.P. Application of the 5′ Fluorogenic Exonuclease Assay (TaqMan) for Quantitative Ribosomal DNA and rRNA Analysis in Sediments. Appl. Environ. Microbiol. 2001, 67, 2781–2789. [Google Scholar] [CrossRef]
- Rhee, S.K.; Liu, X.; Wu, L.; Chong, S.C.; Wan, X.; Zhou, J. Detection of genes involved in biodegradation and biotransformation in microbial communities by using 50-mer oligonucleotide microarrays. Appl. Environ. Microbiol. 2004, 70, 4303–4317. [Google Scholar] [CrossRef]
- López-Gutiérrez, J.C.; Henry, S.; Hallet, S.; Martin-Laurent, F.; Catroux, G.; Philippot, L. Quantification of a novel group of nitrate-reducing bacteria in the environment by real-time PCR. J. Microbiol. Meth. 2004, 57, 399–407. [Google Scholar] [CrossRef]
- Jammer, S.; Voloshenko, A.; Gelman, F.; Lev, O. Chiral and isotope analyses for assessing the degradation of organic contaminants in the environment: Rayleigh dependence. Environ. Sci. Technol. 2014, 48, 3310–3318. [Google Scholar] [CrossRef]
- Bashir, S.; Hitzfeld, K.L.; Gehre, M.; Richnow, H.H.; Fischer, A. Evaluating degradation of hexachlorcyclohexane (HCH) isomers within a contaminated aquifer using compound-specific stable carbon isotope analysis (CSIA). Water Res. 2015, 71, 187–196. [Google Scholar] [CrossRef]
- Rayleigh, L.L. Theoretical considerations respecting the separation of gases by diffusion and similar processes. Lond. Edinb. Dublin Philos. Mag. J. Sci. 1951, 42, 493–498. [Google Scholar] [CrossRef]
- Hoefs, J. Stable Isotope Geochemistry; Springer: Berlin/Heidelberg, Germany, 1980; pp. 102–107. [Google Scholar]
- Meckenstock, R.U.; Morasch, B.; Griebler, C.; Richnow, H.H. Stable isotope fractionation analysis as a tool to monitor biodegradation in contaminated acquifers. J. Contam. Hydrol. 2004, 75, 215–255. [Google Scholar] [CrossRef]
- Hopkins, G.D.; Semprini, L.; Mccarty, P.L. Microcosm and in situ field studies of enhanced biotransformation of trichloroethylene by phenol-utilizing microorganisms. Appl. Environ. Microbiol. 1993, 59, 2277–2285. [Google Scholar] [CrossRef]
- Leahy, J.G.; Byrne, A.M.; Olsen, R.H. Comparison of factors influencing trichloroethylene degradation by toluene-oxidizing bacteria. Appl. Environ. Microbiol. 1996, 62, 825–833. [Google Scholar] [CrossRef]
- Mu, D.Y.; Scow, K.M. Effect of trichloroethylene (TCE) and toluene concentrations on TCE and toluene biodegradation and the population density of TCE and toluene degraders in soil. Appl. Environ. Microbiol. 1994, 60, 2661–2665. [Google Scholar] [CrossRef]
- Palau, J.; Soler, A.; Teixidor, P.; Aravena, R. Compound-specific carbon isotope analysis of volatile organic compounds in water using solid-phase microextraction. J. Chromatogr. A 2007, 1163, 260–268. [Google Scholar] [CrossRef]
- Cichocka, D.; Imfeld, G.; Richnow, H.H.; Nijenhuis, I. Variability in microbial carbon isotope fractionation of tetra- and trichloroethene upon reductive dechlorination. Chemosphere 2008, 71, 639–648. [Google Scholar] [CrossRef]
- Northrop, D.B. The Expression of Isotope Effects on Enzyme-Catalyzed Reactions. Annu. Rev. Biochem. 1981, 50, 103–131. [Google Scholar] [CrossRef]
- Li, Y.; Zhong, J. Cloning and sequencing of a dsrA gene encoding dissimilatory sulfite reductase in a sulfate reducing bacteria consortium (sulfate-reducing bacteriaⅢ). J. Fudan Univ. 2002, 41, 674–678. [Google Scholar]
- Fahrenfeld, N.; Cozzarelli, I.M.; Bailey, Z.; Pruden, A. Insights into Biodegradation Through Depth-Resolved Microbial Community Functional and Structural Profiling of a Crude-Oil Contaminant Plume. Microb. Ecol. 2014, 68, 453–462. [Google Scholar] [CrossRef] [PubMed]
- Kazy, S.K.; Monier, A.L.; Alvarez, P.J. Assessing the correlation between anaerobic toluene degradation activity and bssA concentrations in hydrocarbon-contaminated aquifer material. Biodegradation. 2010, 21, 793–800. [Google Scholar] [CrossRef] [PubMed]
- Temme, H.R.; Sande, K.; Yan, T.; Novak, P.J. Rapid Enrichment of Dehalococcoides-like Bacteria by Partial Hydrophobic Separation. Appl. Environ. Microbiol. 2017, 83, e02946-16. [Google Scholar] [CrossRef]
- Farag, I.F.; Davis, J.P.; Youssef, N.H.; Elshahed, M.S. Global Patterns of Abundance, Diversity and Community Structure of the Aminicenantes (Candidate Phylum OP8). PLoS ONE 2014, 9, e92139. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, X.; Hu, M.; Li, P. Effect of hydraulic retention time (HRT) on the biodegradation of trichloroethylene wastewater and anaerobic bacterial community in the UASB reactor. Appl. Microbiol. Biotechnol. 2014, 99, 1977–1987. [Google Scholar] [CrossRef]
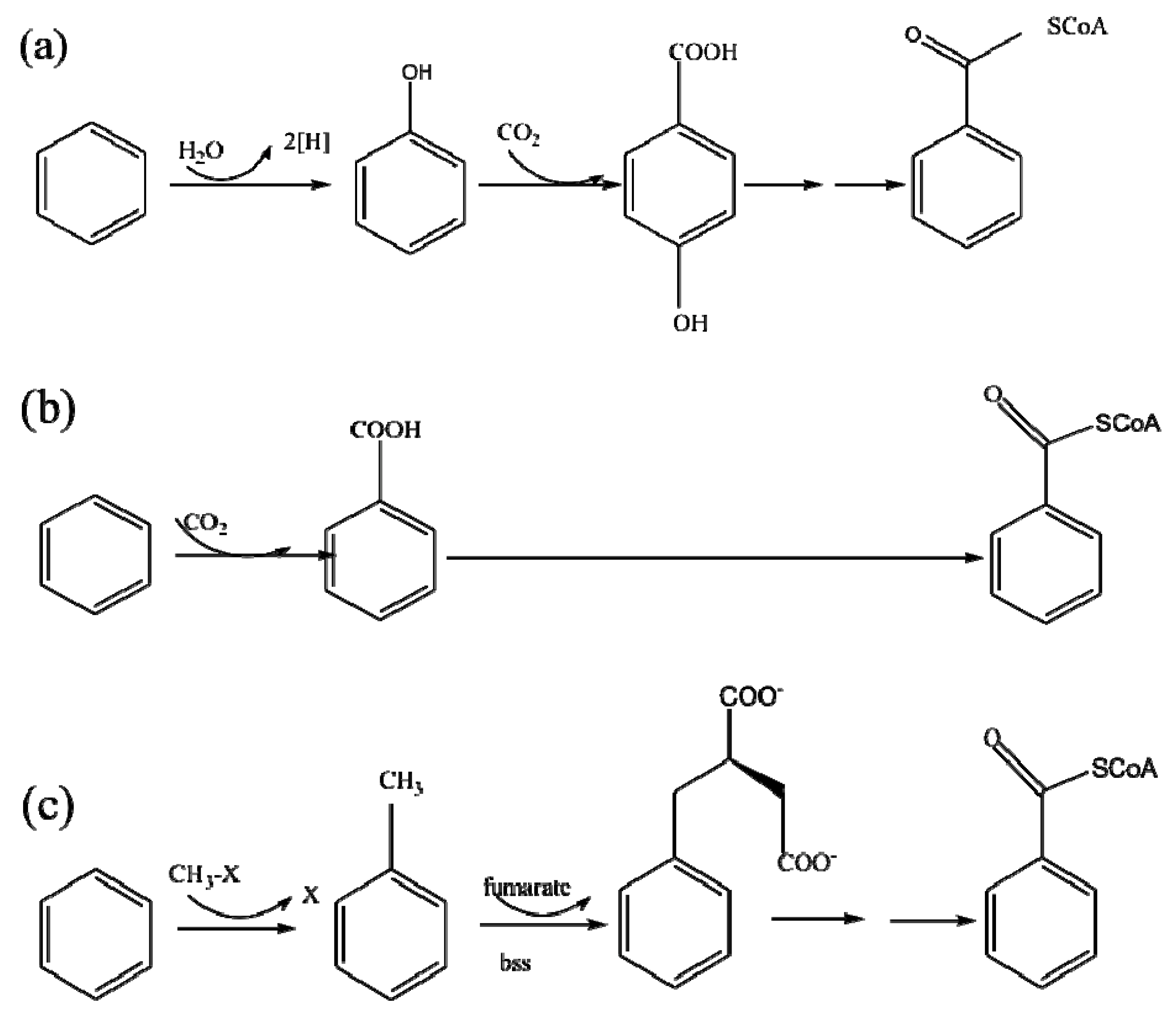
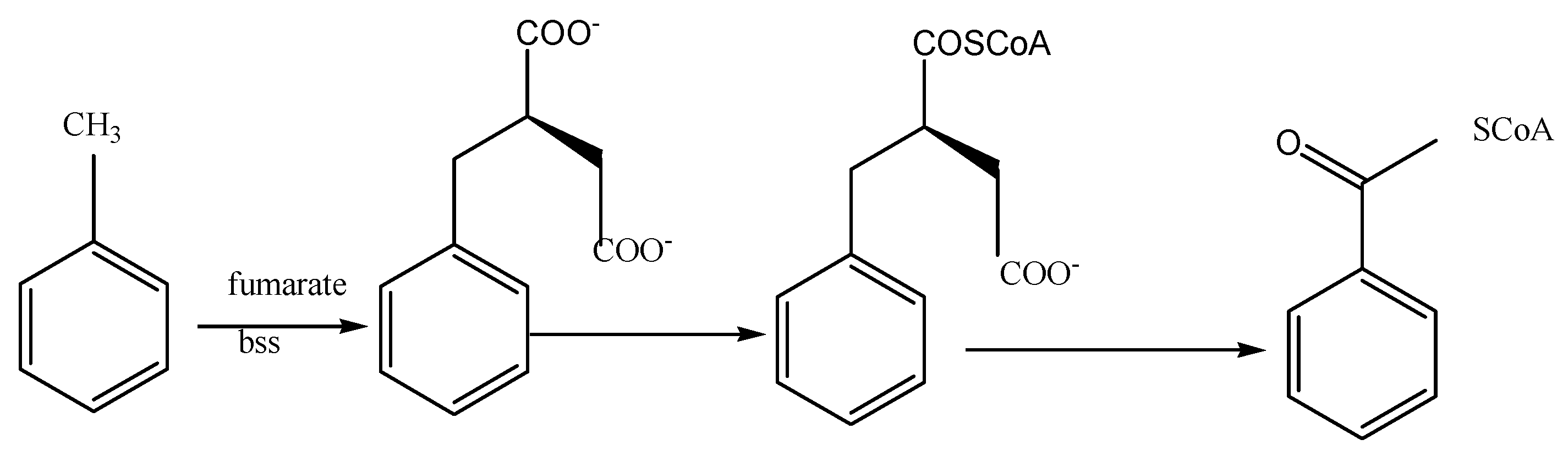
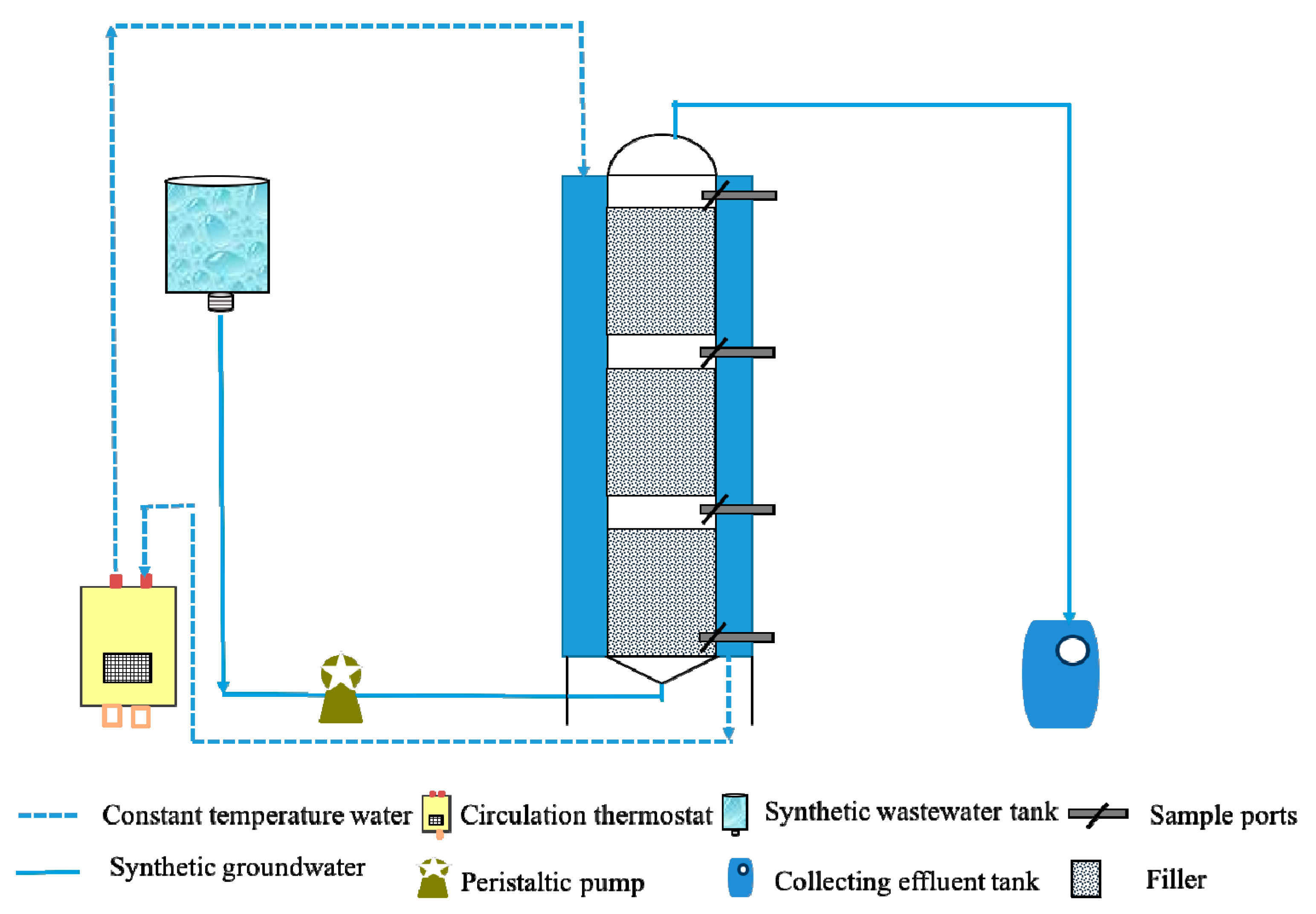
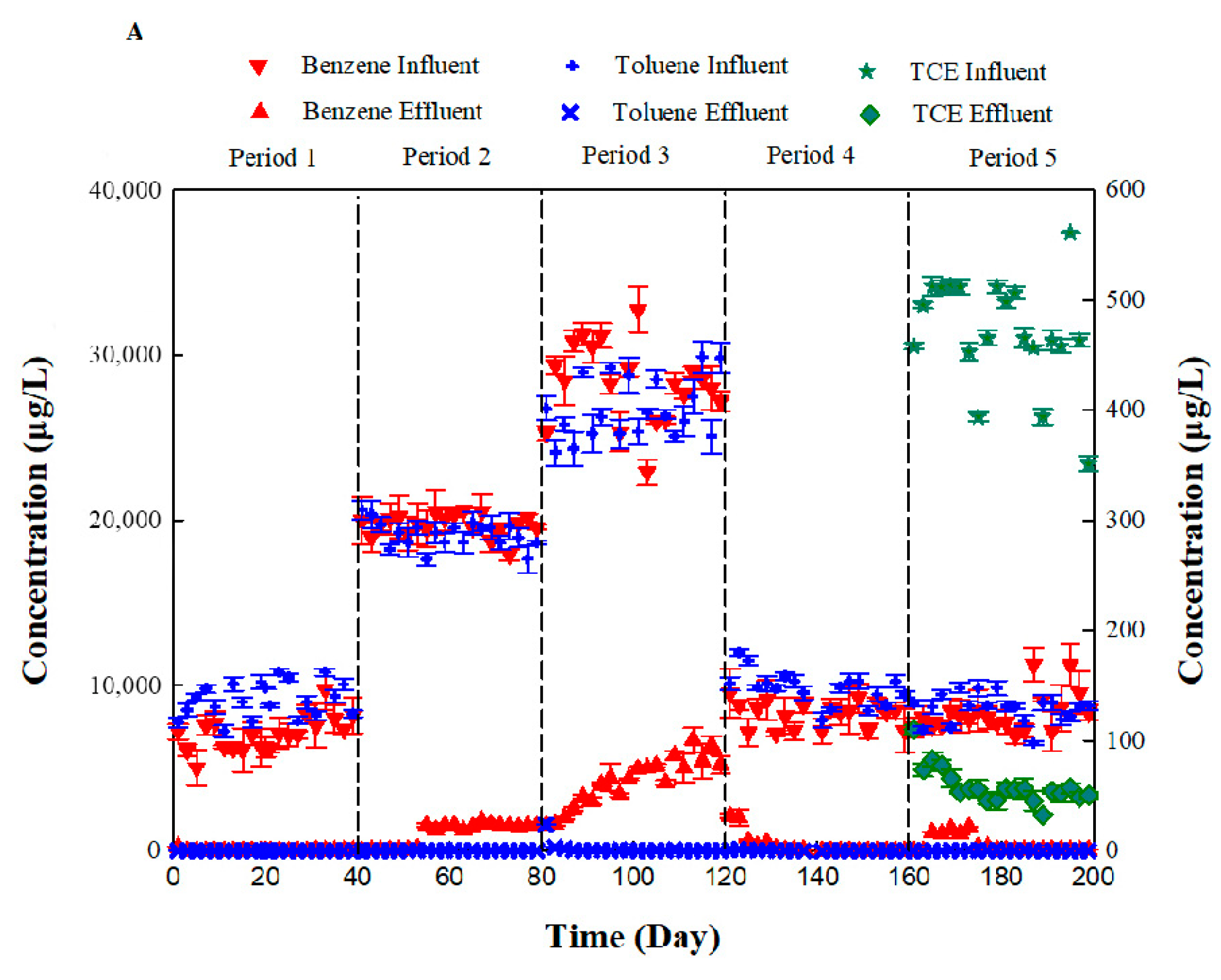
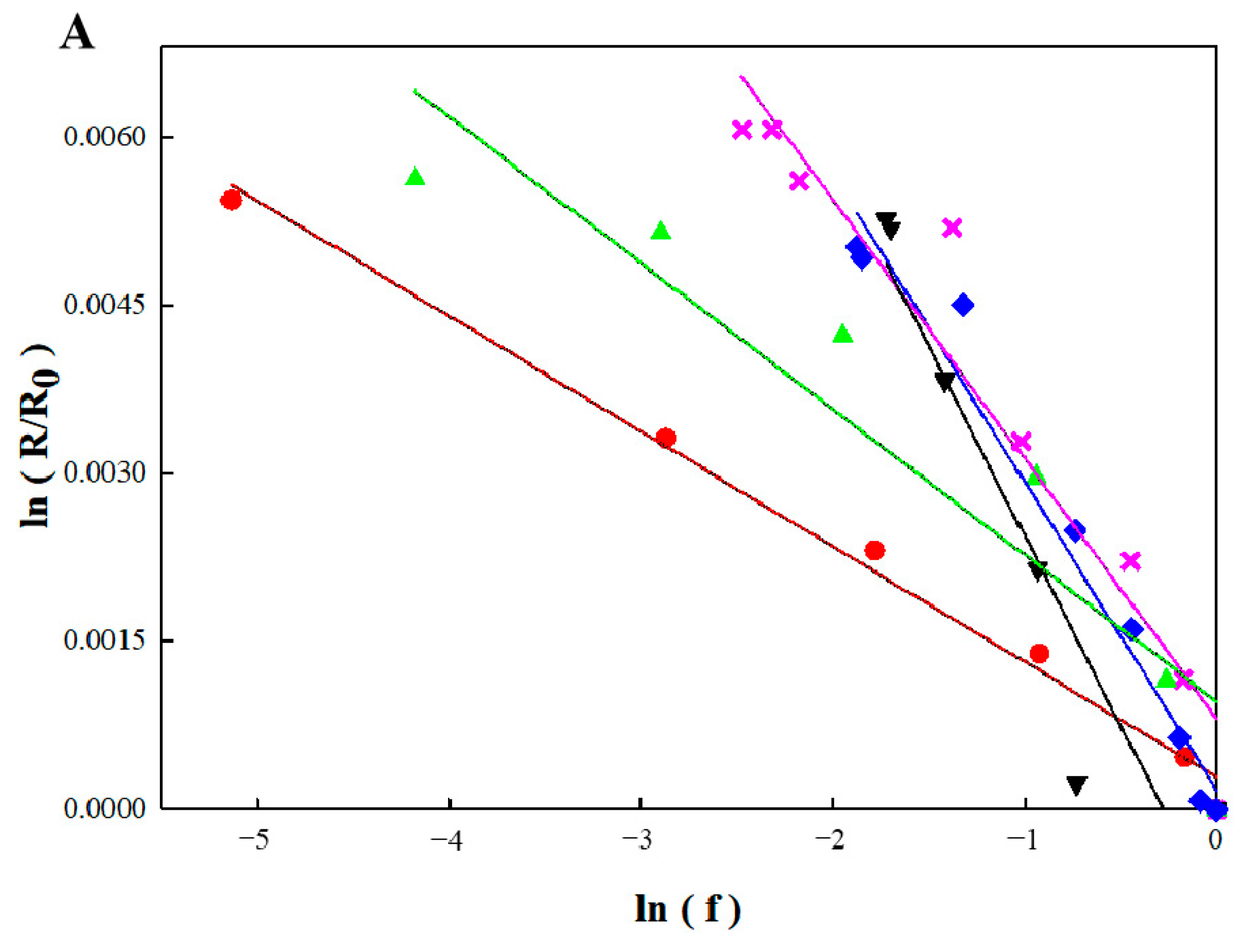
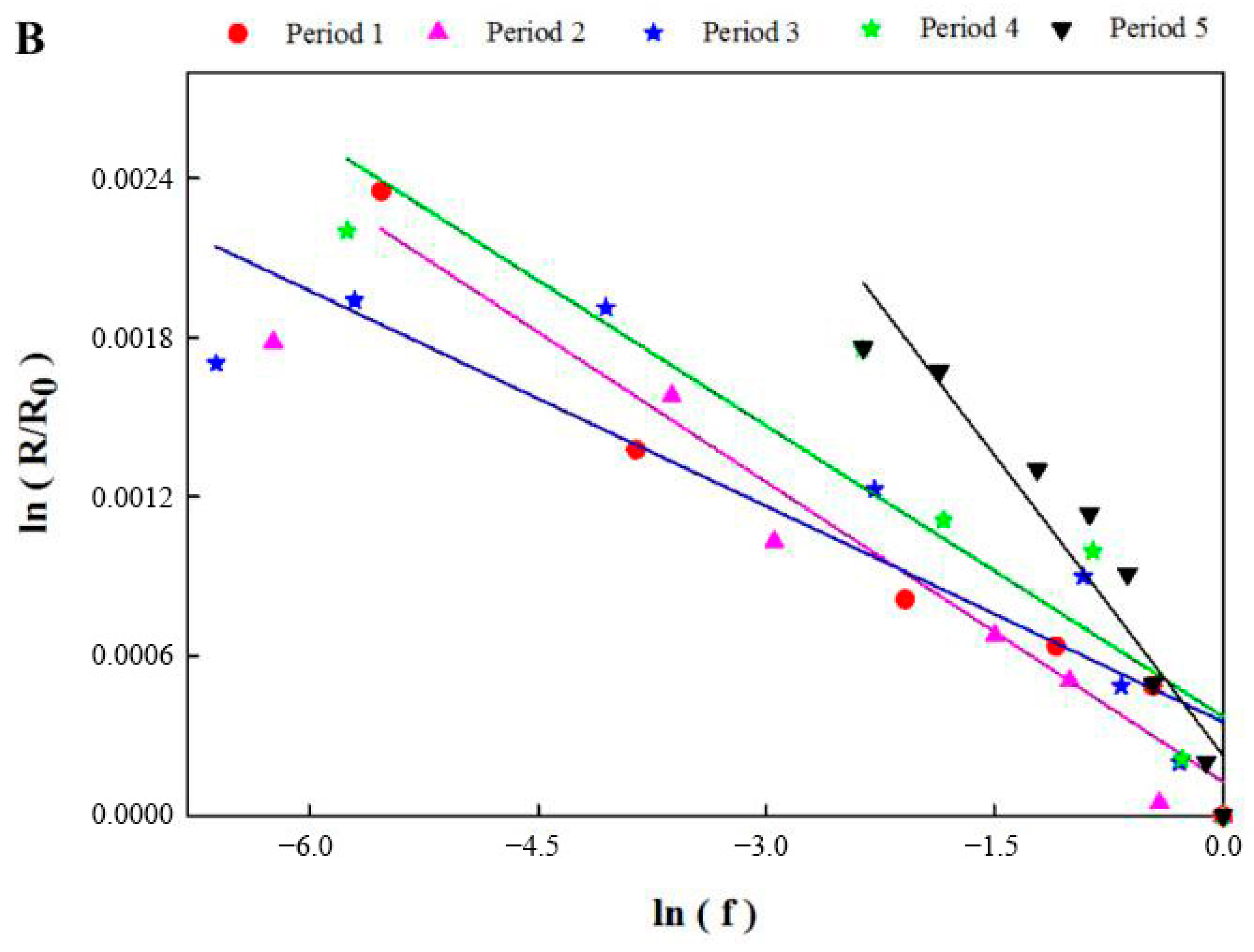


| Period | εc of Benzene (‰) | R2 | εc of Toluene (‰) | R2 |
|---|---|---|---|---|
| 1 | −1.0 | 0.9922 | −0.4 | 0.9615 |
| 2 | −2.3 | 0.9372 | −0.4 | 0.9803 |
| 3 | −3.4 | 0.9099 | −0.3 | 0.9061 |
| 4 | −1.3 | 0.8878 | −0.4 | 0.9062 |
| 5 | −2.8 | 0.9710 | −0.8 | 0.9559 |
| Period | δ13C of Remaining Benzene | Actual Biodegradation Rate of Benzene (%) | Apparent Removal Rate of Benzene (%) | δ13C of Remaining Toluene | Actual Biodegradation Rate of Toluene (%) | Apparent Removal Rate of Toluene (%) |
|---|---|---|---|---|---|---|
| 1 | −21.4689 | 99.62 | 99.83 | −26.0785 | 99.59 | 99.97 |
| 2 | −22.0532 | 89.40 | 92.37 | −26.5468 | 98.37 | 99.88 |
| 3 | −22.1076 | 77.81 | 82.00 | −27.2187 | 98.10 | 99.55 |
| 4 | −21.2965 | 98.74 | 99.36 | −26.3108 | 99.27 | 99.85 |
| 5 | −22.4165 | 80.26 | 95.18 | −26.6258 | 85.75 | 99.89 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, T.; Wu, Y.; Wang, J.; Philippe, C.F.-X. Assessing the Biodegradation of BTEX and Stress Response in a Bio-Permeable Reactive Barrier Using Compound-Specific Isotope Analysis. Int. J. Environ. Res. Public Health 2022, 19, 8800. https://doi.org/10.3390/ijerph19148800
Chen T, Wu Y, Wang J, Philippe CF-X. Assessing the Biodegradation of BTEX and Stress Response in a Bio-Permeable Reactive Barrier Using Compound-Specific Isotope Analysis. International Journal of Environmental Research and Public Health. 2022; 19(14):8800. https://doi.org/10.3390/ijerph19148800
Chicago/Turabian StyleChen, Tianyu, Yan Wu, Jinnan Wang, and Corvini François-Xavier Philippe. 2022. "Assessing the Biodegradation of BTEX and Stress Response in a Bio-Permeable Reactive Barrier Using Compound-Specific Isotope Analysis" International Journal of Environmental Research and Public Health 19, no. 14: 8800. https://doi.org/10.3390/ijerph19148800
APA StyleChen, T., Wu, Y., Wang, J., & Philippe, C. F.-X. (2022). Assessing the Biodegradation of BTEX and Stress Response in a Bio-Permeable Reactive Barrier Using Compound-Specific Isotope Analysis. International Journal of Environmental Research and Public Health, 19(14), 8800. https://doi.org/10.3390/ijerph19148800






